We narrowed to 4,504 results for: golden gate
-
Plasmid#209983PurposeContains Level 0* Part: 5' Connector (5C_07_5C1C) for the construction of Level 2 plasmidsDepositorInsert5' Connector (5C_07_5C1C)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
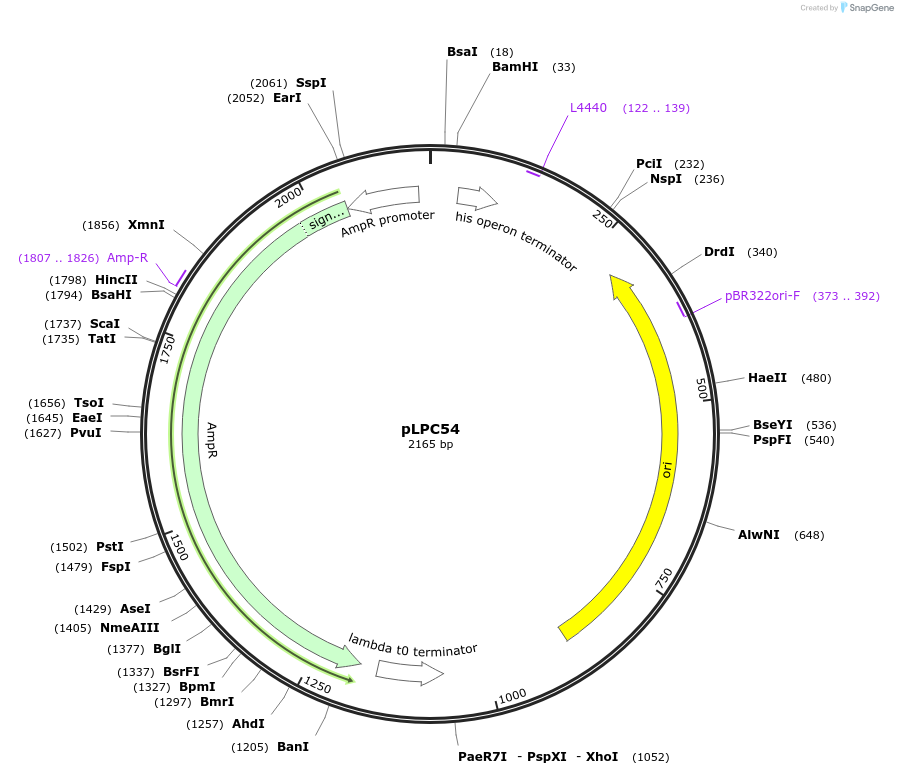
pLPC33
Plasmid#209984PurposeContains Level 0* Part: 3' Connector (3C_08_3C3) for the construction of Level 2 plasmidsDepositorInsert3' Connector (3C_08_3C3)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
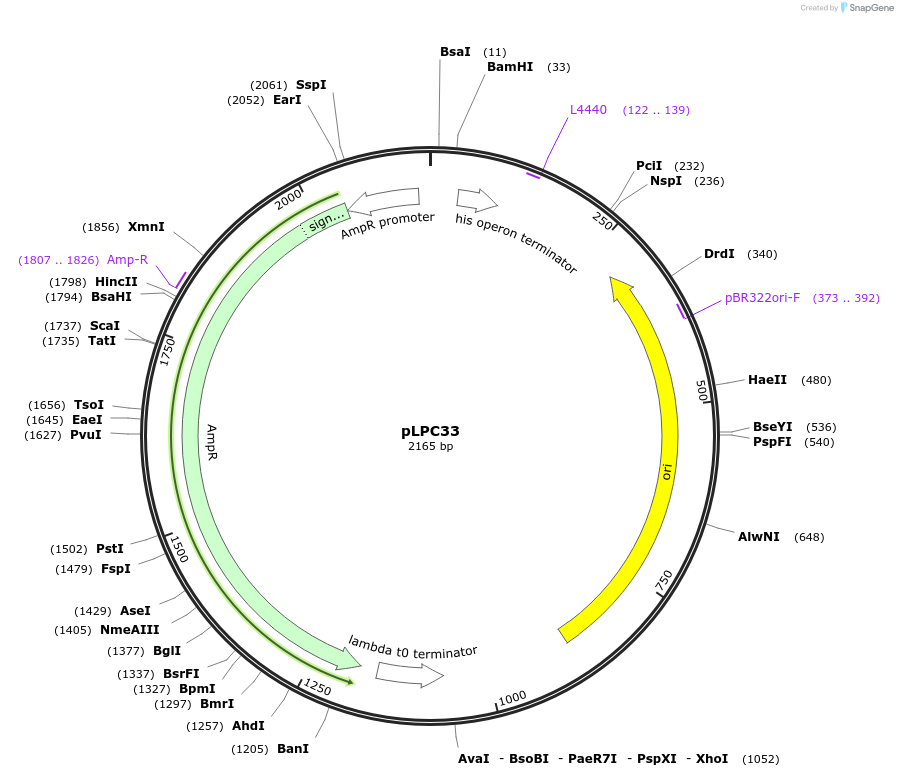
pLPR3
Plasmid#209986PurposeContains Level 0* Part: resistance cassette (ermBL) for the construction of Level 2 plasmidsDepositorInsertresistance cassette (ermBL)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
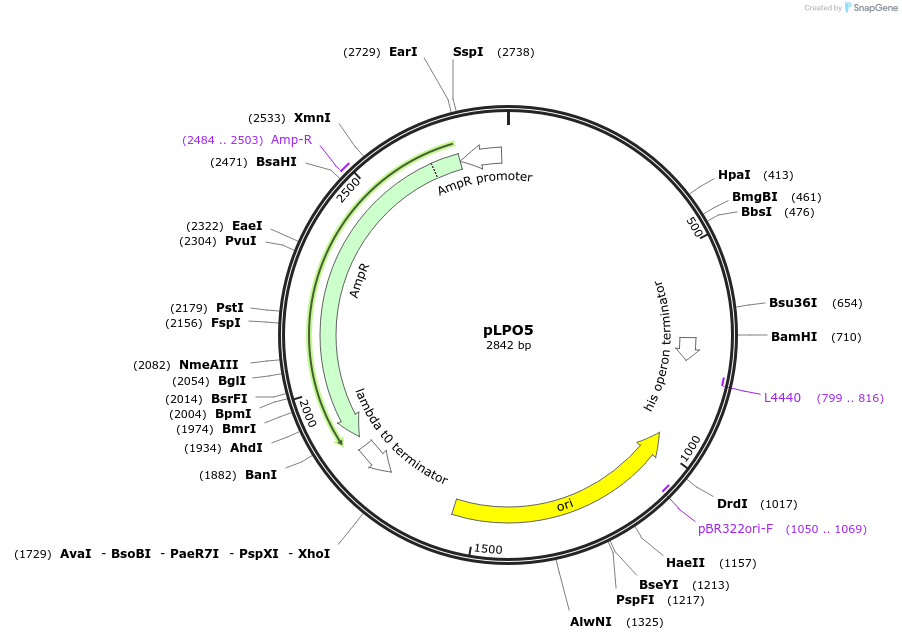
pLPO5
Plasmid#209987PurposeContains Level 0* Part: replication origin (256rep) for the construction of Level 2 plasmidsDepositorInsertreplication origin (256_rep)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
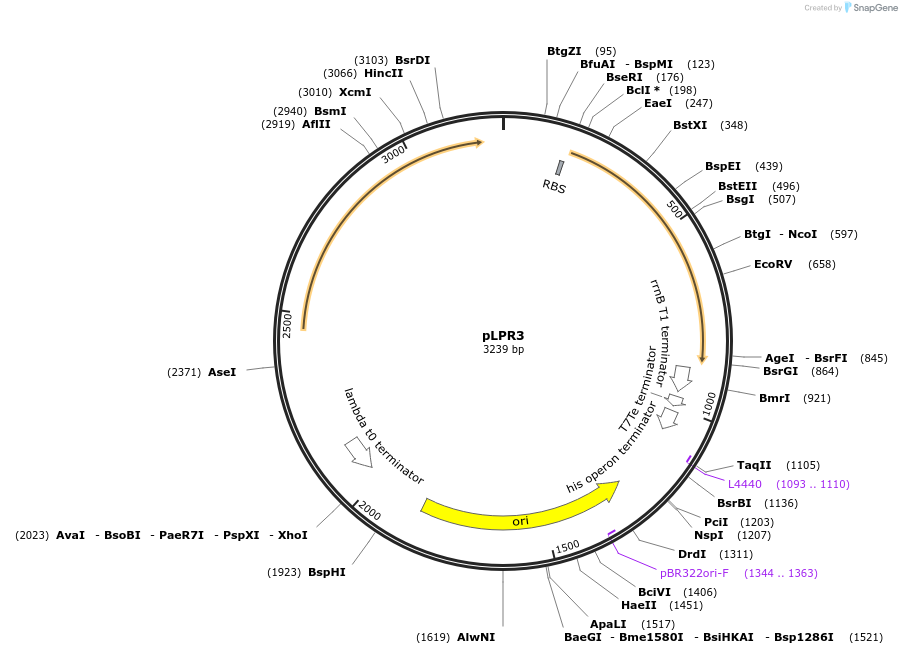
pLPP3
Plasmid#209965PurposeContains Level 0 Part: constitutive Promoter (P25) for the construction of Level 1 plasmidsDepositorInsertPromoter (P25)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pLPP7
Plasmid#209969PurposeContains Level 0 Part: autoinducible Promoter (PfabZ) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_fabZ)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pLPP8
Plasmid#209970PurposeContains Level 0 Part: autoinducible Promoter (PgadB) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_gadB)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pLPP9
Plasmid#209971PurposeContains Level 0 Part: constitutive Promoter (PldhD) for the construction of Level 1 plasmidsDepositorInsertPromoter (P_ldhD)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
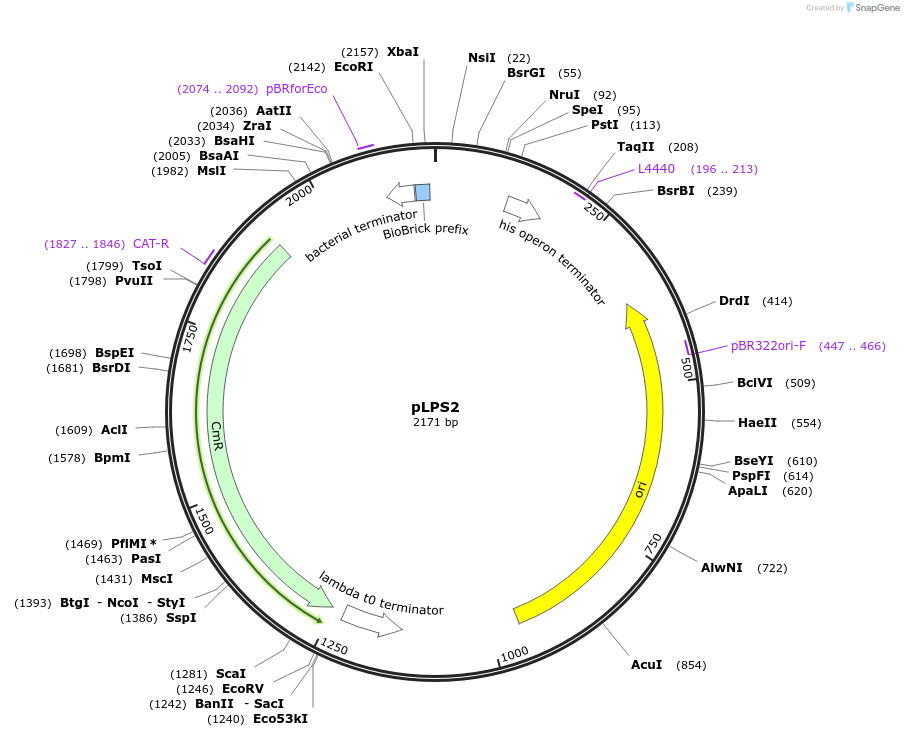
pLPS2
Plasmid#209972PurposeContains Level 0 Part: ribosome binding site (L. plantarum optimized) for the construction of Level 1 plasmidsDepositorInsertRBS (Lp)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
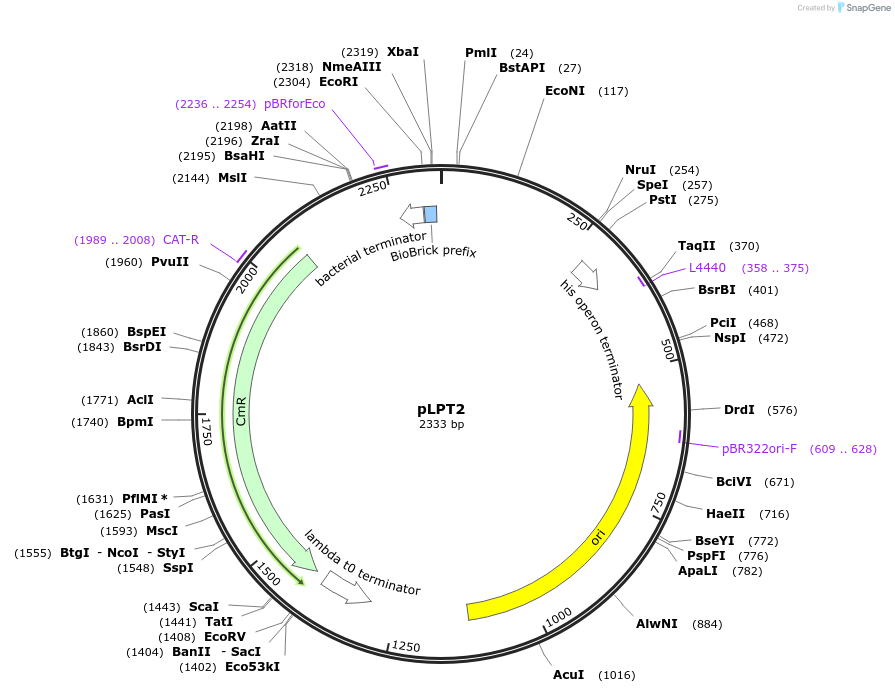
pLPT2
Plasmid#209974PurposeContains Level 0 Part: Terminator (TpepN) for the construction of Level 1 plasmidsDepositorInsertTerminator (T_pepN)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
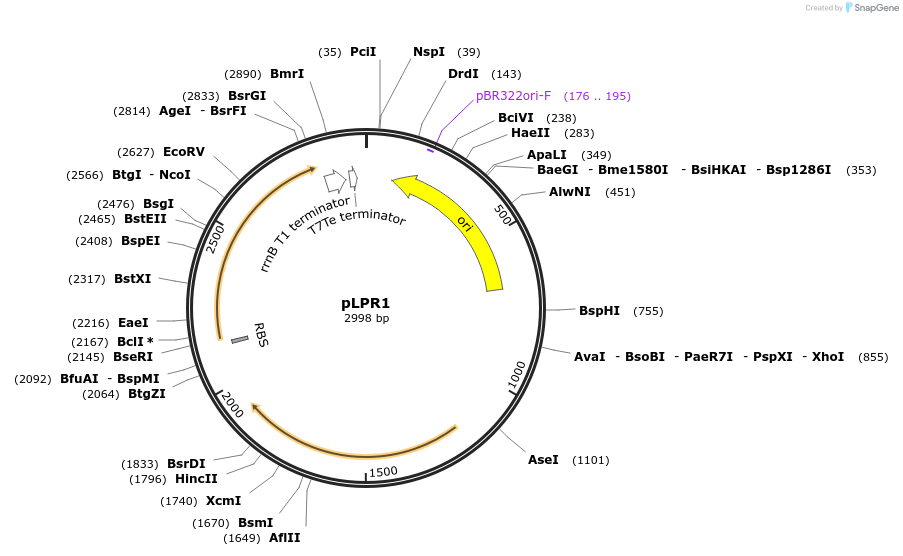
pLPR1
Plasmid#209979PurposeContains Level 0 Part: resistance cassette (ermBL) for the construction of Level 1 plasmidsDepositorInsertresistance cassette (ermBL)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
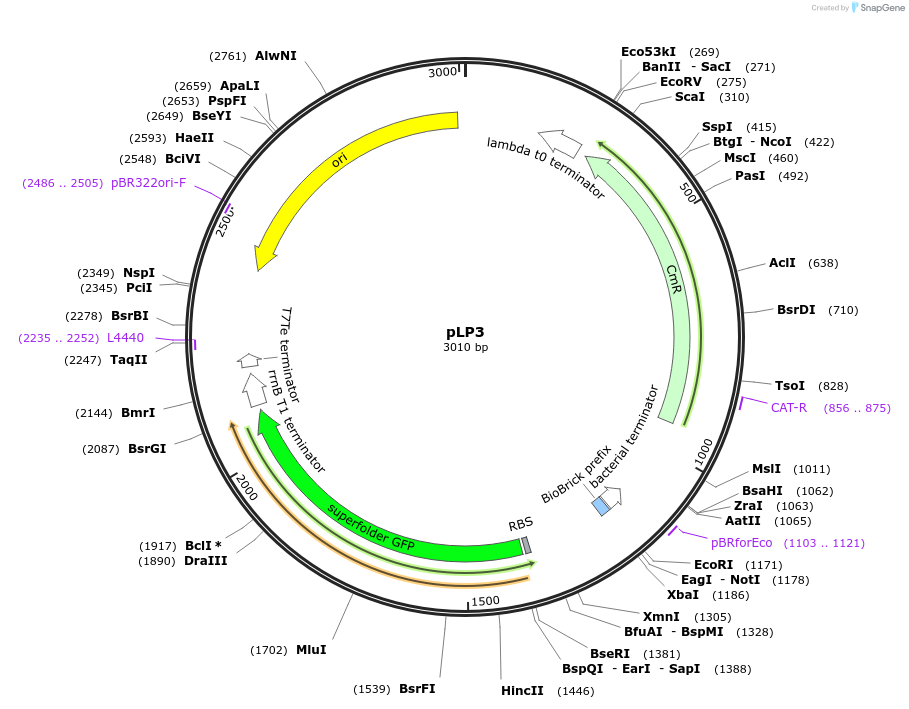
pLP3
Plasmid#209958PurposeEntry vector to store Level 0 broad-host range L. plantarum replication origin partsDepositorTypeEmpty backboneExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
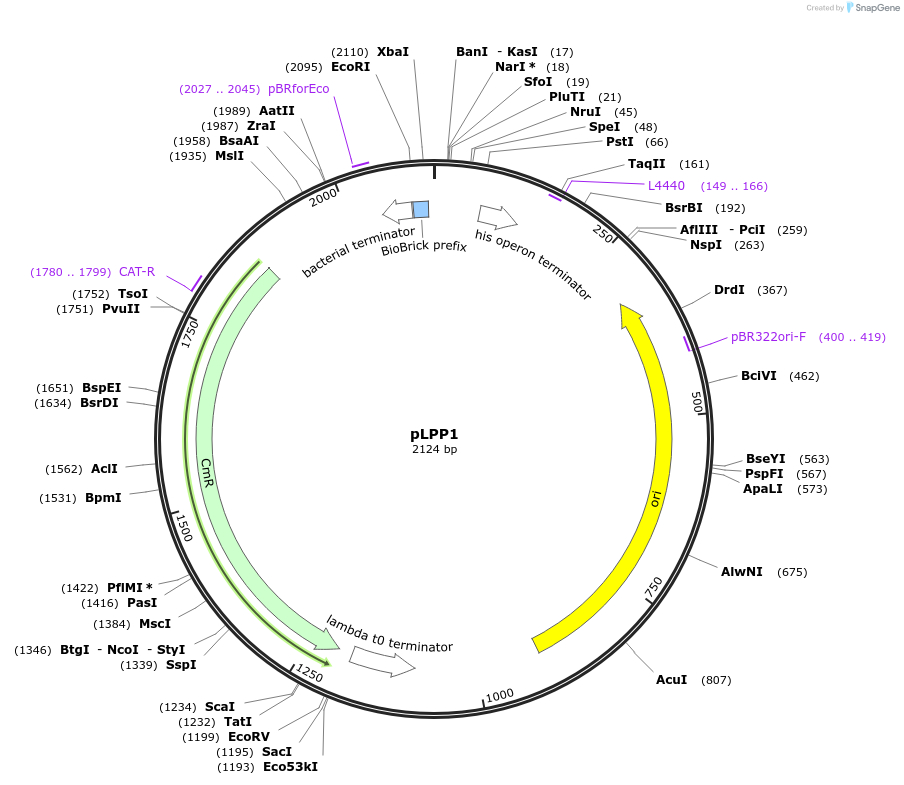
pLPP1
Plasmid#209963PurposeContains Level 0 Part: mock / negative control Promoter (P-nc) for the construction of Level 1 plasmidsDepositorInsertPromoter (mock)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
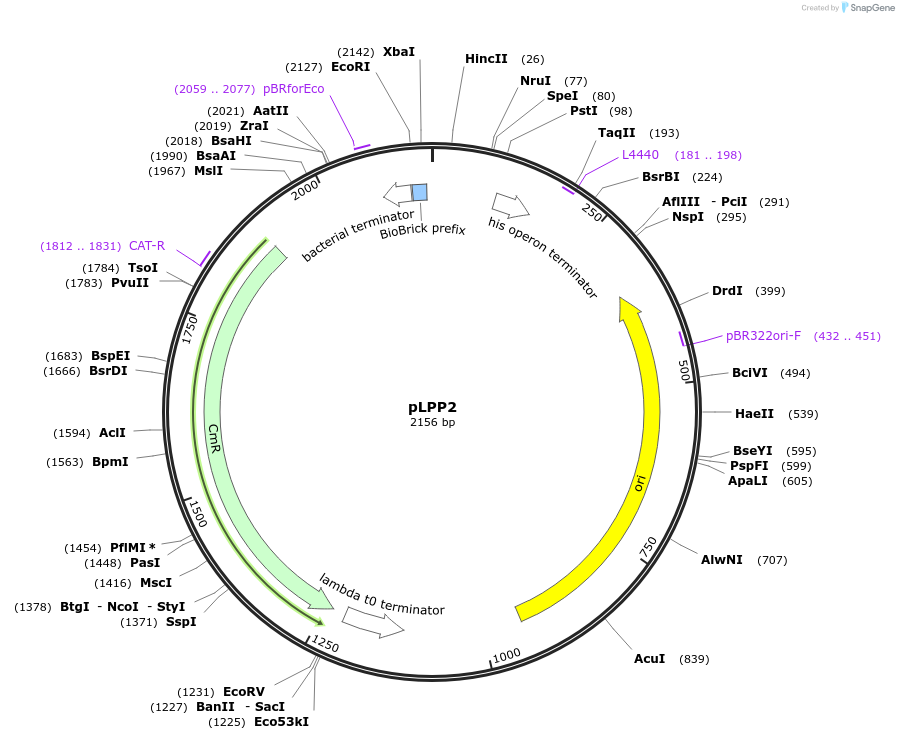
pLPP2
Plasmid#209964PurposeContains Level 0 Part: constitutive Promoter (P48) for the construction of Level 1 plasmidsDepositorInsertPromoter (P48)
ExpressionBacterialAvailable SinceSept. 13, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
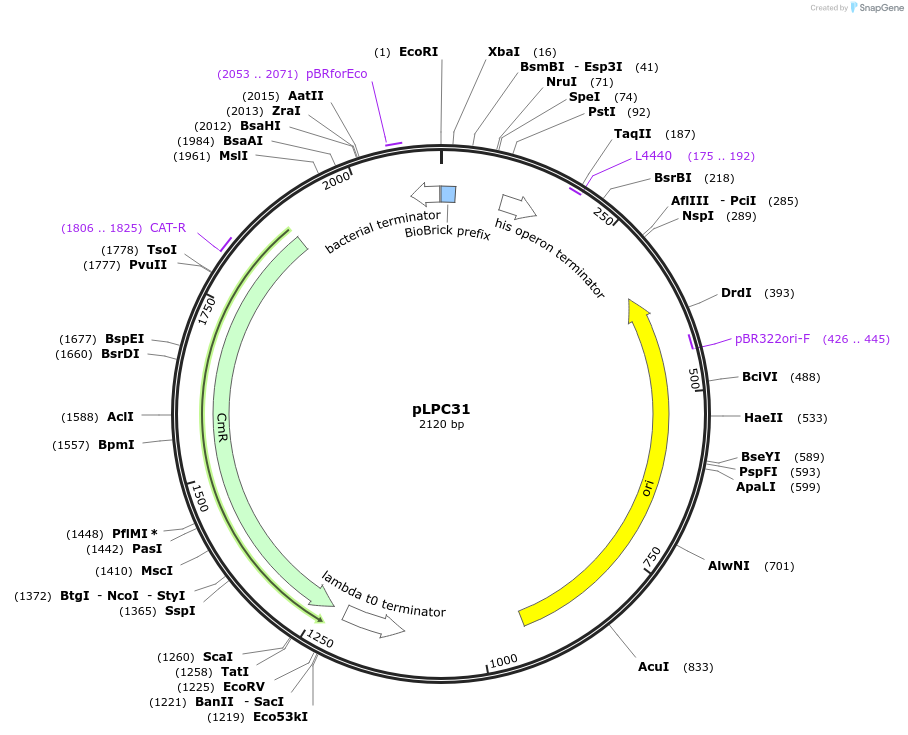
pLPC31
Plasmid#209996PurposeContains Level 0 Part: 3' Connector (3C2SN) for the construction of Level 1 plasmidsDepositorInsert3' Connector (3C2SN)
ExpressionBacterialAvailable SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
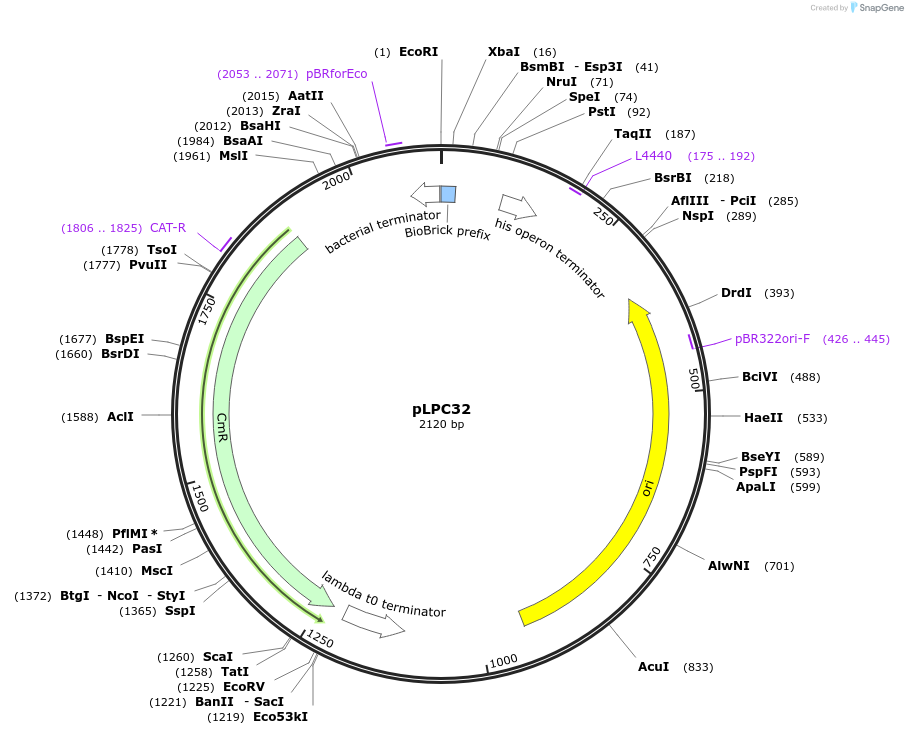
pLPC32
Plasmid#209997PurposeContains Level 0 Part: 3' Connector (3C5CSN) for the construction of Level 1 plasmidsDepositorInsert3' Connector (3C5CSN)
ExpressionBacterialAvailable SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
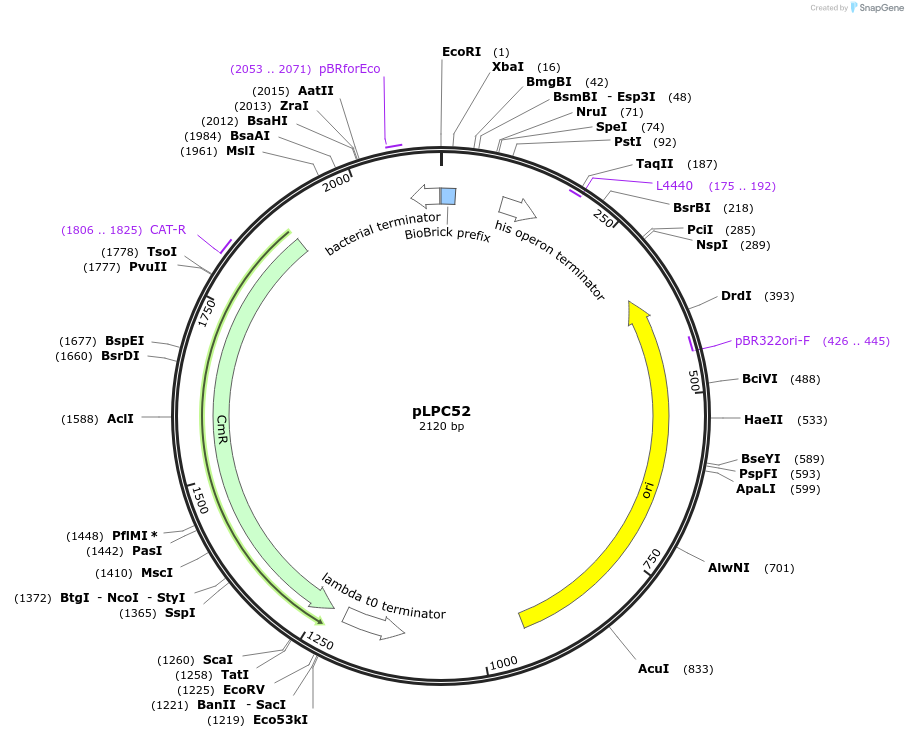
pLPC52
Plasmid#209991PurposeContains Level 0 Part: 5' Connector (5C2SN) for the construction of Level 1 plasmidsDepositorInsert5' Connector (5C2SN)
ExpressionBacterialAvailable SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
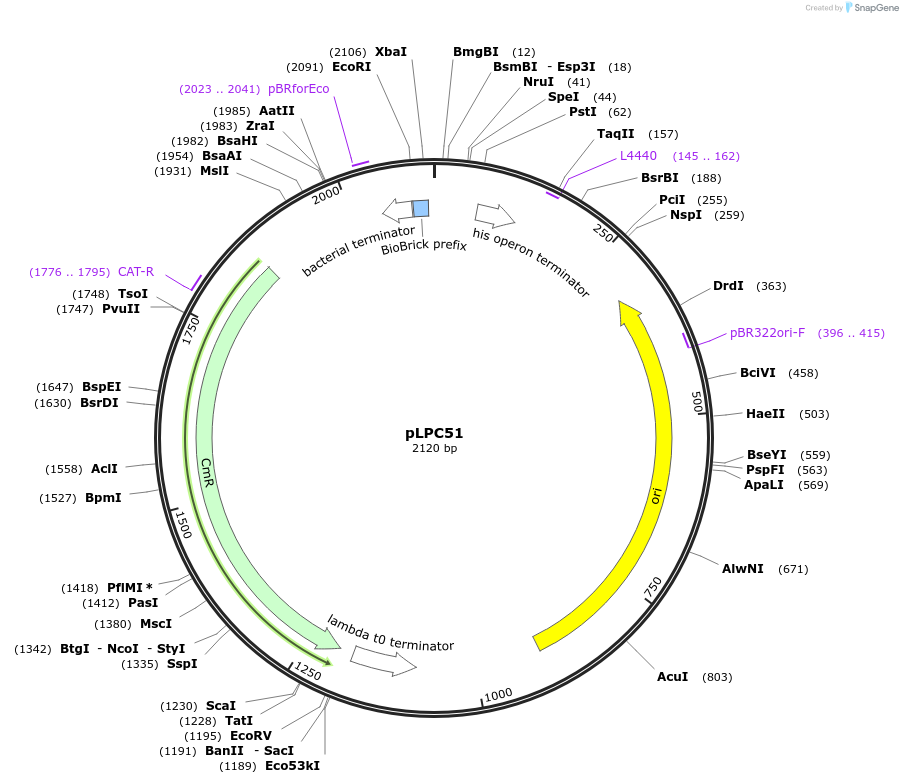
pLPC51
Plasmid#209990PurposeContains Level 0 Part: 5' Connector (5C1CSN) for the construction of Level 1 plasmidsDepositorInsert5' Connector (5C1CSN)
ExpressionBacterialAvailable SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
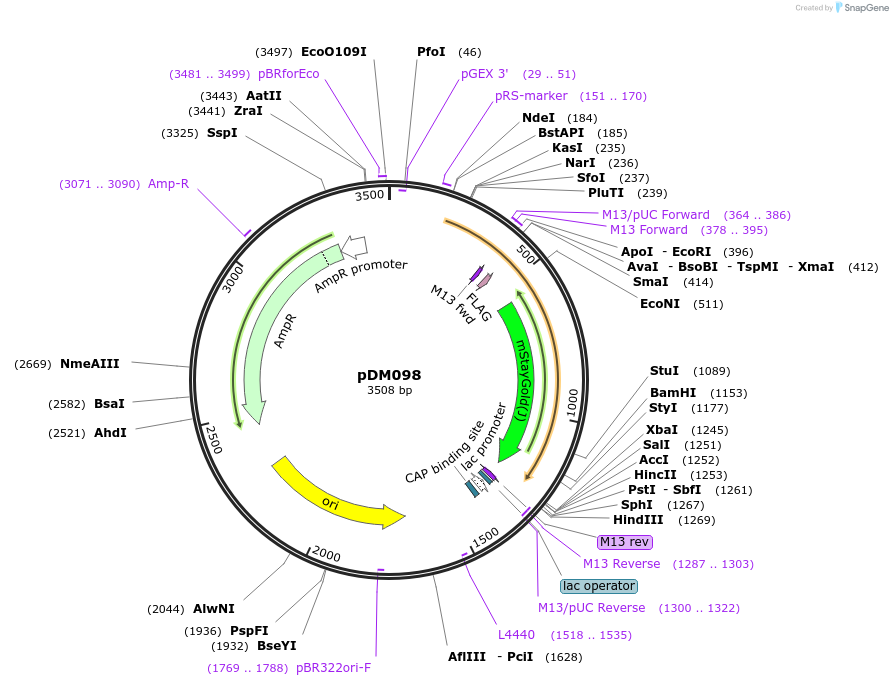
pDM098
Plasmid#216818PurposeAspergillus nidulans codon-adjusted mStayGold fluorescent protein, includes linker for C-terminal tagging.DepositorInsertmStayGold
TagsFLAG-(SGGS)x2-XTEN16-(GGGGS)x3ExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
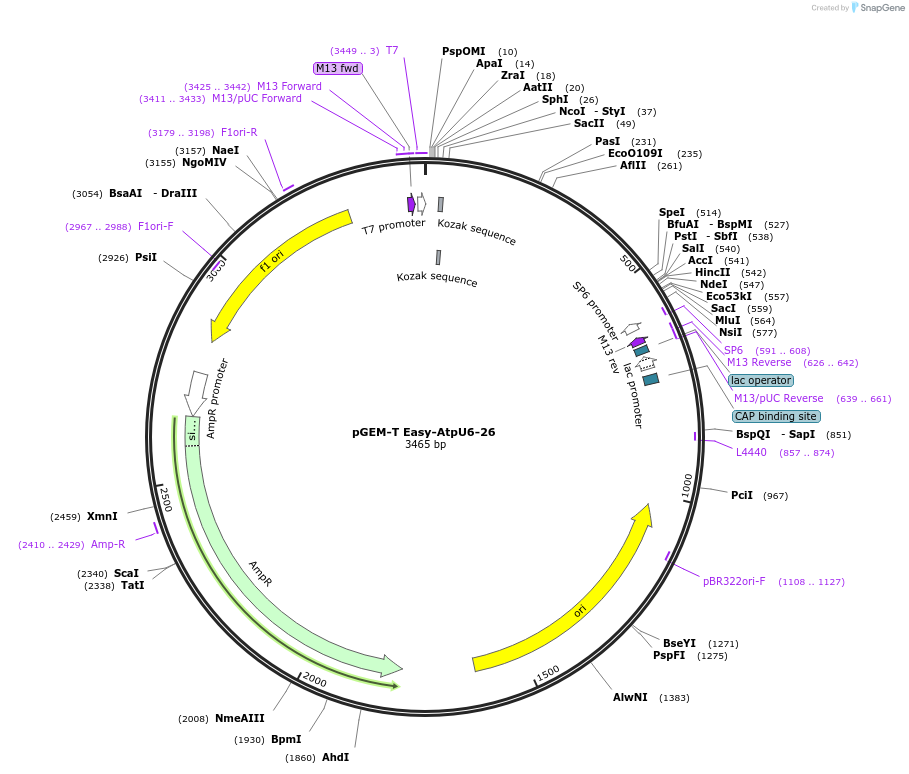
pGEM-T Easy-AtpU6-26
Plasmid#160218PurposeAtpU6-26 Golden Gate level 0 piece to express gRNAsDepositorInsertpAtU6-26 promoter
UseGolden gate level 0 piece to express grnasAvailable SinceAug. 15, 2022AvailabilityAcademic Institutions and Nonprofits only