We narrowed to 28,450 results for: tat
-
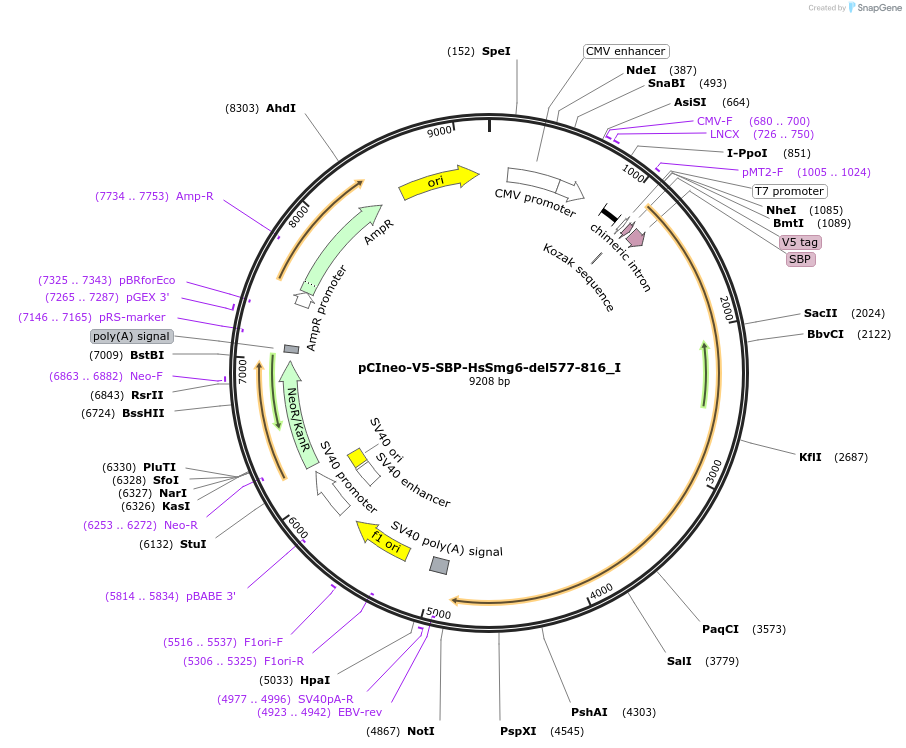
Plasmid#146554PurposeMammalian Expression of HsSmg6-del577-816DepositorInsertHsSmg6-del577-816 (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pCIneo-V5-SBP-HsSmg6-siRNAres_I
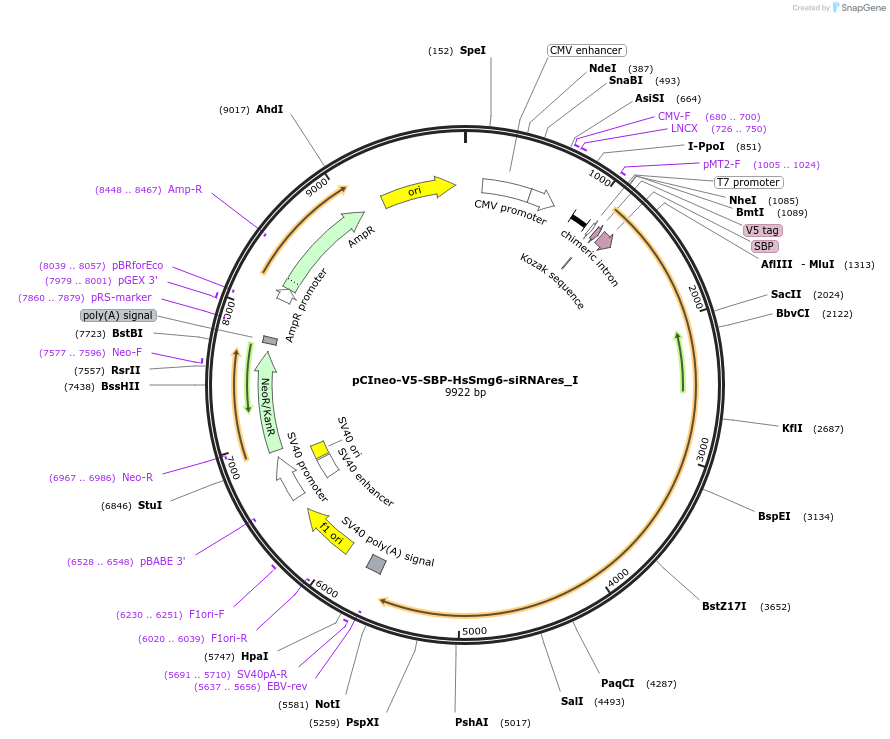
Plasmid#146556PurposeMammalian Expression of HsSmg6-siRNAresDepositorInsertHsSmg6-siRNAres (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
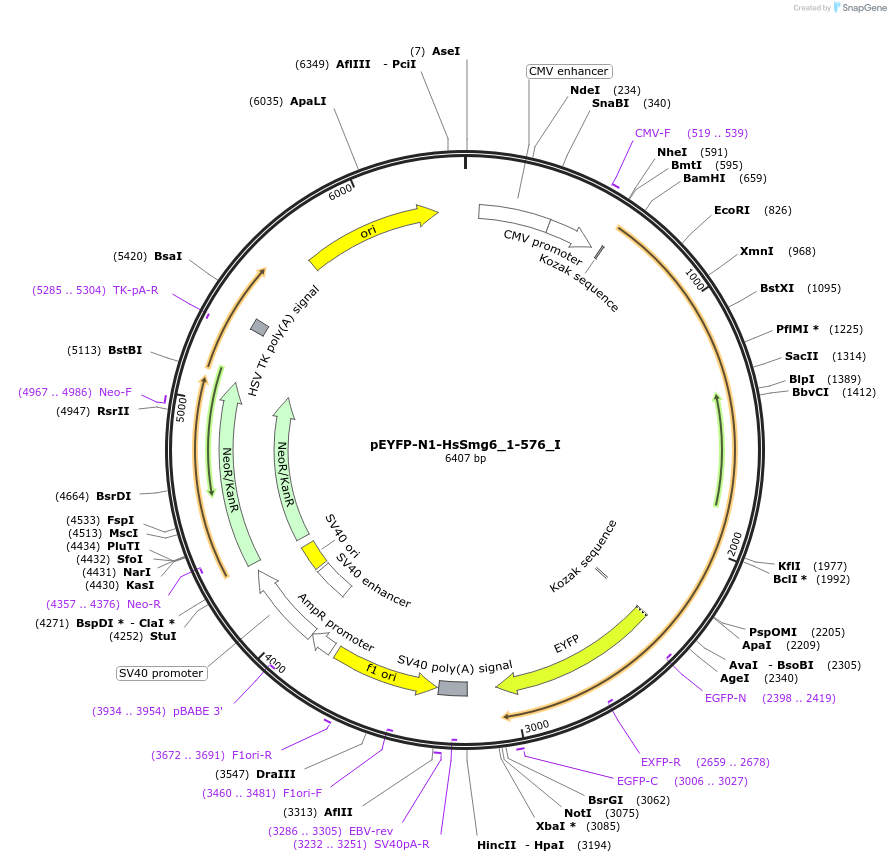
pEYFP-N1-HsSmg6_1-576_I
Plasmid#146557PurposeMammalian Expression of HsSmg6_1-576DepositorInsertHsSmg6_1-576 (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
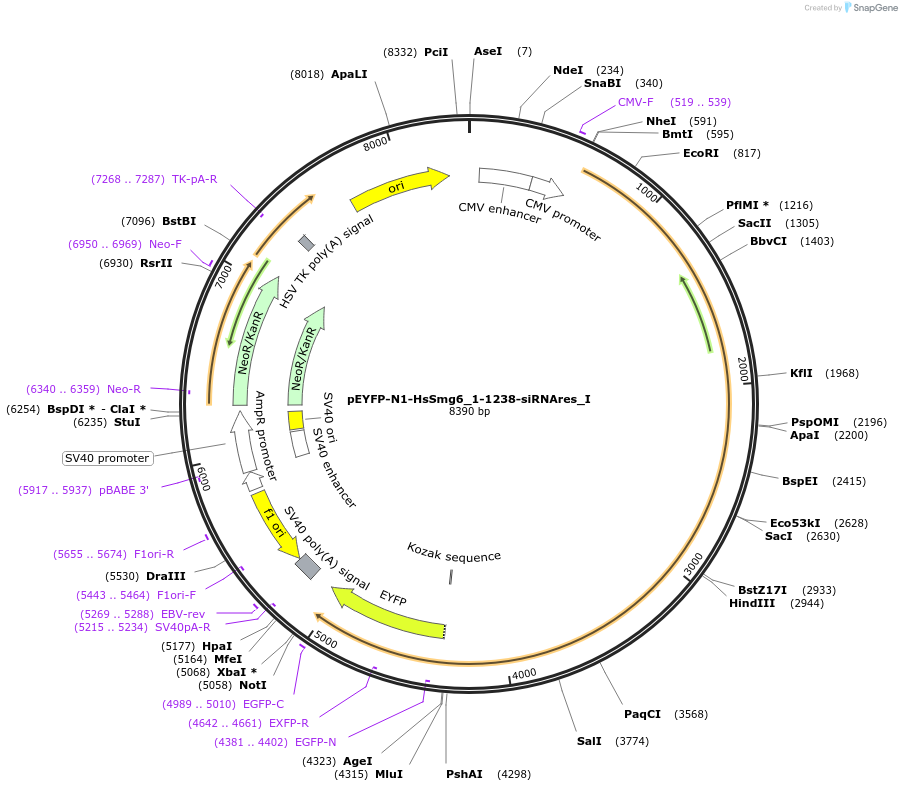
pEYFP-N1-HsSmg6_1-1238-siRNAres_I
Plasmid#146558PurposeMammalian Expression of HsSmg6_1-1238-siRNAresDepositorInsertHsSmg6_1-1238-siRNAres (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and one non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
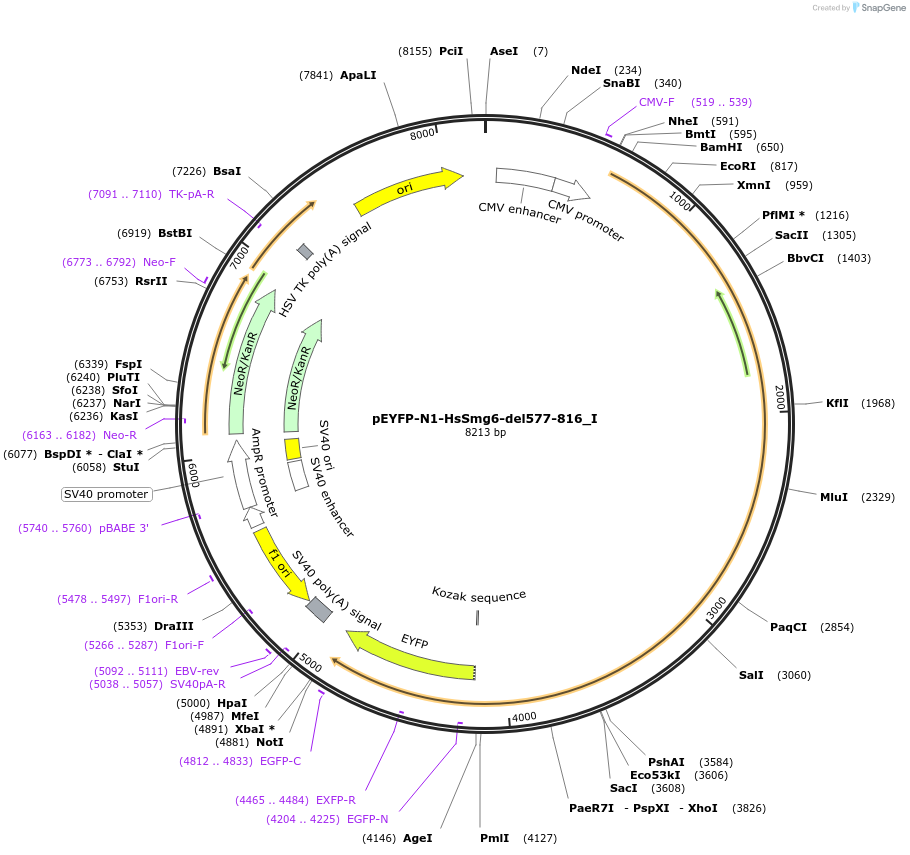
pEYFP-N1-HsSmg6-del577-816_I
Plasmid#146563PurposeMammalian Expression of HsSmg6-del577-816DepositorInsertHsSmg6-del577-816 (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
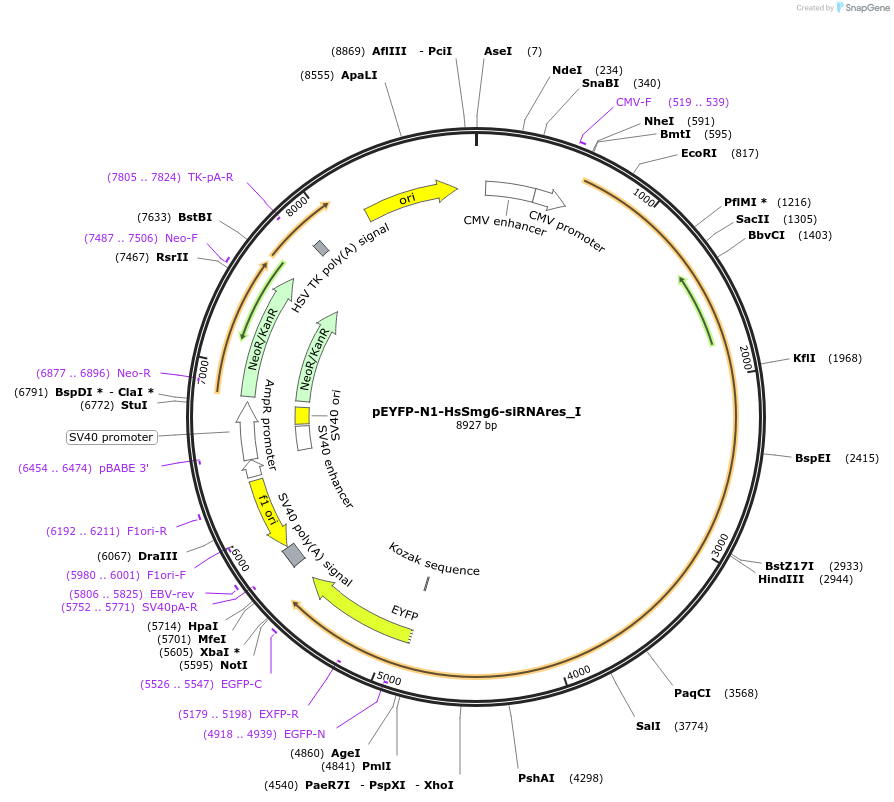
pEYFP-N1-HsSmg6-siRNAres_I
Plasmid#146565PurposeMammalian Expression of HsSmg6-siRNAresDepositorInsertHsSmg6-siRNAres (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
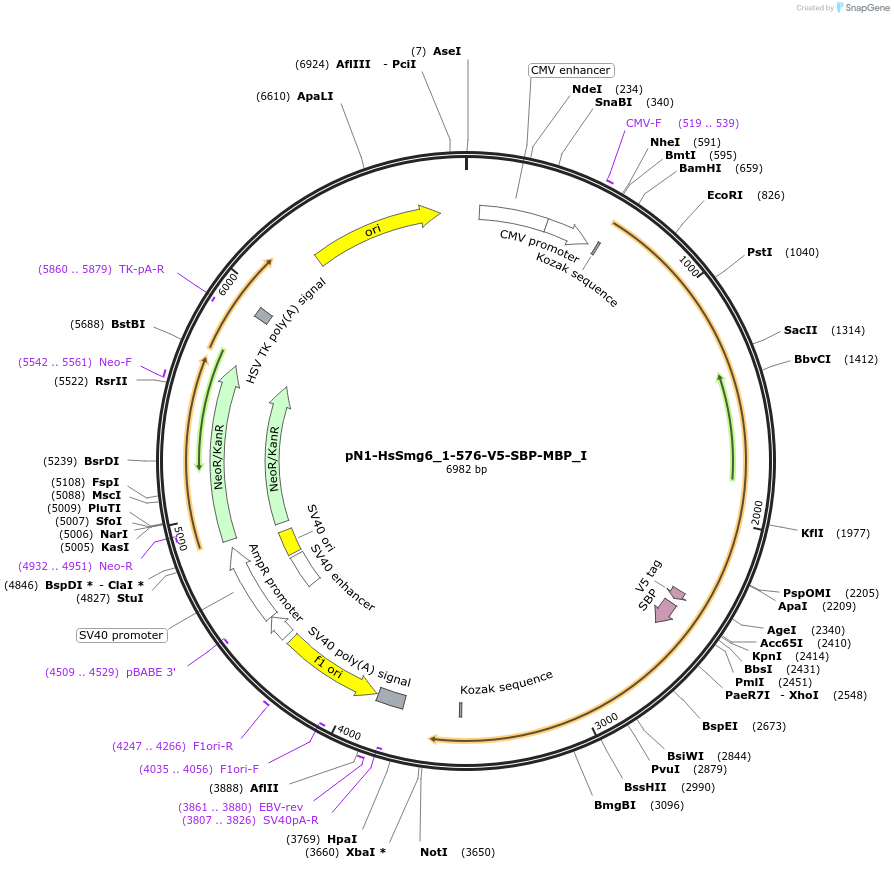
pN1-HsSmg6_1-576-V5-SBP-MBP_I
Plasmid#146569PurposeMammalian Expression of HsSmg6_1-576DepositorInsertHsSmg6_1-576 (SMG6 Human)
ExpressionMammalianMutationtwo silent mutations T594C, A699G and two non sil…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
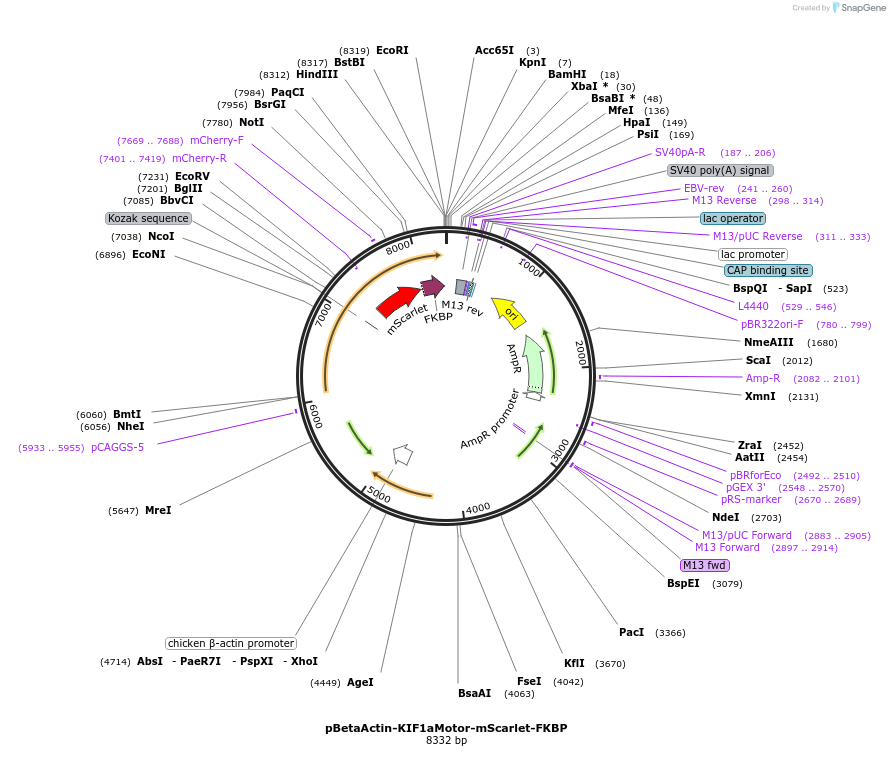
pBetaActin-KIF1aMotor-mScarlet-FKBP
Plasmid#191336PurposeExpresses the fluorescently labeled plus-end directed KIF1a motor domain fused to the dimerization domain FKBP to recruit it to the plasma membrane by using the chemical dimerization system FKBP-FRBDepositorAvailable SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
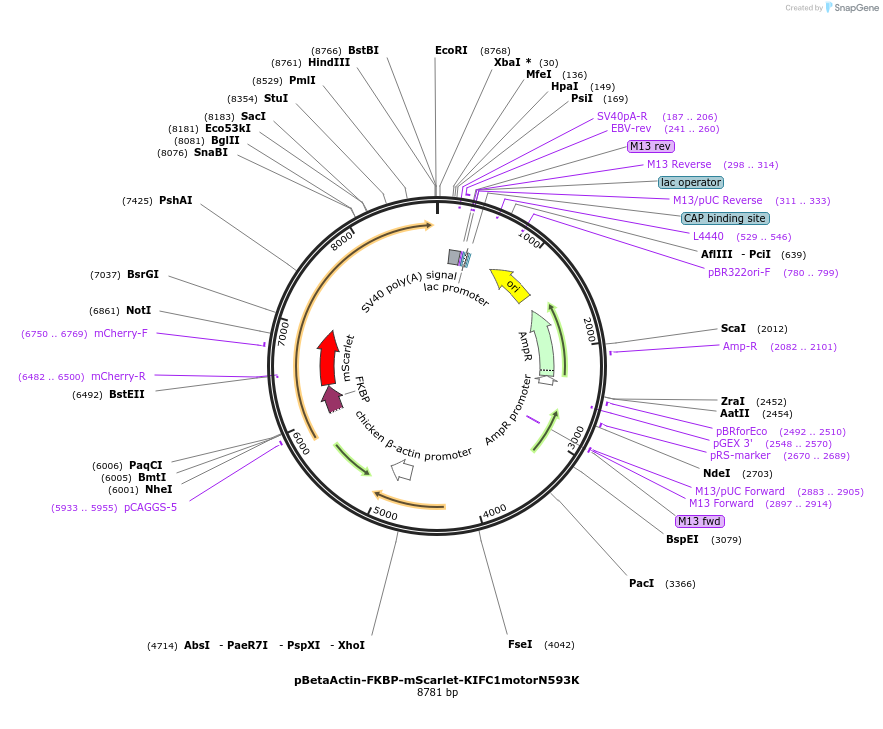
pBetaActin-FKBP-mScarlet-KIFC1motorN593K
Plasmid#191335PurposeExpresses the fluorescently labeled minus-end directed KIFC1 motor-deficient motor domain fused to FKBP to recruit it to the plasma membrane by using the chemical dimerization system FKBP-FRBDepositorInsertFKBP-mScarlet-KIFC1N593K (KIFC1 Human)
ExpressionMammalianMutationmotor domain aa 125-673; N593KAvailable SinceJan. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
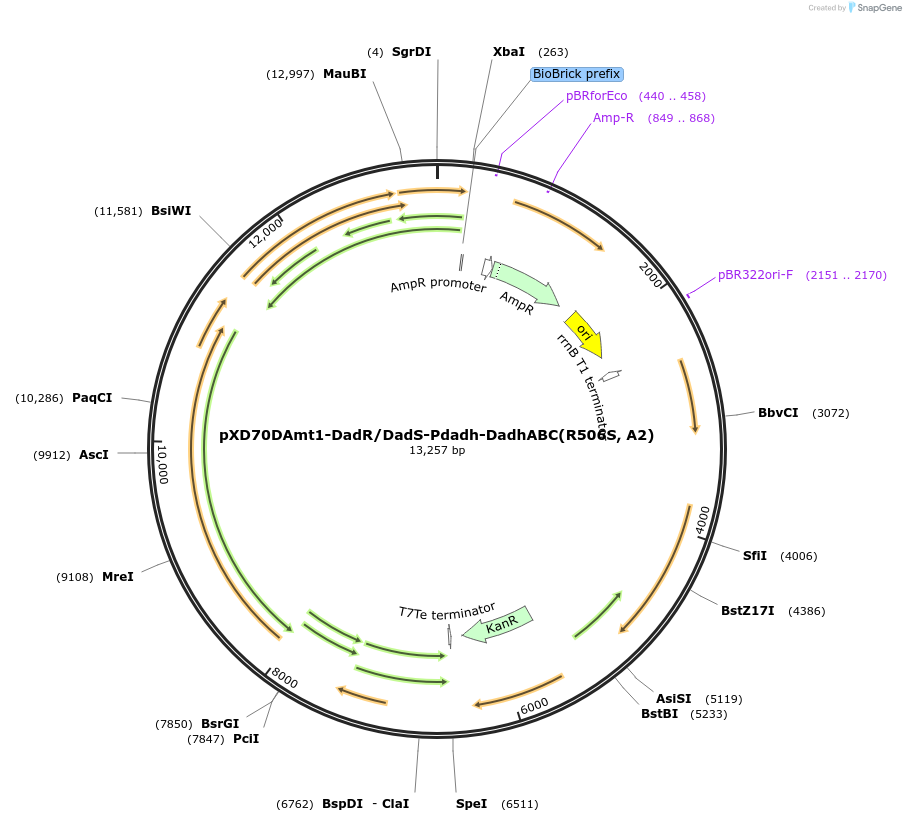
pXD70DAmt1-DadR/DadS-Pdadh-DadhABC(R506S, A2)
Plasmid#191631PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), native promoter-dopamine dehydroxylase from dopamine-metabolizing E. lenta A2 strain (R506S mutation) with transcriptional regulators DadR/DadSDepositorInsertDopamine dehydroxylase (R506S mutation, inactive form)
ExpressionBacterialAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
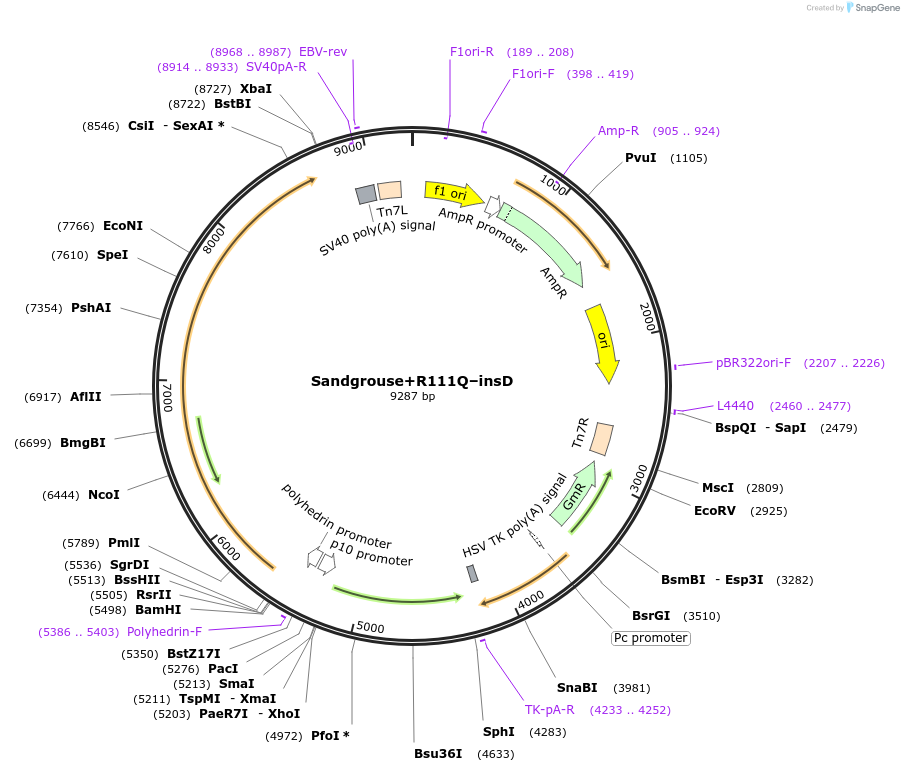
Sandgrouse+R111Q–insD
Plasmid#191229PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationremoved aspartic acid at 121 and replaced arginin…PromoterPH and p10Available SinceDec. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
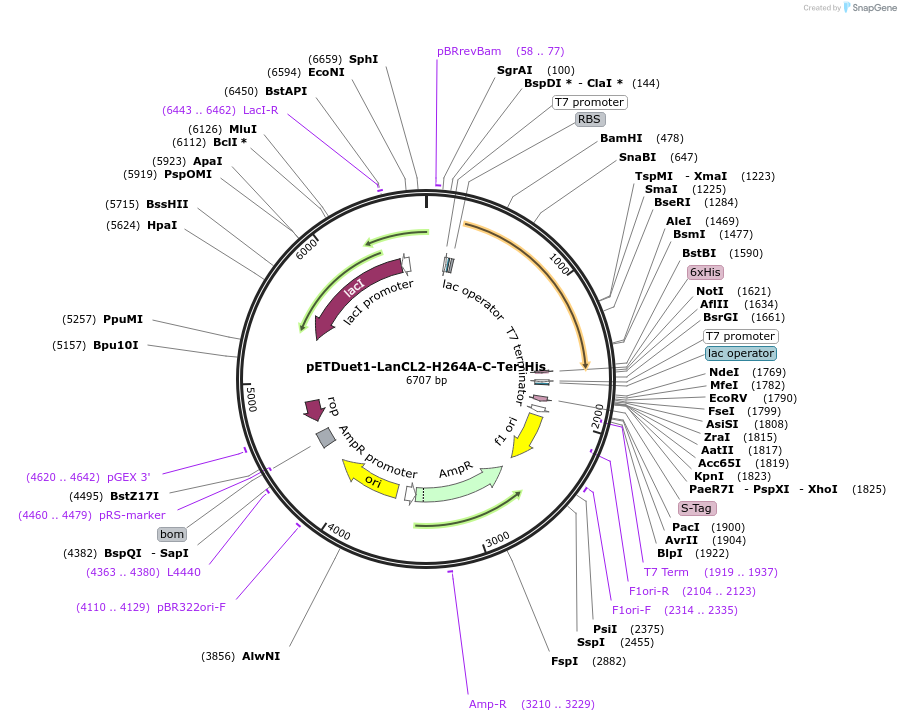
pETDuet1-LanCL2-H264A-C-Ter-His
Plasmid#154187PurposeTo express LanCL2-H264A in bacterial cellsDepositorInsertLanCL2
TagsHexahistidineExpressionBacterialMutationHis264->Ala264Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
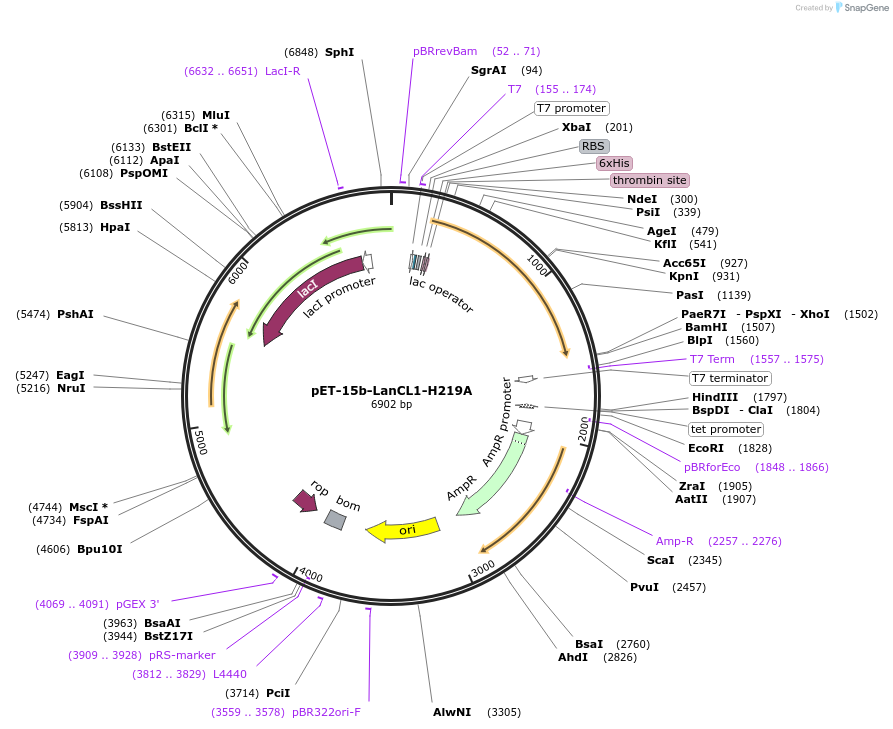
pET-15b-LanCL1-H219A
Plasmid#154190PurposeTo express LanCL1-H219A in bacterial cellsDepositorInsertLanCL1
TagsHexahistidineExpressionBacterialMutationHis219->Ala219Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
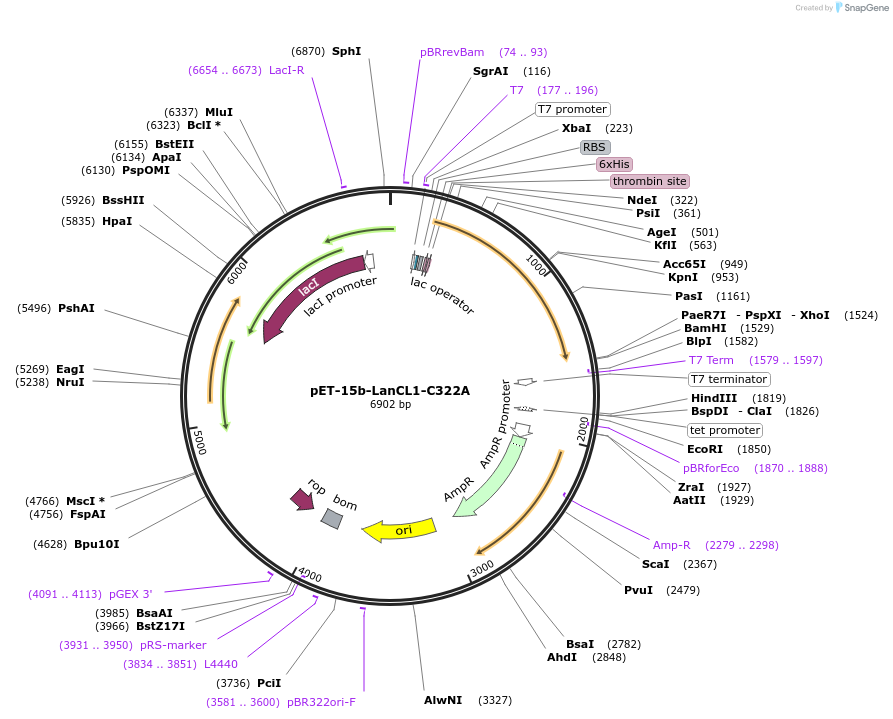
pET-15b-LanCL1-C322A
Plasmid#154191PurposeTo express LanCL1-C322A in bacterial cellsDepositorInsertLanCL1
TagsHexahistidineExpressionBacterialMutationCys322->Ala322Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
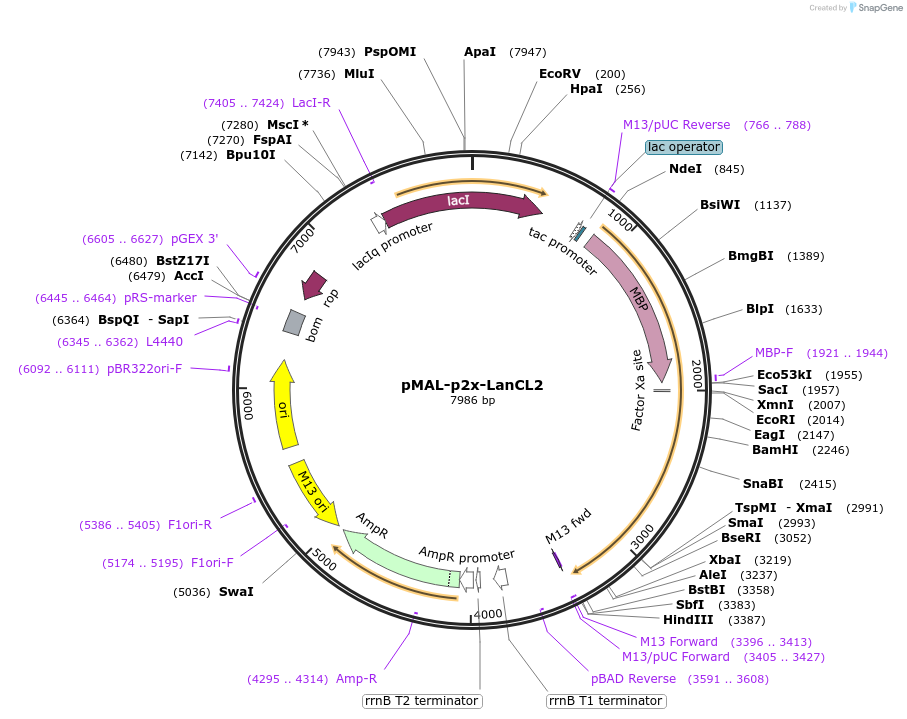
pMAL-p2x-LanCL2
Plasmid#154192PurposeTo express MBP-LanCL2 in bacterial cellsDepositorInsertLanCL2
TagsMaltose binding protein (MBP)ExpressionBacterialAvailable SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
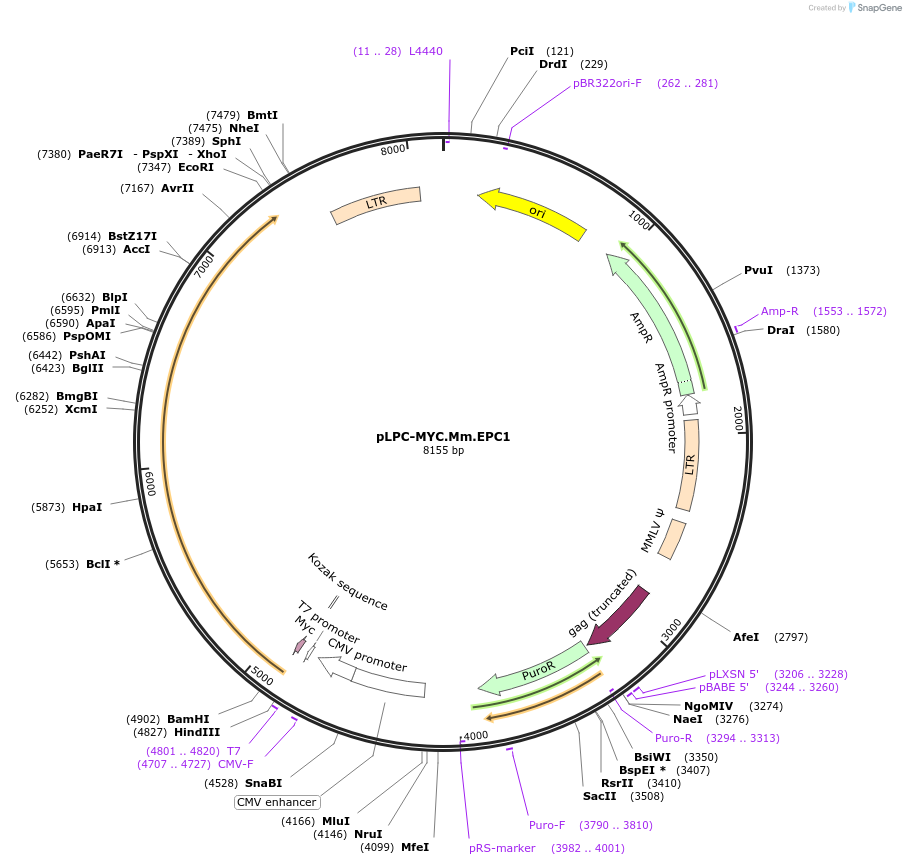
pLPC-MYC.Mm.EPC1
Plasmid#184653PurposeRetroviral expression of mouse Epc1DepositorAvailable SinceMay 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
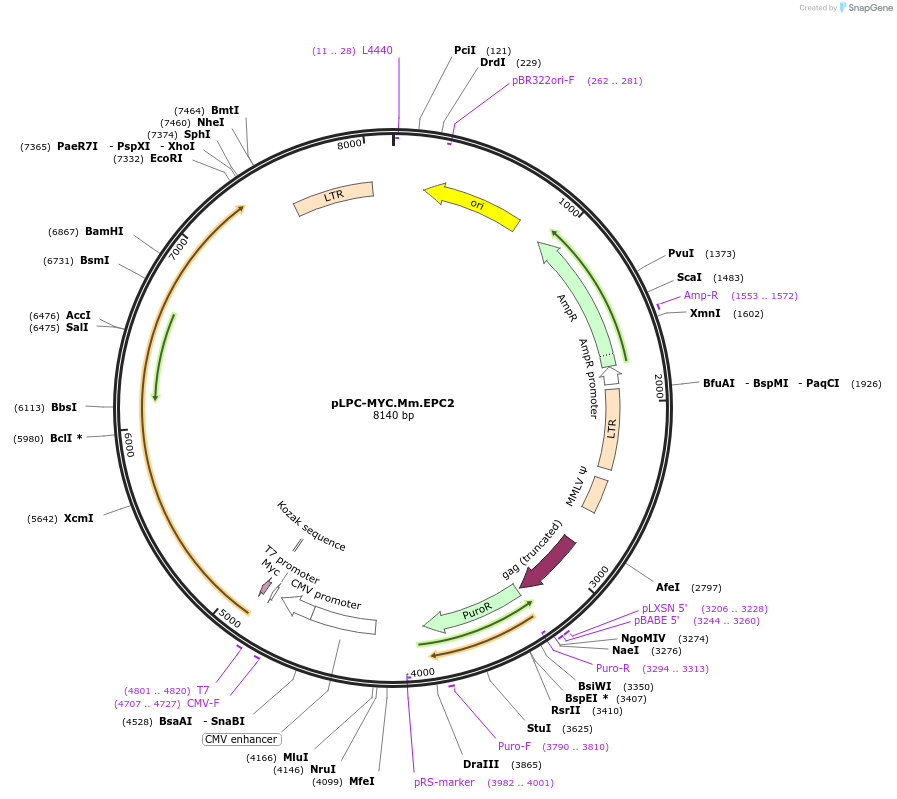
pLPC-MYC.Mm.EPC2
Plasmid#184654PurposeRetroviral expression of mouse Epc2DepositorAvailable SinceMay 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
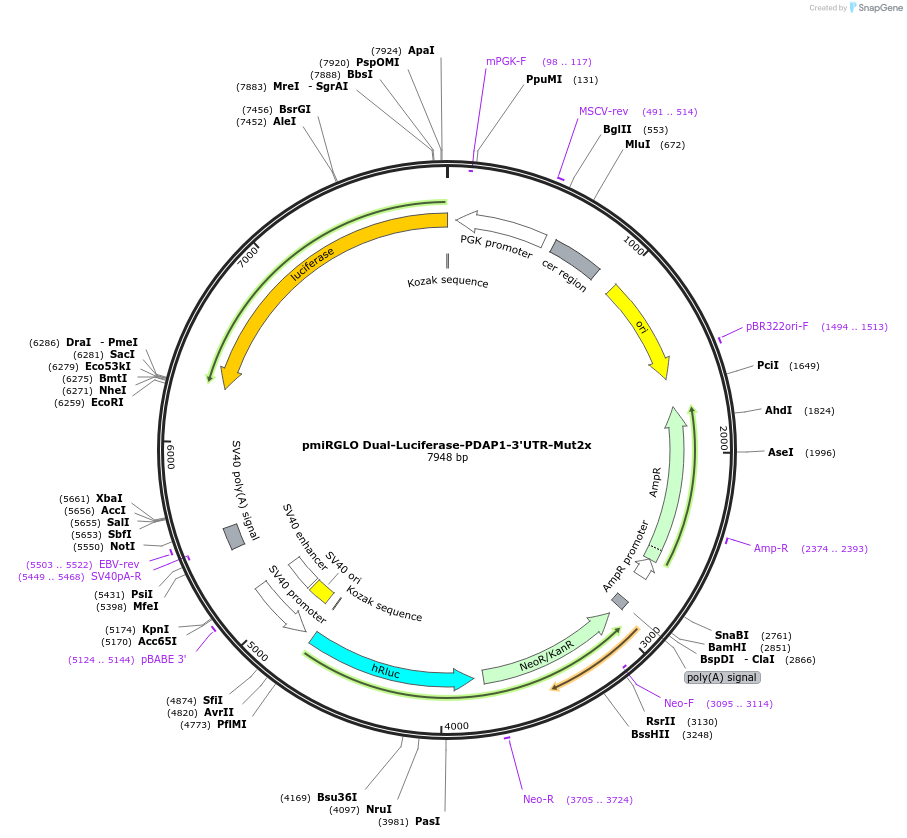
pmiRGLO Dual-Luciferase-PDAP1-3'UTR-Mut2x
Plasmid#182254PurposeHuman PDAP1 3` UTR region mutated in 2x binding site for miR-150 cloned downstream of luciferase reporter geneDepositorInsert3' UTR region of PDAP1 gene mutated in 2 binding sites for mir-150
UseLuciferaseExpressionMammalianAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
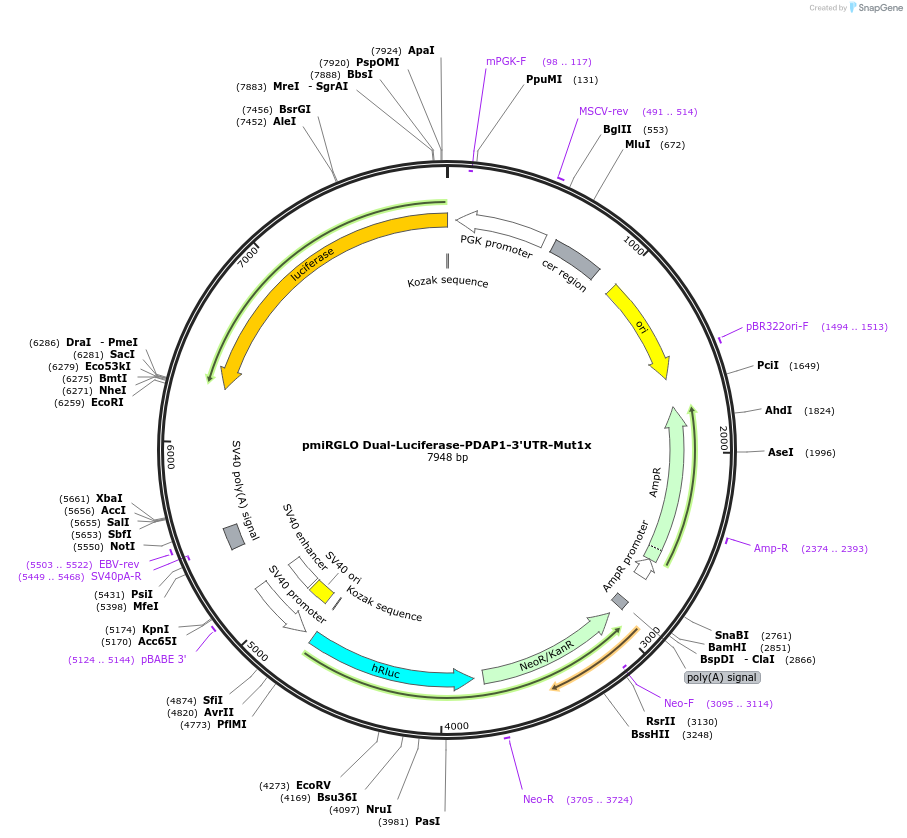
pmiRGLO Dual-Luciferase-PDAP1-3'UTR-Mut1x
Plasmid#182253PurposeHuman PDAP1 3` UTR region mutated in 1x binding site for miR-150 cloned downstream of luciferase reporter geneDepositorInsert3' UTR region of PDAP1 gene mutated in 1 binding site for mir-150
UseLuciferaseExpressionMammalianAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
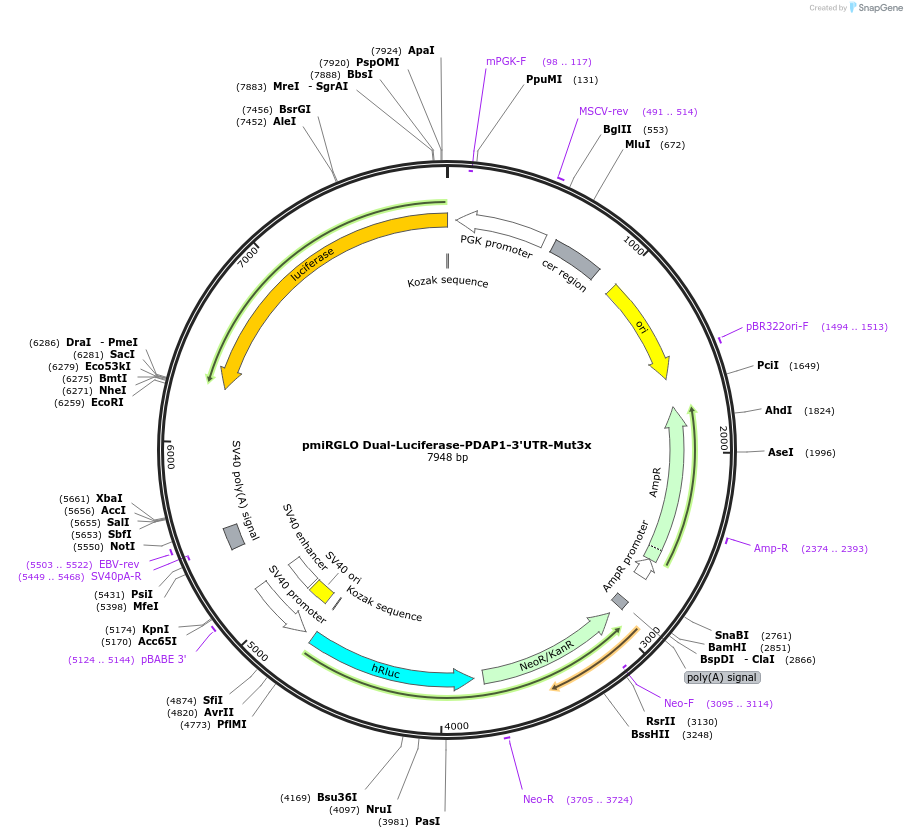
pmiRGLO Dual-Luciferase-PDAP1-3'UTR-Mut3x
Plasmid#182255PurposeHuman PDAP1 3` UTR region mutated in 3x binding site for miR-150 cloned downstream of luciferase reporter geneDepositorInsert3' UTR region of PDAP1 gene mutated in 3 binding sites for mir-150
UseLuciferaseExpressionMammalianAvailable SinceApril 14, 2022AvailabilityAcademic Institutions and Nonprofits only