We narrowed to 7,686 results for: CCH
-
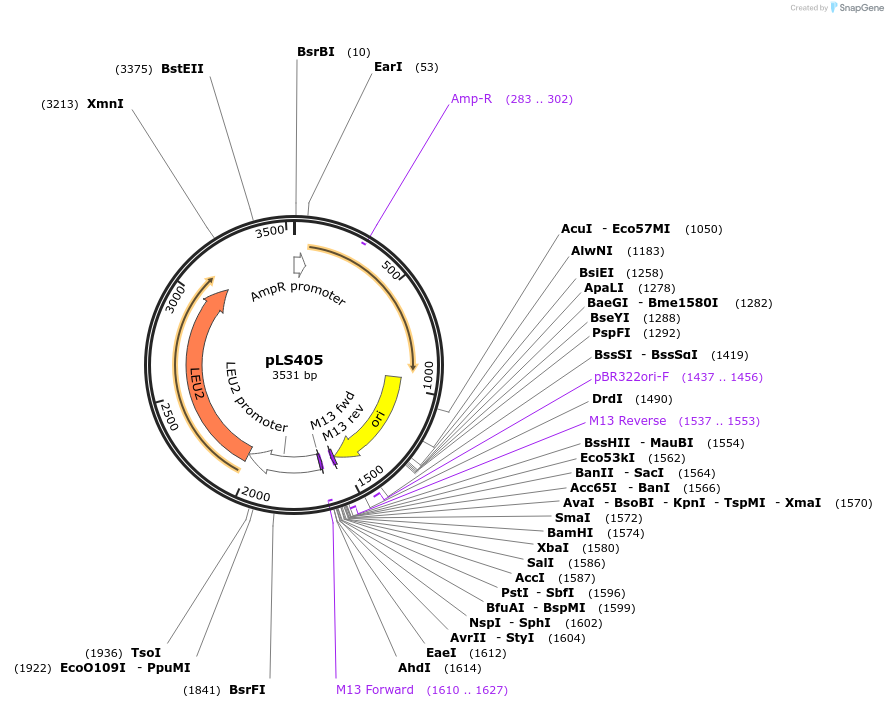
Plasmid#231077PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker LEU2DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only
-
pLS404
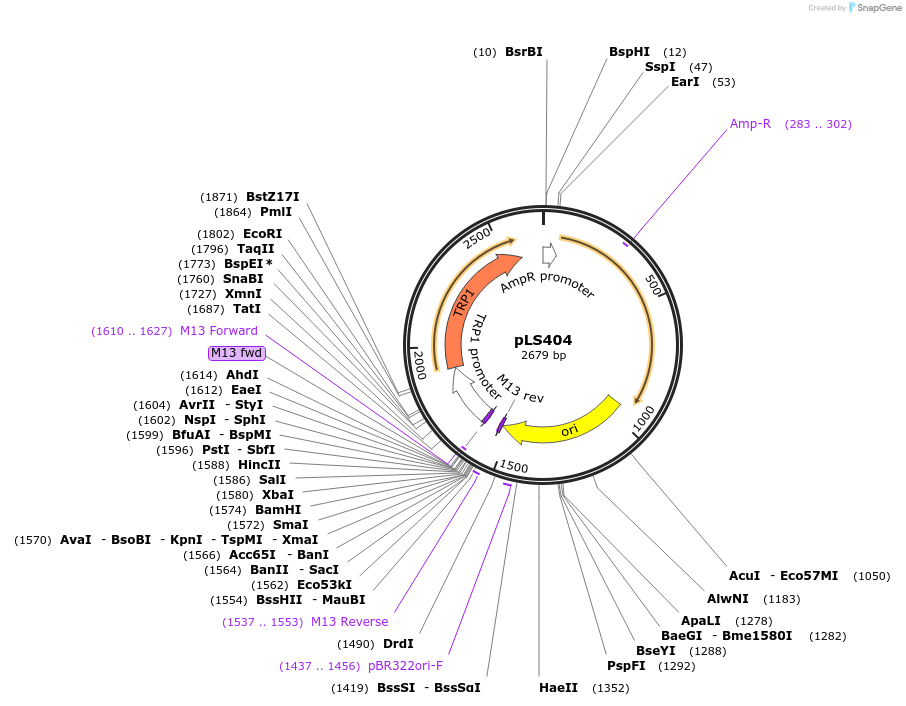
Plasmid#231076PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker TRP1DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
pLS408
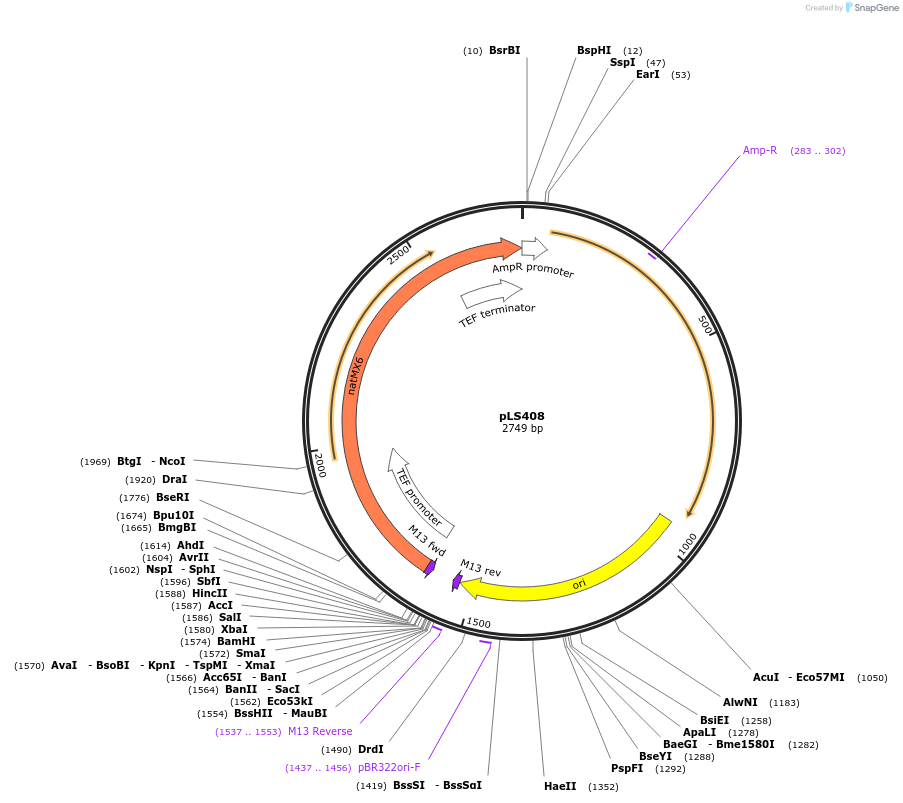
Plasmid#231079PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker natMX6DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
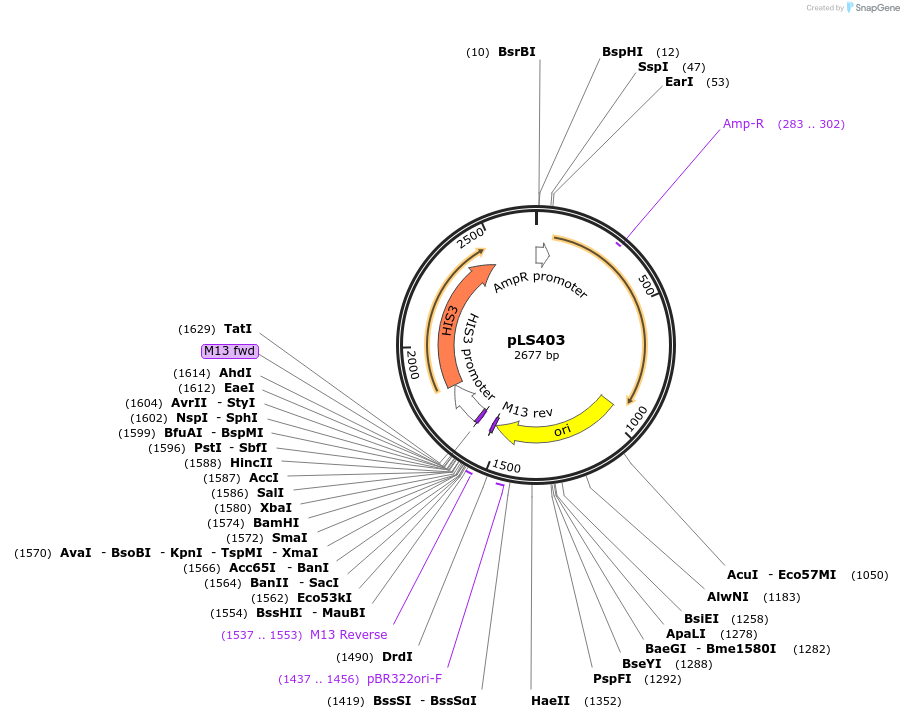
pLS403
Plasmid#231075PurposeMinimal integrating shuttle vector depleted of restriction sites outside the polylinker region, yeast selection marker HIS3DepositorTypeEmpty backboneUseIntegratingExpressionBacterial and YeastAvailable SinceJan. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
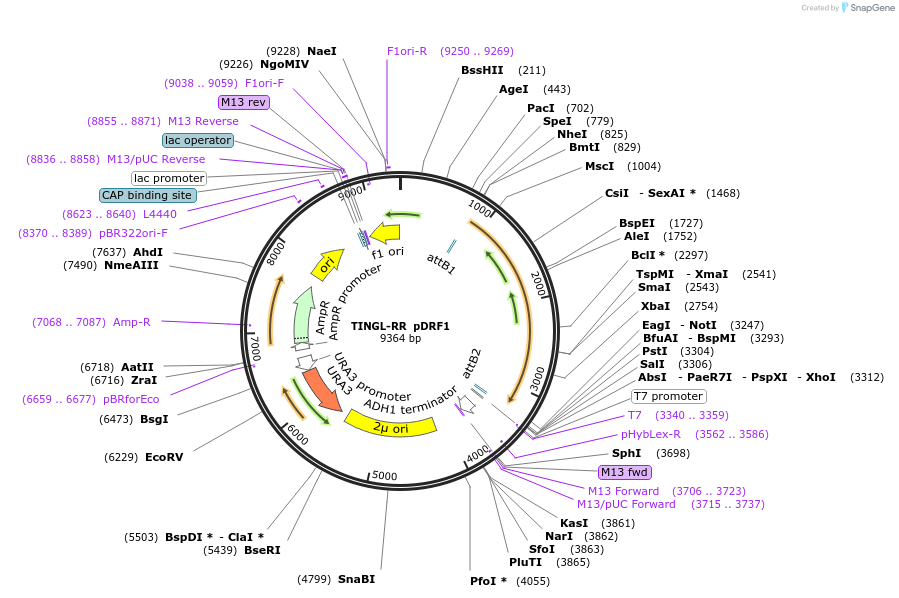
TINGL-RR pDRF1
Plasmid#226435PurposeExpresses TINGL-RR (mTq2-based glucose sensor - O2A - ymScarletI) in yeastDepositorInsertTINGL-O2A-ymScarletI
ExpressionYeastPromoterPMA1Available SinceDec. 2, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
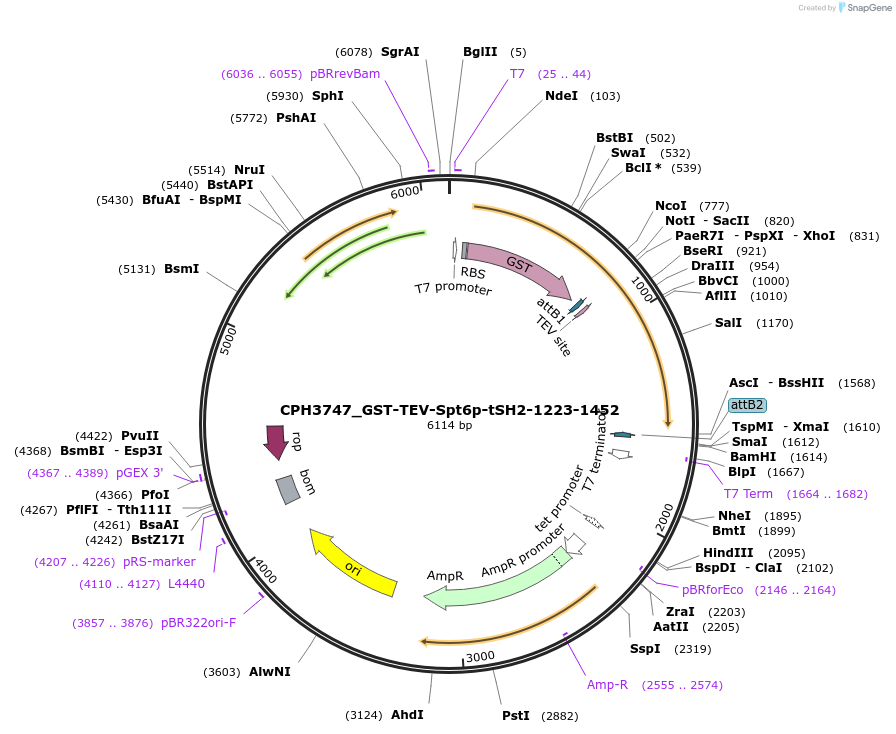
CPH3747_GST-TEV-Spt6p-tSH2-1223-1452
Plasmid#227086PurposeBacterial expression of R1282H single-mutant GST-tagged Spt6 tSH2 domain (1223-1452)DepositorInsertspt6-tsh2 (SPT6 Budding Yeast)
TagsGST-TEV_cleavage_siteExpressionBacterialMutationR1282HPromoterT7Available SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
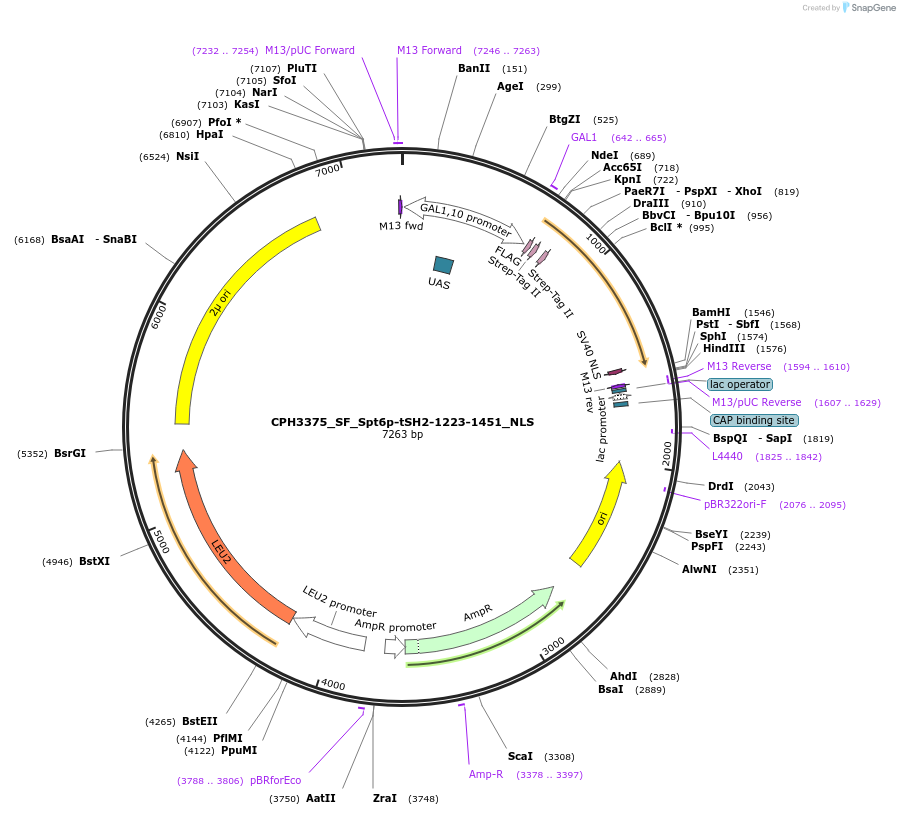
CPH3375_SF_Spt6p-tSH2-1223-1451_NLS
Plasmid#227084PurposeGal1p dependent budding yeast expression of tandem-affinity tagged spt6p tSH2 domain (1123-1451) localized to nucleusDepositorInsertspt6-tsh2 (SPT6 Budding Yeast)
TagsSTREPii-FLAG and SV40 NLS 1ExpressionYeastPromoterGal1-10Available SinceNov. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
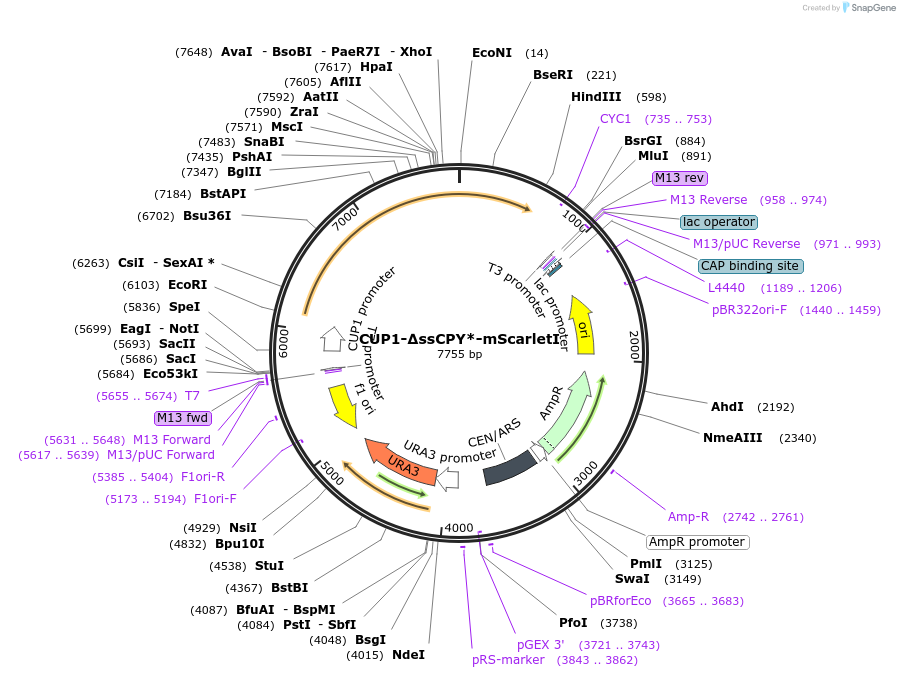
CUP1-∆ssCPY*-mScarletI
Plasmid#221157PurposeFluorescent reporter for cytoplasmic aggregatesDepositorInsertCarboxypeptidase Y (PRC1 Budding Yeast)
TagsmScarletIExpressionYeastMutationDeletion signal peptide, G255A mutationAvailable SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
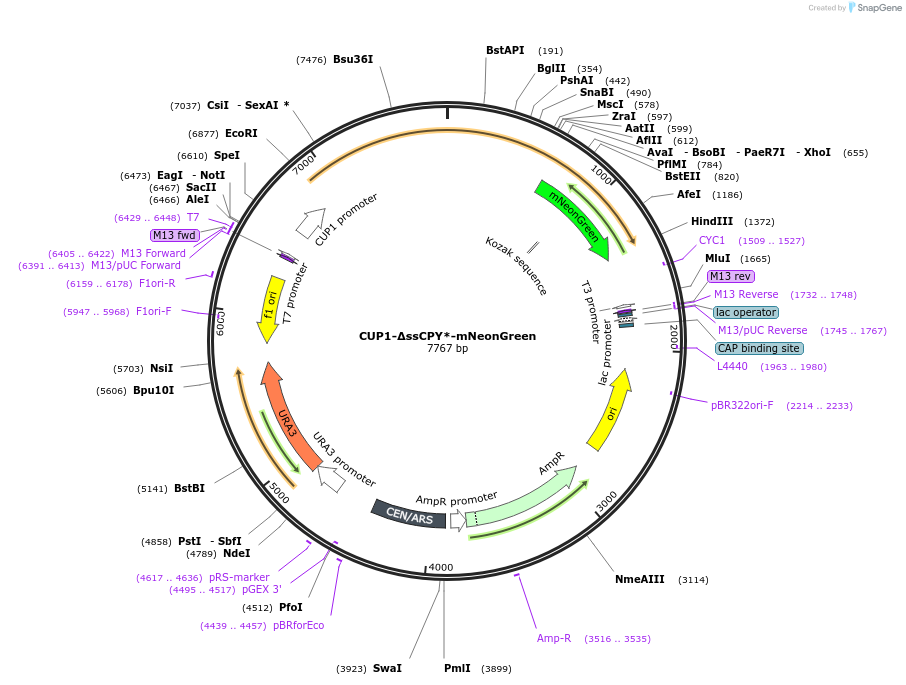
CUP1-∆ssCPY*-mNeonGreen
Plasmid#221156PurposeFluorescent reporter for cytoplasmic aggregatesDepositorInsertCarboxypeptidase Y (PRC1 Budding Yeast)
TagsmNeonGreenExpressionYeastMutationDeletion signal peptide, G255A mutationAvailable SinceNov. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
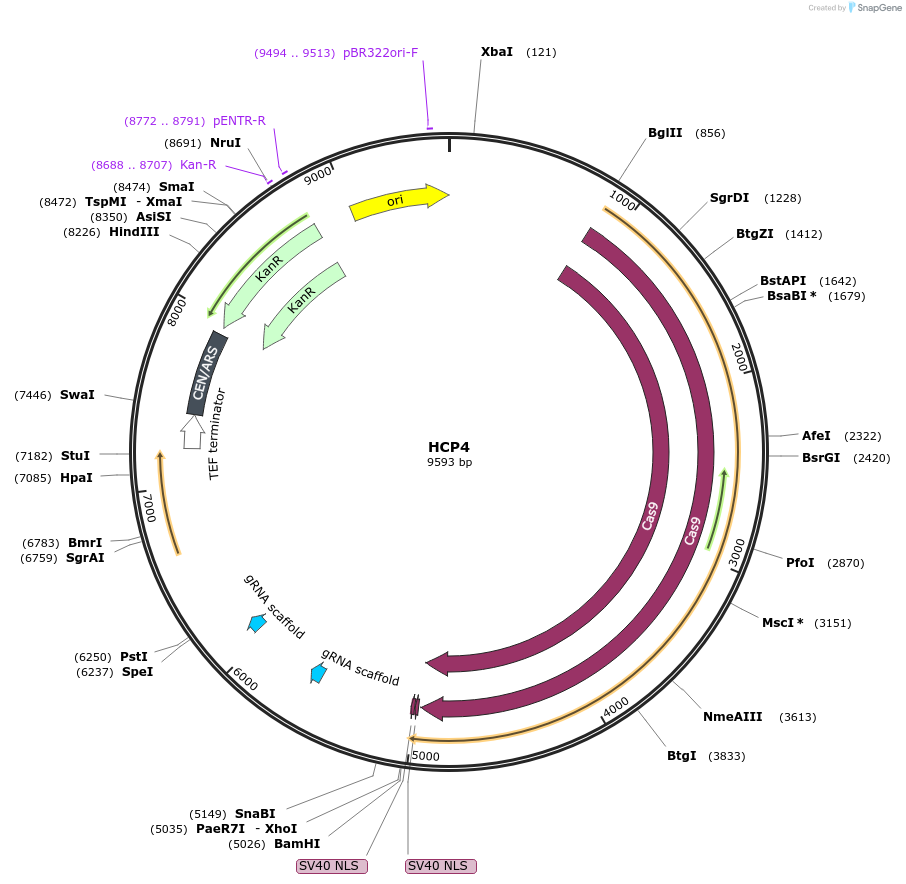
HCP4
Plasmid#166106PurposeThis plasmid encodes a Cas9 protein as well as two sgRNAs, one targets the center of Cyr1 and the other targets a non-coding region on chromosome X (location X1)DepositorAvailable SinceApril 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
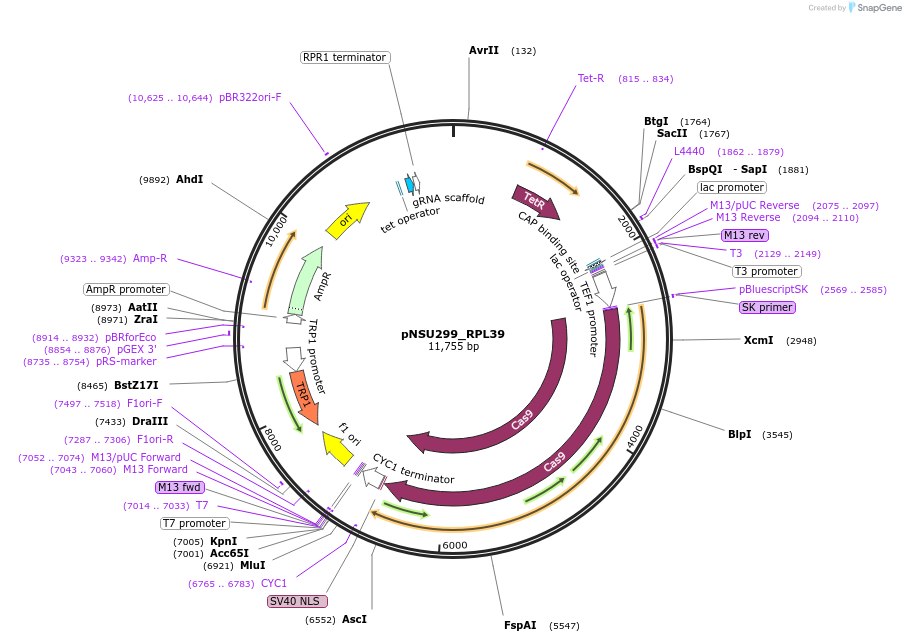
pNSU299_RPL39
Plasmid#166078PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA binding to the promoter of RPL39 for double stranded break formation in yeast.DepositorInsertPromoter of RPL39 (Overlaps with 5'UTR and first base of gene) (RPL39 Budding Yeast)
UseCRISPRExpressionYeastPromoterTet-inducibleAvailable SinceApril 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
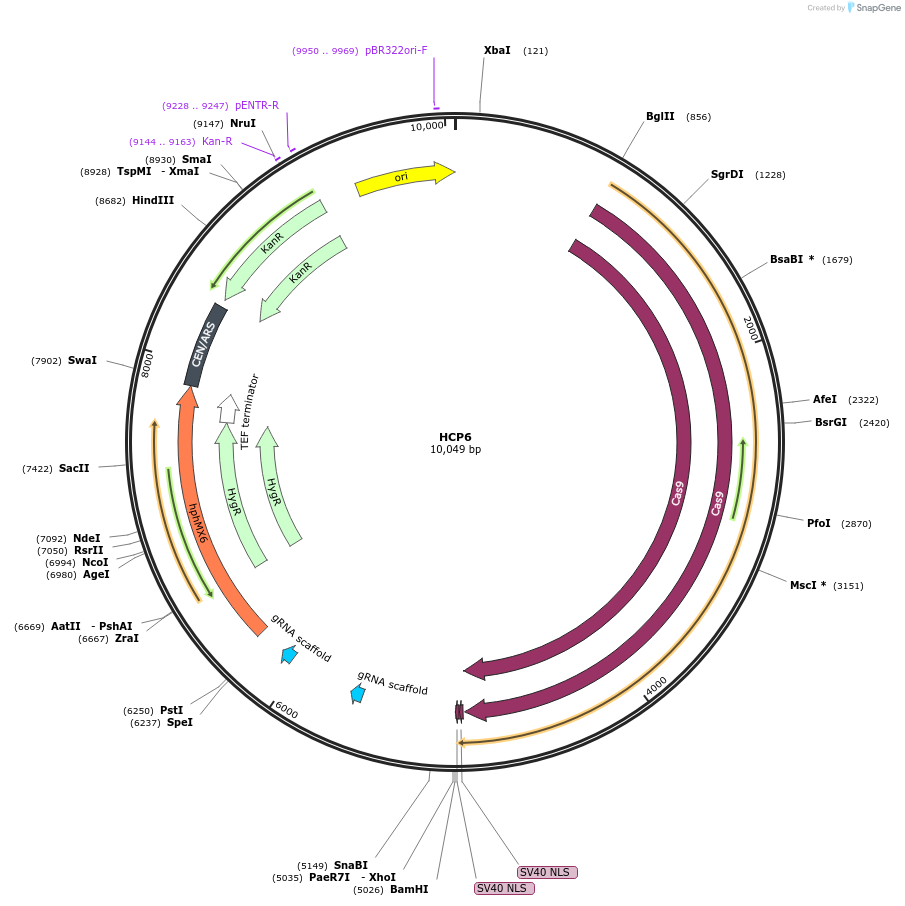
HCP6
Plasmid#166108PurposeThis plasmid encodes a Cas9 protein as well as two sgRNAs, one targets the C-terminus of Cyr1 and the other targets a non-coding region on chromosome X (location X1)DepositorAvailable SinceApril 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
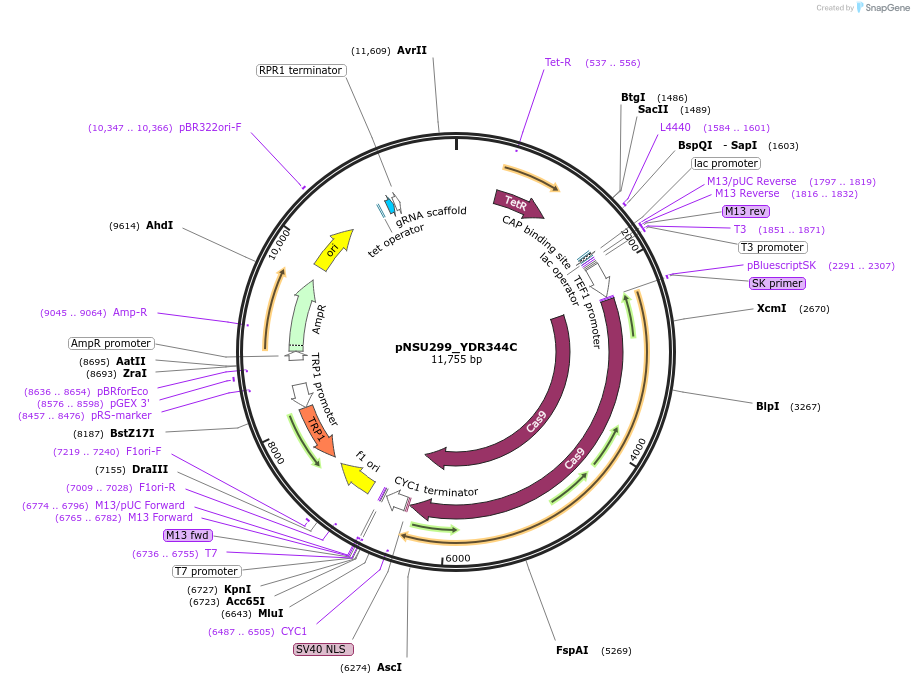
pNSU299_YDR344C
Plasmid#166072PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA targeting intergenic site near HXT6 (within YDR344C) for double stranded break formation in yeast.DepositorInsertIntergenic site near HXT6 (in YDR344C, possible dubious ORF)
UseCRISPRExpressionYeastPromoterTet-inducibleAvailable SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
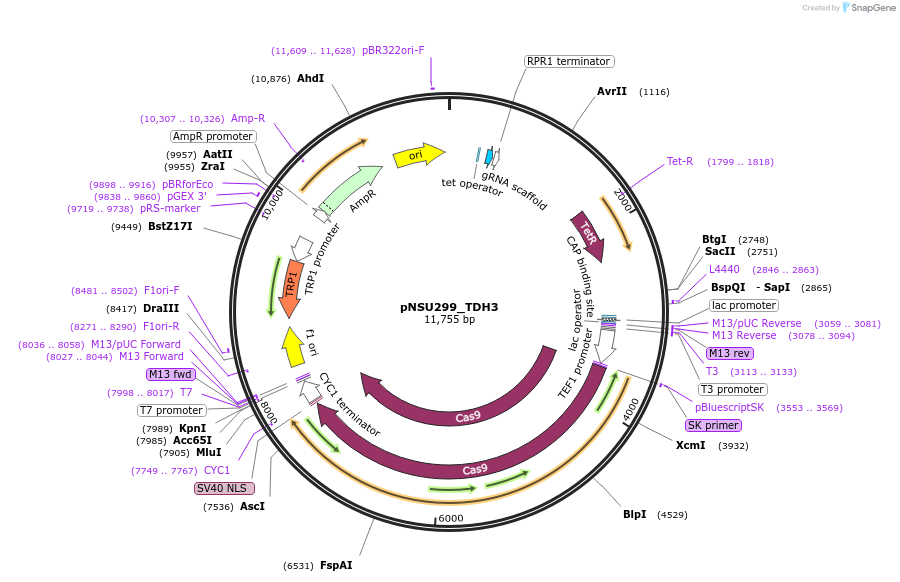
pNSU299_TDH3
Plasmid#166084PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA binding to the promoter of TDH3 for double stranded break formation in yeast.DepositorAvailable SinceMarch 31, 2021AvailabilityAcademic Institutions and Nonprofits only -
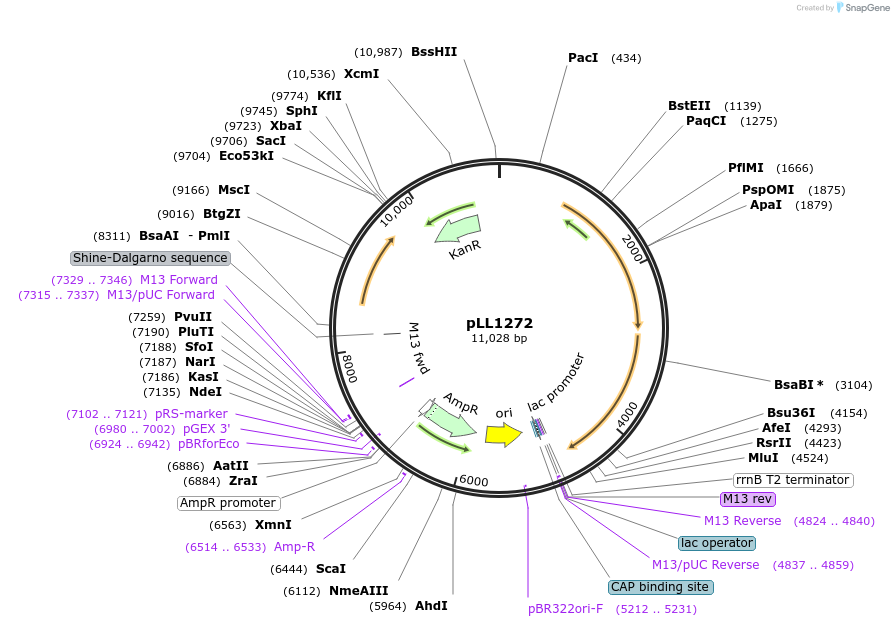
pLL1272
Plasmid#129287PurposeExpression plasmid with pLL1270 as backbone and alpha-Xylp_1211 and alpha-L-Araf_1120 as insertsDepositorInsertsalpha-Xylp_1211 (Ga0256695_ 1211)
alpha-L-Araf_1120
ExpressionBacterialPromoterClostridium thermocellum DSM1313 Cellobiose Phosp…Available SinceMarch 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
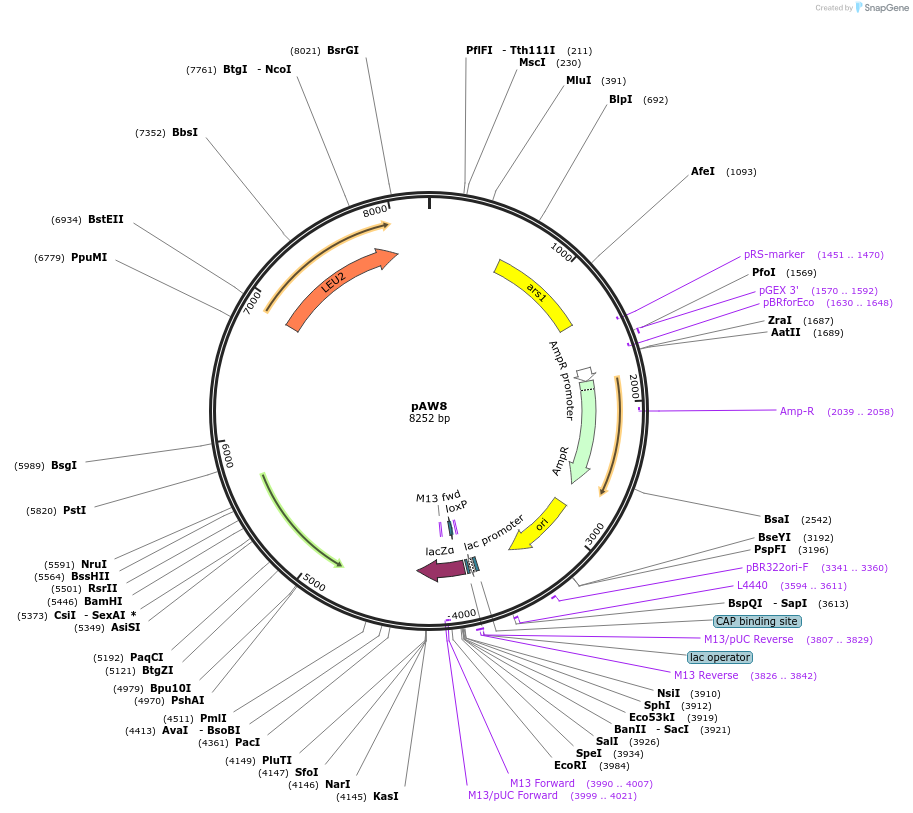
pAW8
Plasmid#110222PurposeCre-expression plasmid for cassette exchangeDepositorTypeEmpty backboneUseCre/LoxAvailable SinceOct. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
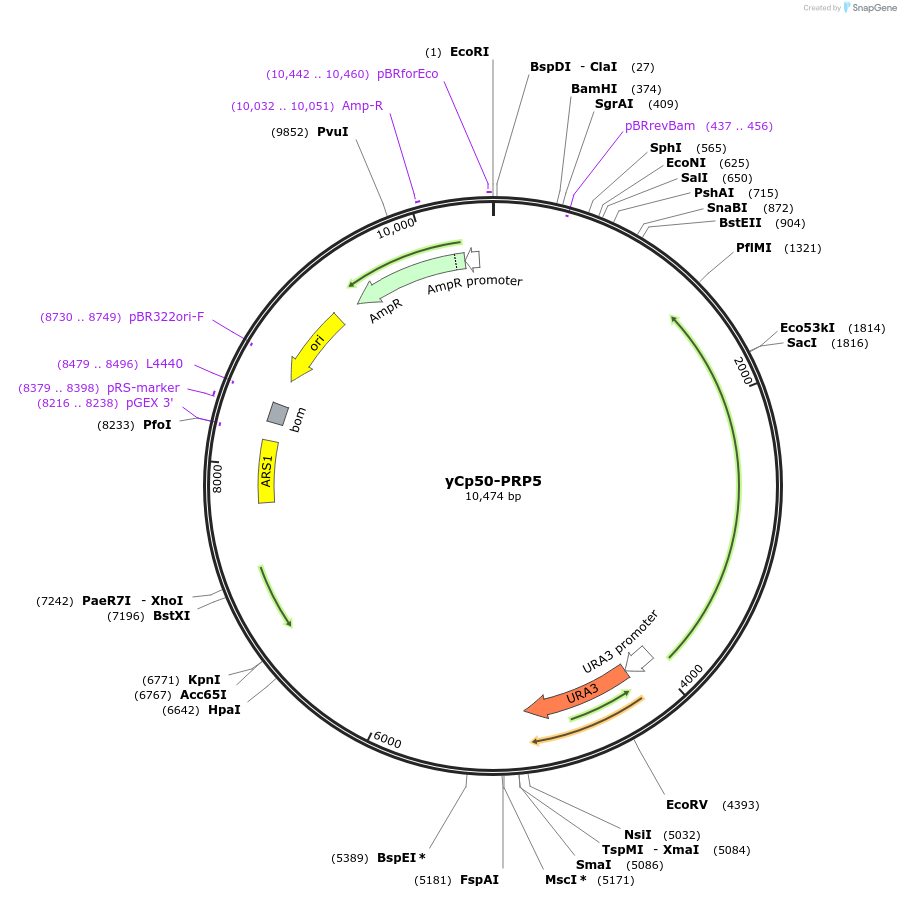
yCp50-PRP5
Plasmid#111303PurposePrp5 in yCp50 plasmidDepositorInsertPRP5
ExpressionYeastAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
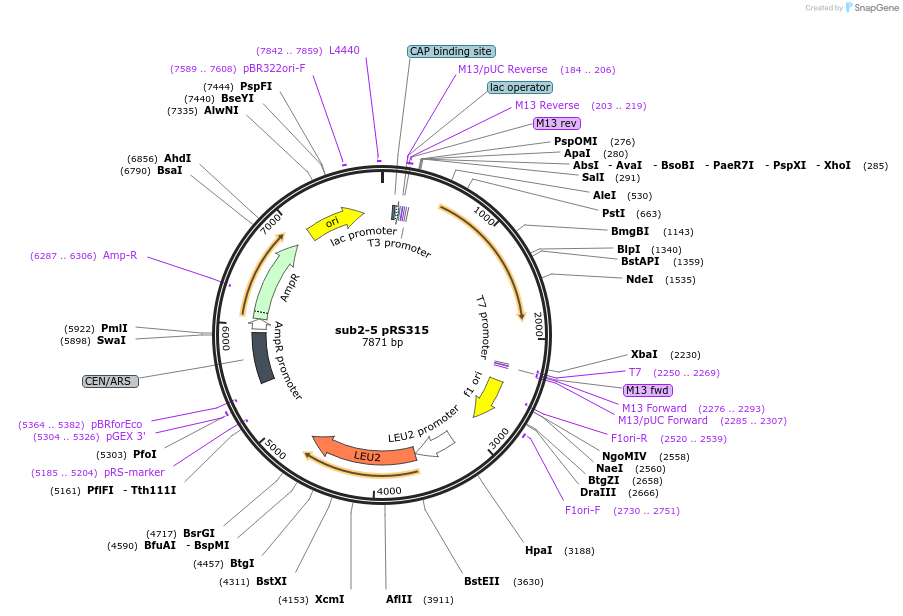
sub2-5 pRS315
Plasmid#111302Purposesub2-5 in pRS315DepositorInsertSUB2
ExpressionYeastMutationsub2-5Available SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
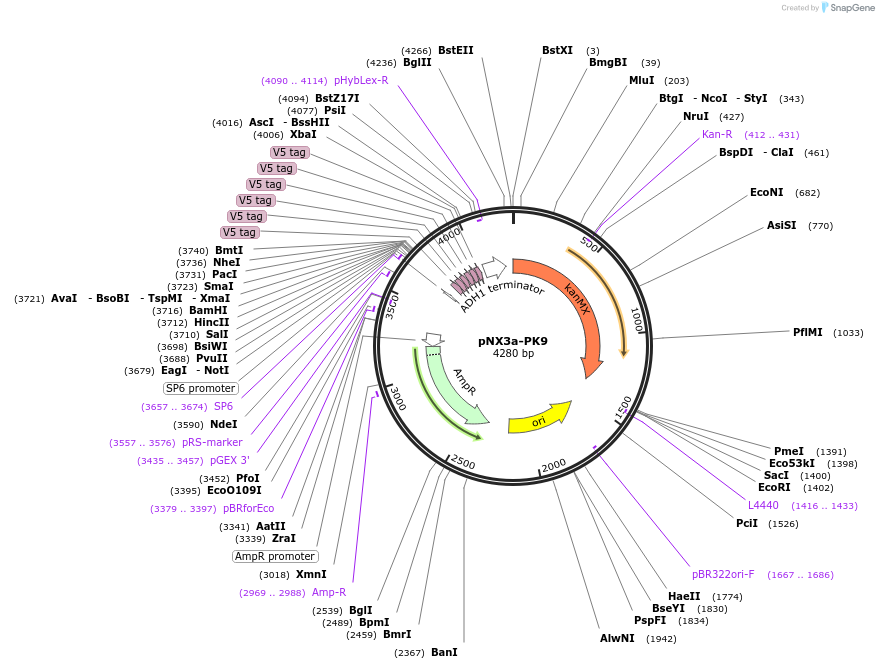
pNX3a-PK9
Plasmid#78356Purposefission yeast epitope taggingDepositorInserts9xPK
kanMX6
ExpressionYeastAvailable SinceMarch 10, 2017AvailabilityAcademic Institutions and Nonprofits only -
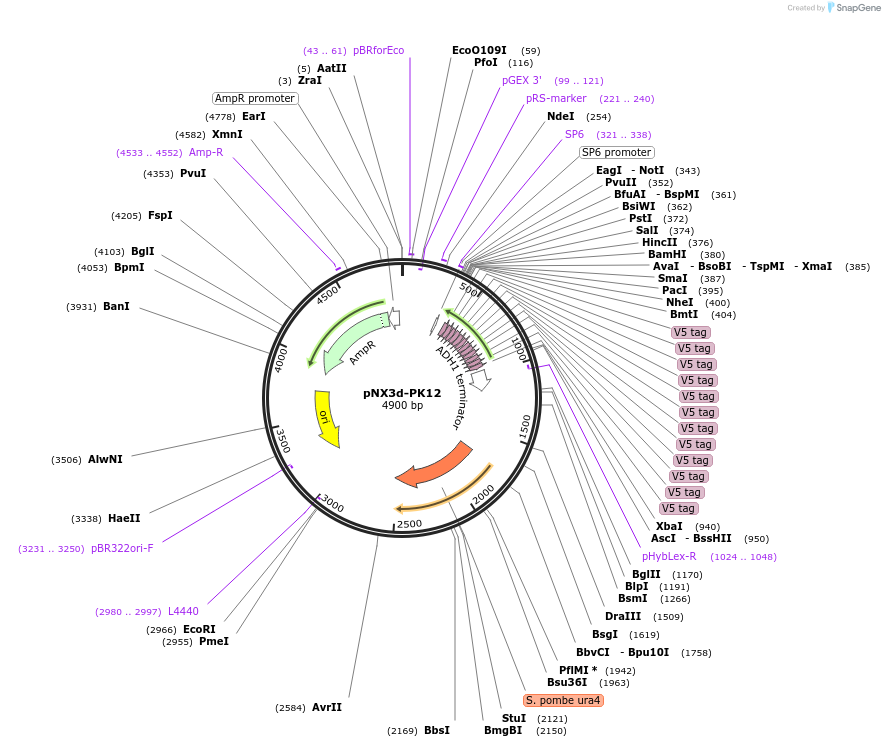
pNX3d-PK12
Plasmid#78378Purposefission yeast epitope taggingDepositorInsertsAvailable SinceNov. 17, 2016AvailabilityAcademic Institutions and Nonprofits only -
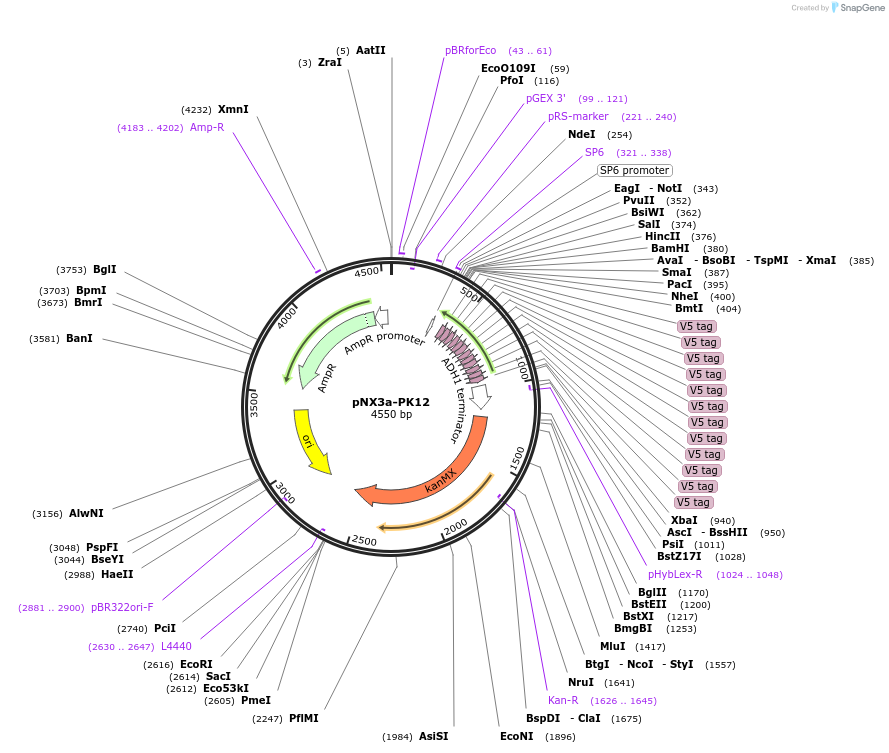
pNX3a-PK12
Plasmid#78357Purposefission yeast epitope taggingDepositorInserts12xPK
kanMX6
ExpressionYeastAvailable SinceNov. 17, 2016AvailabilityAcademic Institutions and Nonprofits only -
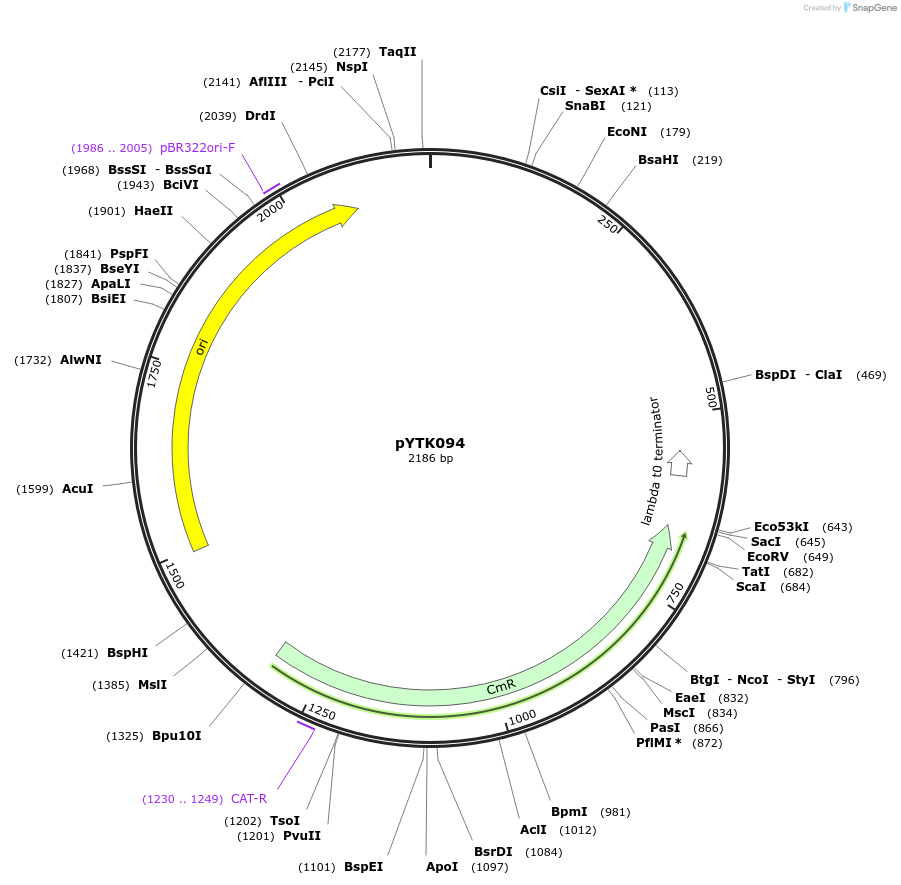
pYTK094
Plasmid#65201PurposeEncodes HO 5' Homology as a Type 8b part to be used in the Dueber YTK systemDepositorInsertHO 5' Homology
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
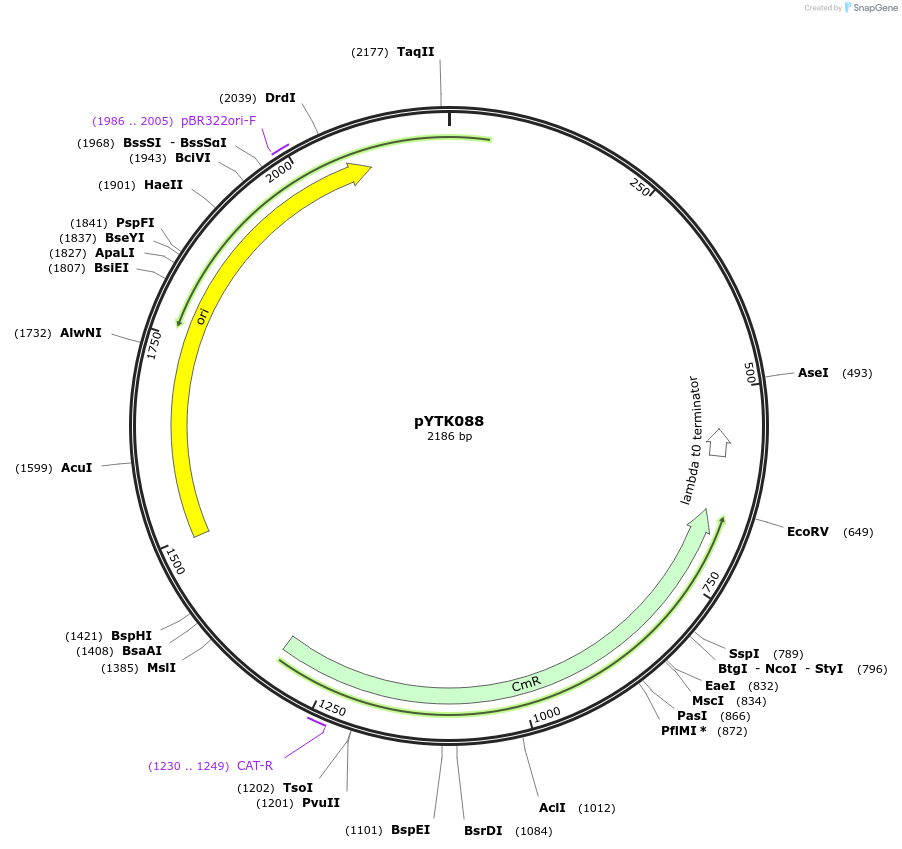
pYTK088
Plasmid#65195PurposeEncodes HO 3' Homology as a Type 7 part to be used in the Dueber YTK systemDepositorInsertHO 3' Homology
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
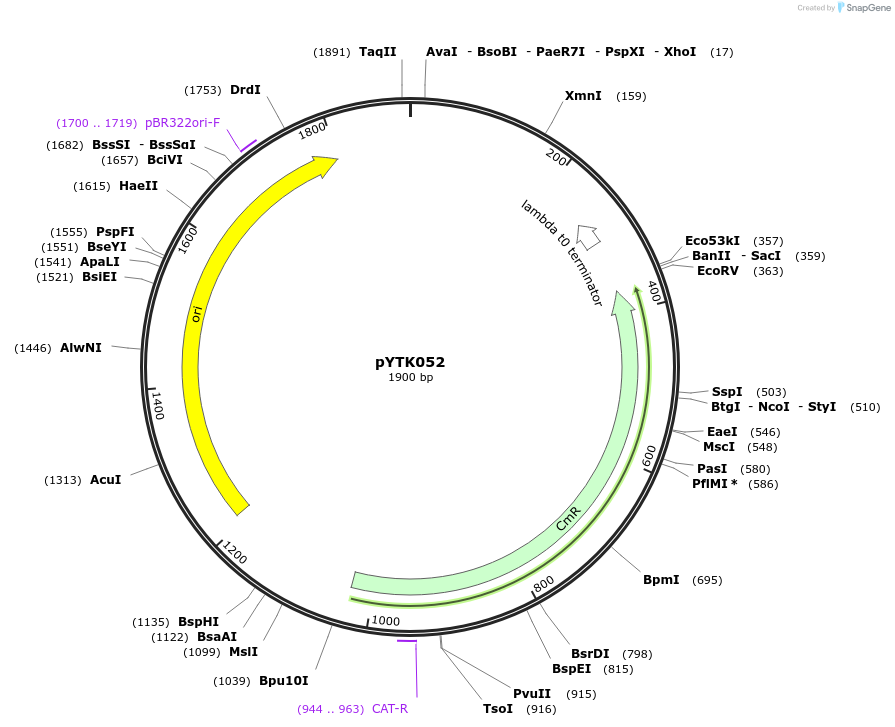
pYTK052
Plasmid#65159PurposeEncodes tSSA1 as a Type 4 part to be used in the Dueber YTK systemDepositorInserttSSA1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
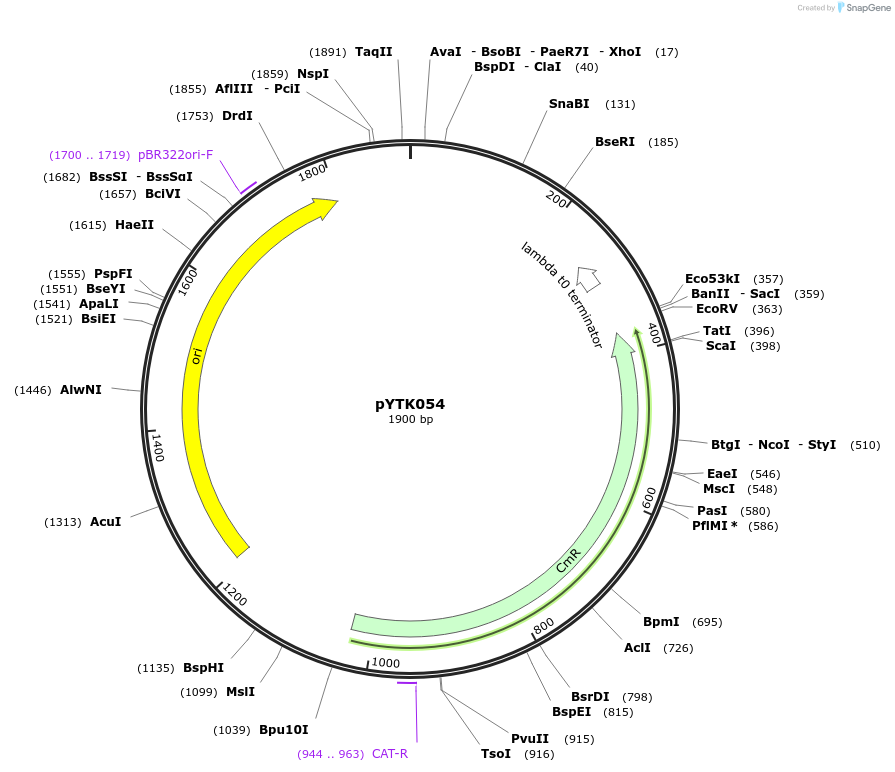
pYTK054
Plasmid#65161PurposeEncodes tPGK1 as a Type 4 part to be used in the Dueber YTK systemDepositorInserttPGK1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
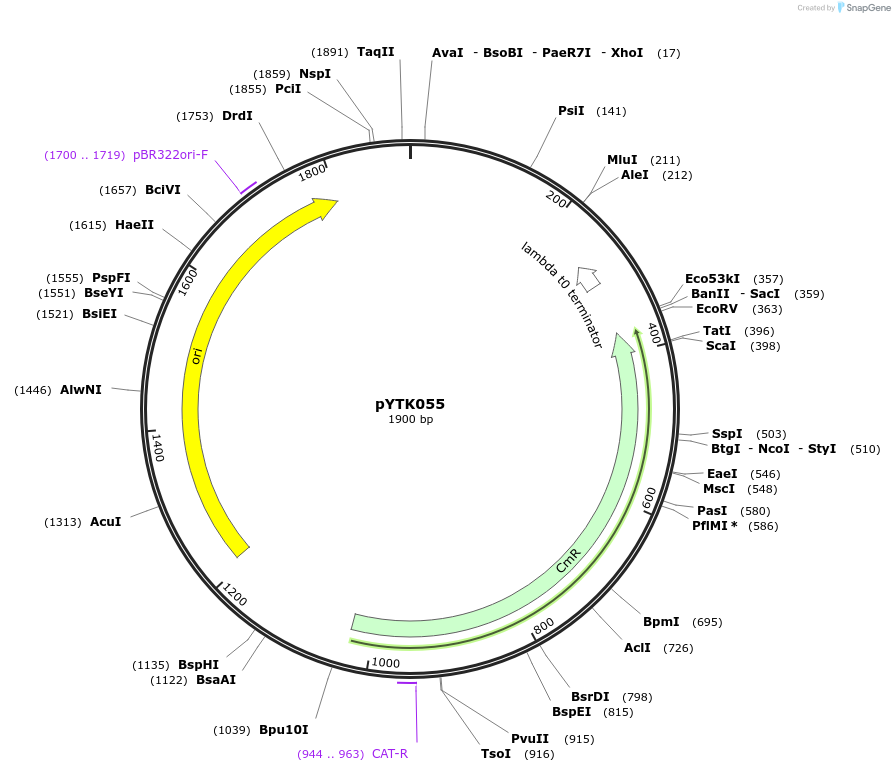
pYTK055
Plasmid#65162PurposeEncodes tENO2 as a Type 4 part to be used in the Dueber YTK systemDepositorInserttENO2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
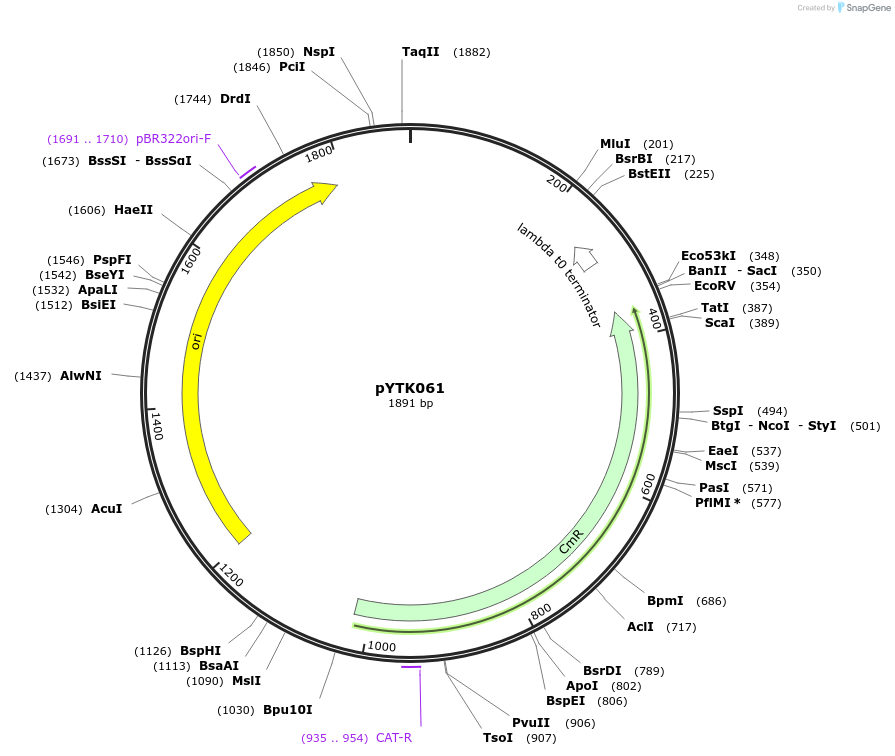
pYTK061
Plasmid#65168PurposeEncodes tENO1 as a Type 4b part to be used in the Dueber YTK systemDepositorInserttENO1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
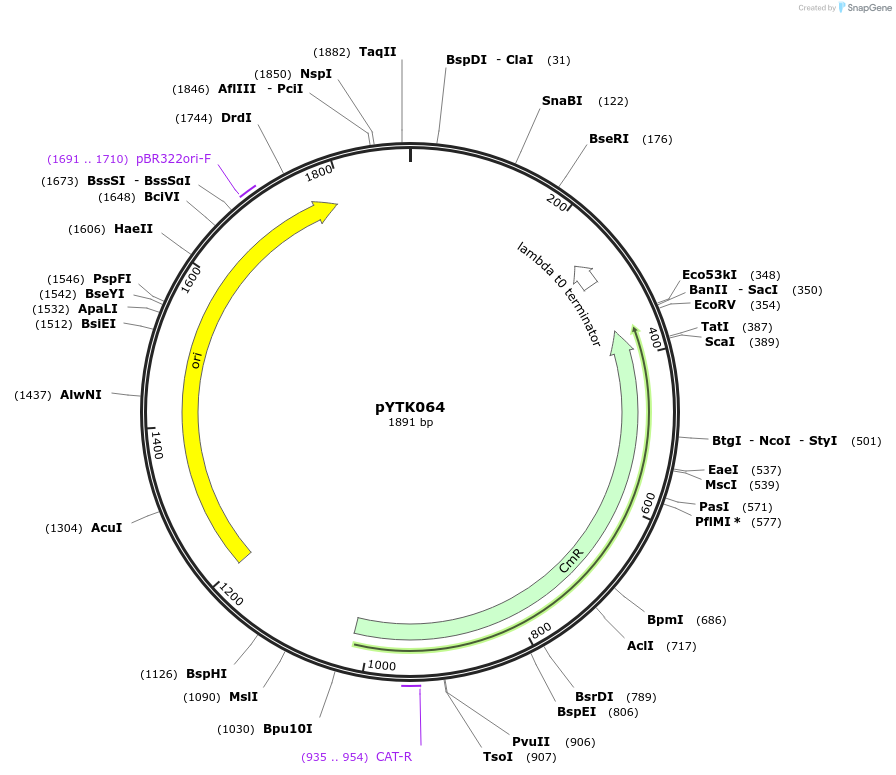
pYTK064
Plasmid#65171PurposeEncodes tPGK1 as a Type 4b part to be used in the Dueber YTK systemDepositorInserttPGK1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
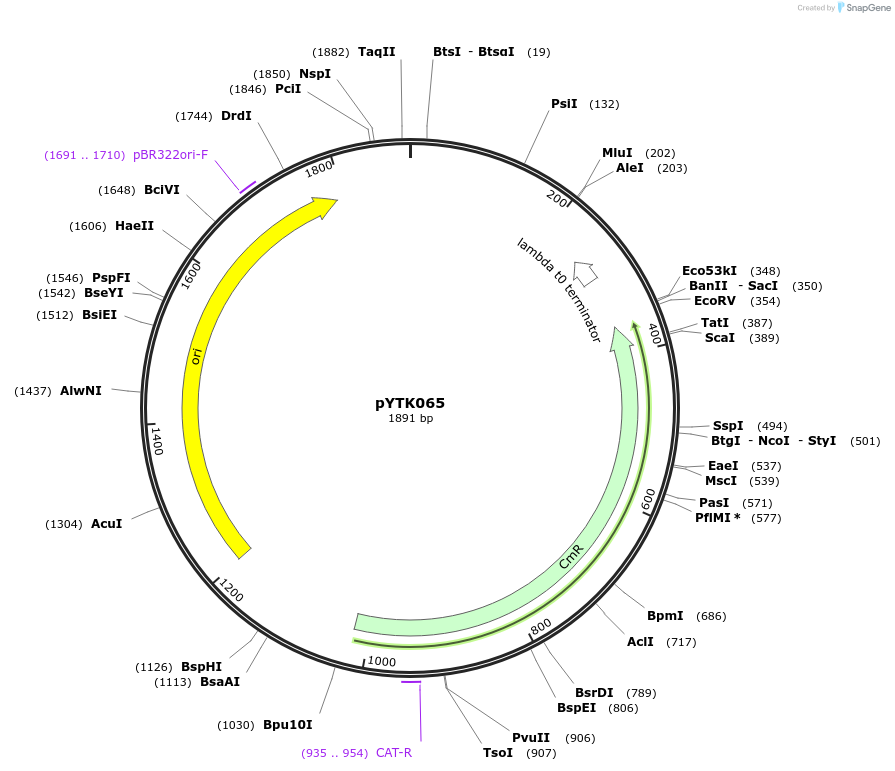
pYTK065
Plasmid#65172PurposeEncodes tENO2 as a Type 4b part to be used in the Dueber YTK systemDepositorInserttENO2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
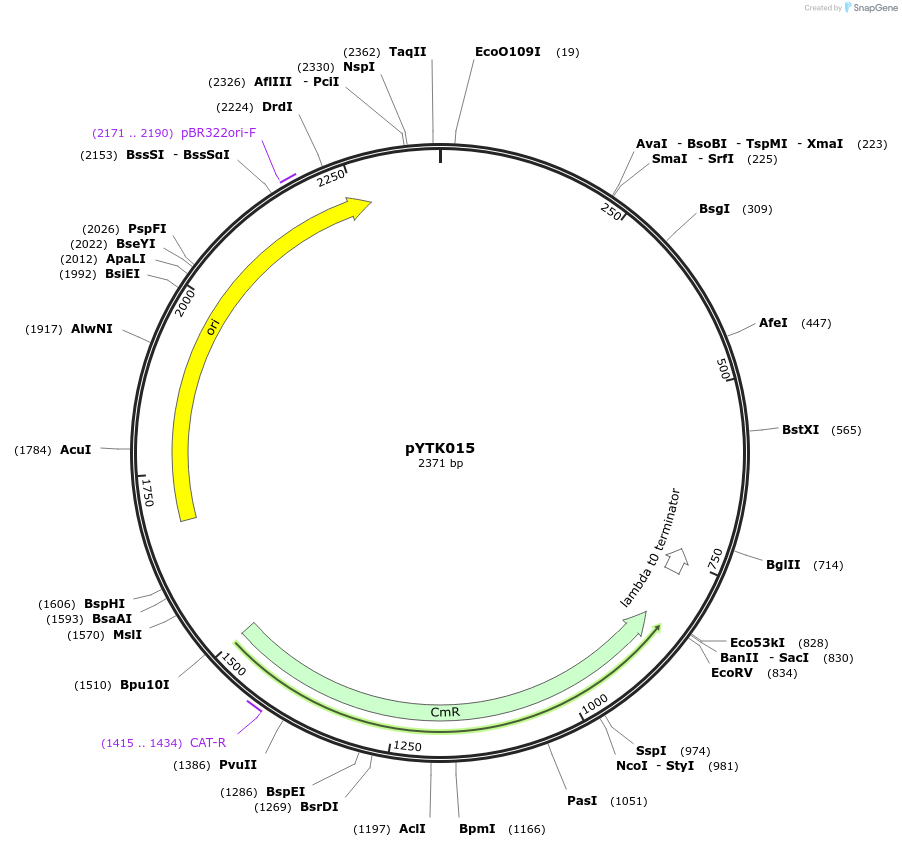
pYTK015
Plasmid#65122PurposeEncodes pHHF1 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpHHF1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
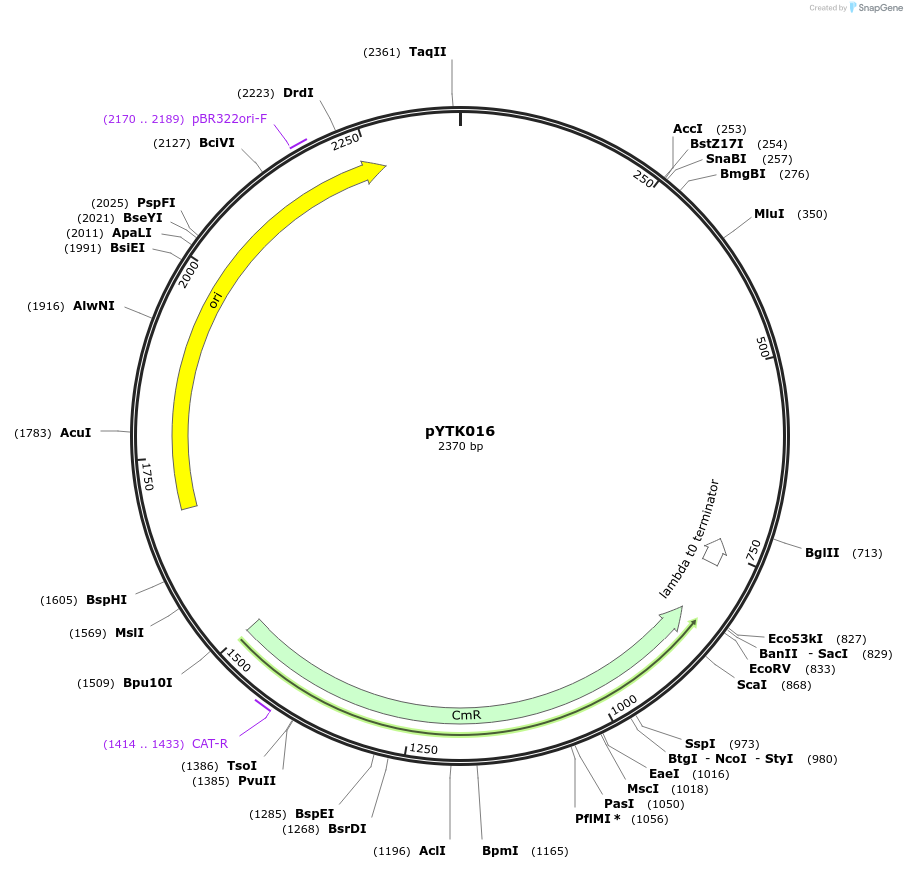
pYTK016
Plasmid#65123PurposeEncodes pHTB2 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpHTB2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
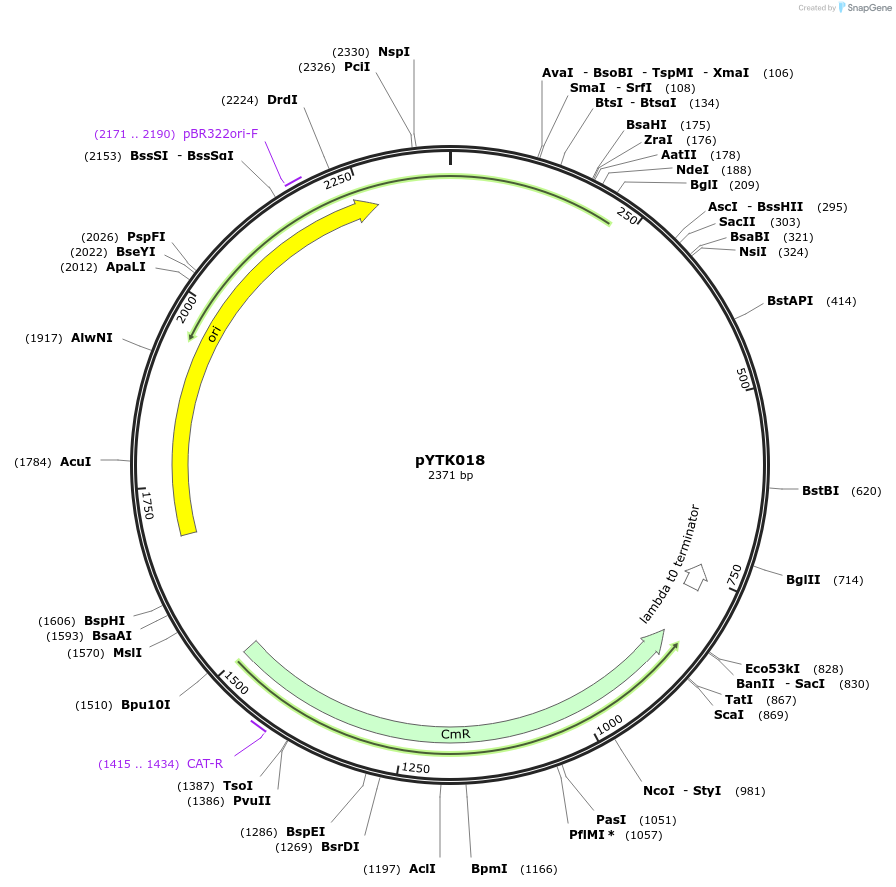
pYTK018
Plasmid#65125PurposeEncodes pALD6 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpALD6
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
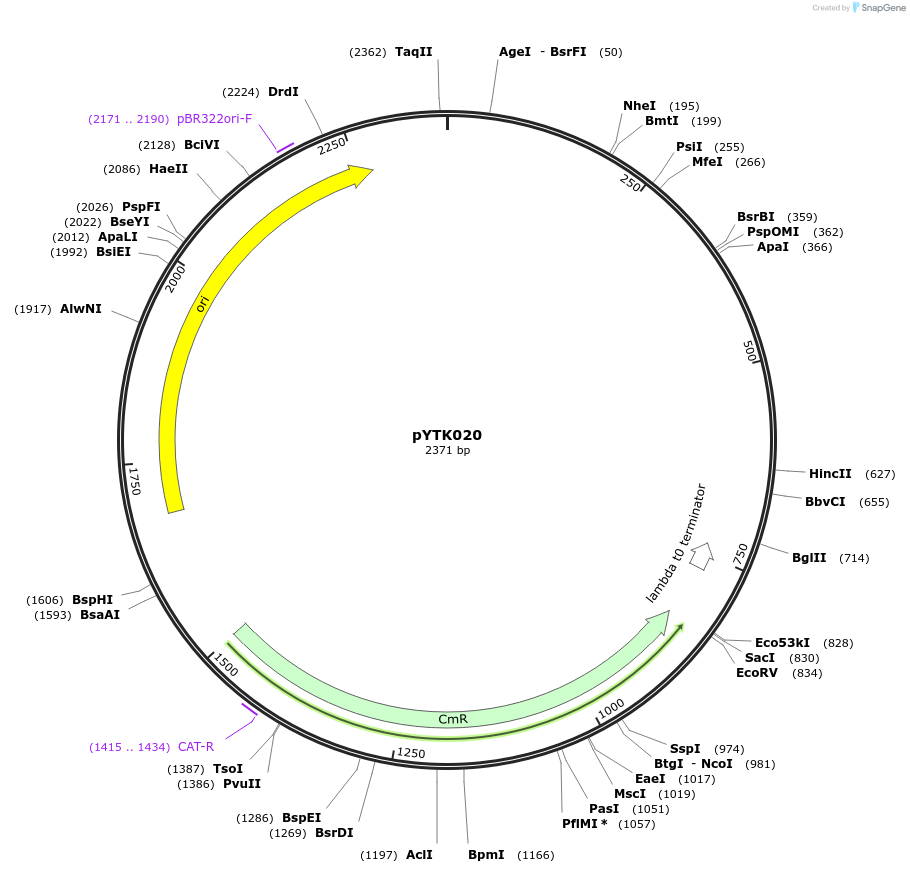
pYTK020
Plasmid#65127PurposeEncodes pRET2 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpRET2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
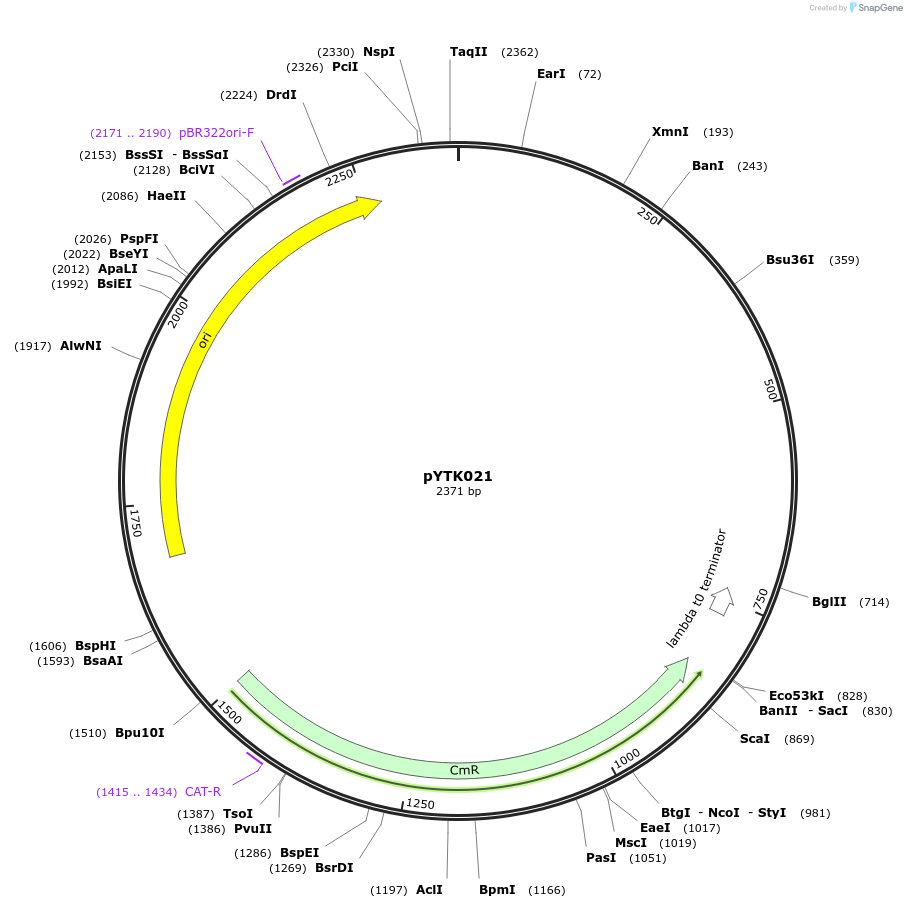
pYTK021
Plasmid#65128PurposeEncodes pRNR1 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpRNR1
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
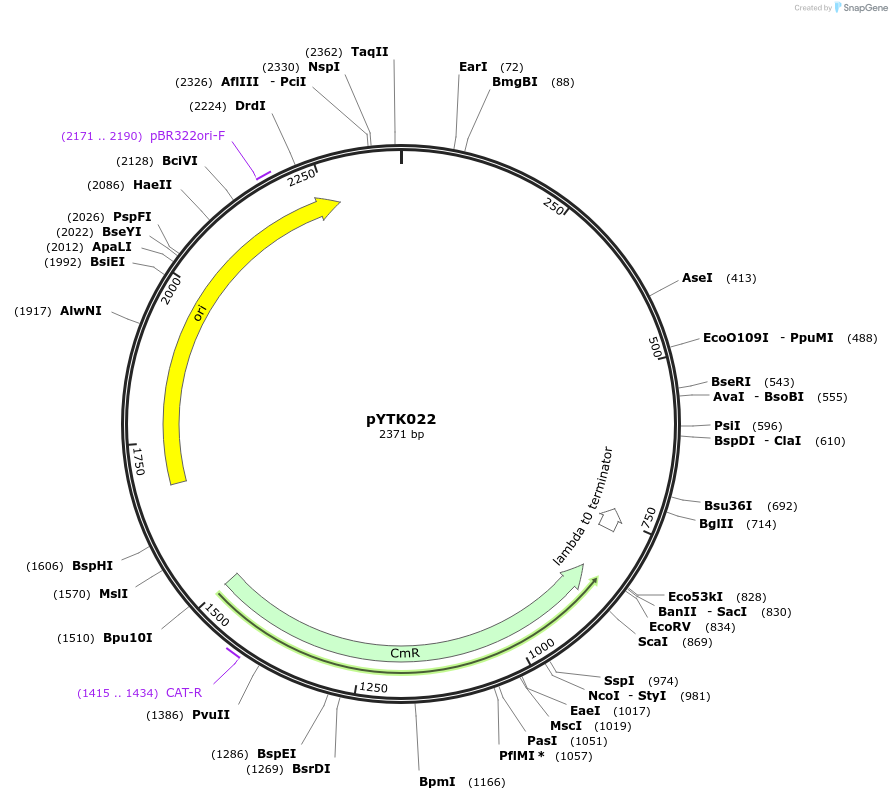
pYTK022
Plasmid#65129PurposeEncodes pSAC6 as a Type 2 part to be used in the Dueber YTK systemDepositorInsertpSAC6
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRPR1_g2gRNA_RPR1t
Plasmid#64388Purposeencodes g2 gRNADepositorInsertg2 gRNA
ExpressionYeastPromoterpRPR1Available SinceMay 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFX05
Plasmid#44531DepositorInsertHHT2
ExpressionYeastMutationK56QAvailable SinceMay 23, 2013AvailabilityAcademic Institutions and Nonprofits only -
pCM350
Plasmid#44549DepositorInsertHA-htz1
TagsHAExpressionYeastMutationdeleted amino acids 1-22Available SinceMay 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
pFX06
Plasmid#44532DepositorInsertHHT2
ExpressionYeastMutationK56RAvailable SinceMay 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
pTH672-SUP35-Y351C
Plasmid#29733DepositorInsertSUP35 gene encoding translation release factor 3 from S. cerevisiae (SUP35 Budding Yeast)
ExpressionYeastMutationY351C mutant gene of SUP35 including genomic sequ…Available SinceSept. 6, 2011AvailabilityAcademic Institutions and Nonprofits only -
FC511
Plasmid#27330DepositorAvailable SinceFeb. 28, 2011AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F22
Plasmid#23080DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 5 to Arginine, H4 K5R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F24
Plasmid#23081DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 16 to Arginine, H4 K16R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F25
Plasmid#23082DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed lysine 16 to Glutamine, H4 K16…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F26
Plasmid#23083DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone 4 changed Lysine 16 to glycine, H4 K16G.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
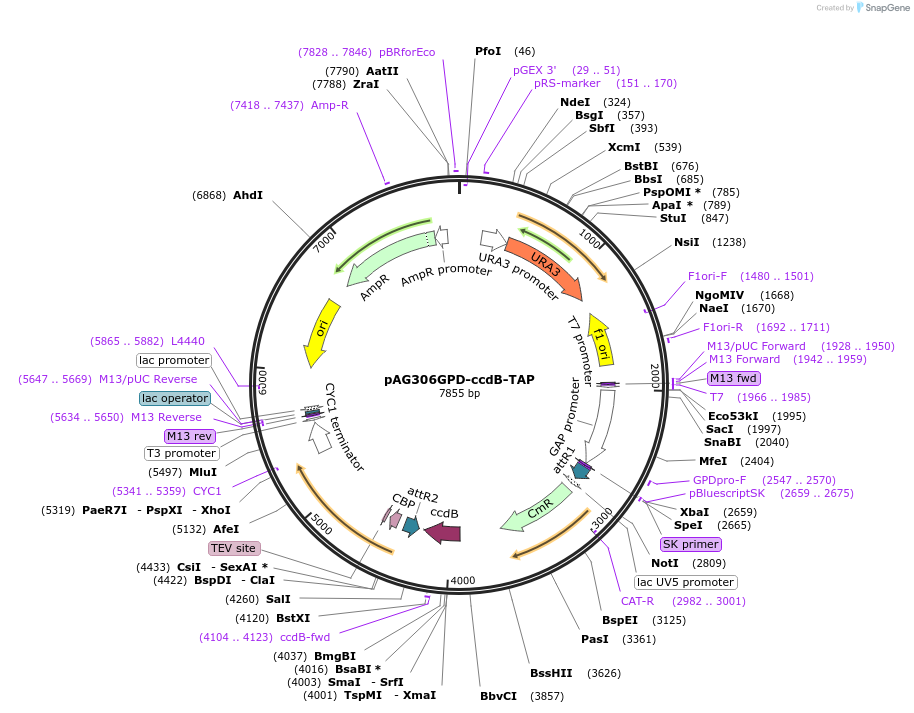
pAG306GPD-ccdB-TAP
Plasmid#14260DepositorTypeEmpty backboneUseGateway destinationTagsTAPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
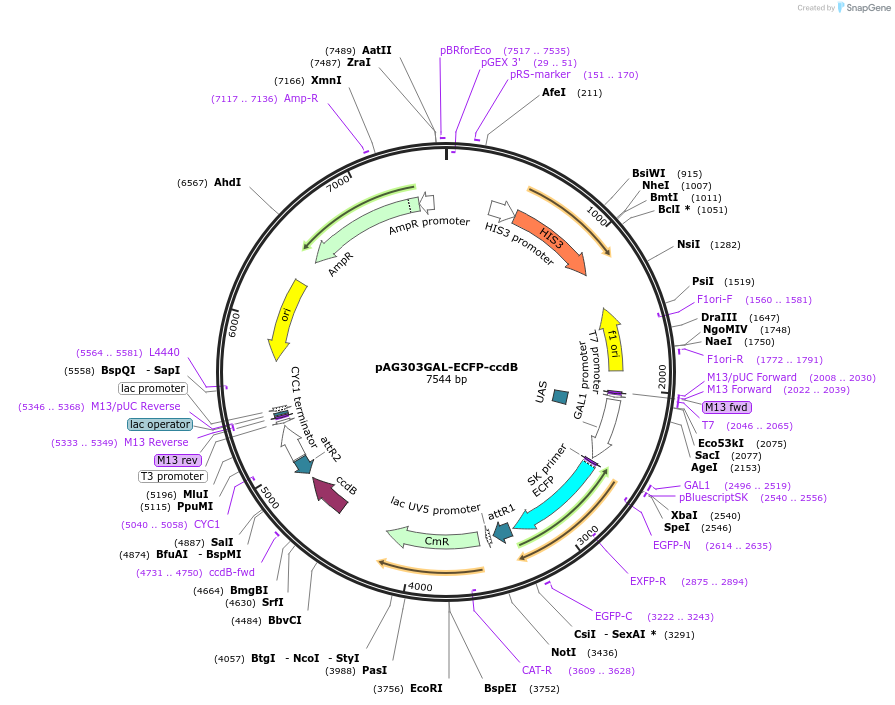
pAG303GAL-ECFP-ccdB
Plasmid#14277DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
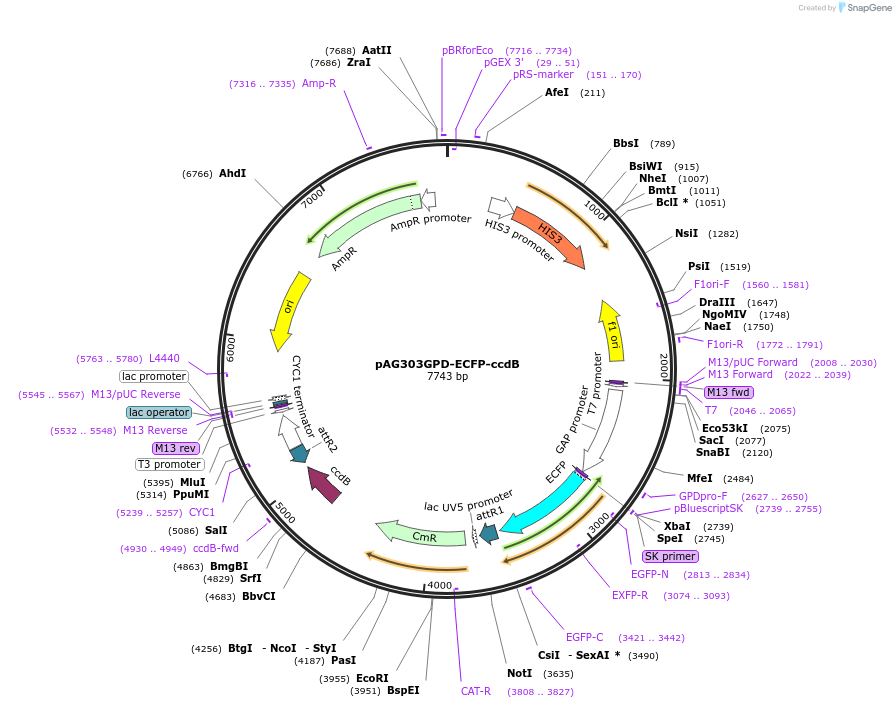
pAG303GPD-ECFP-ccdB
Plasmid#14278DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
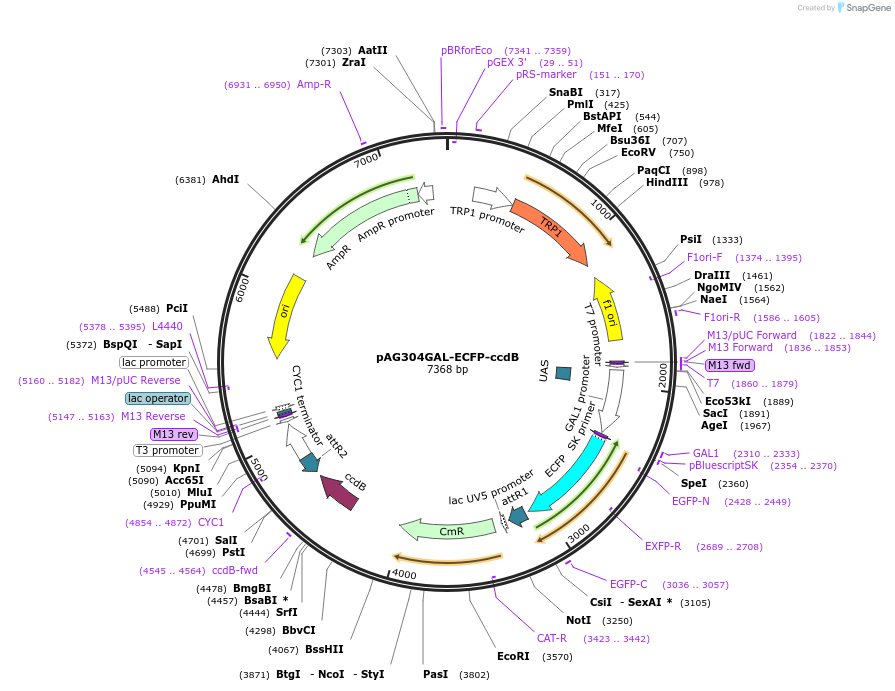
pAG304GAL-ECFP-ccdB
Plasmid#14279DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
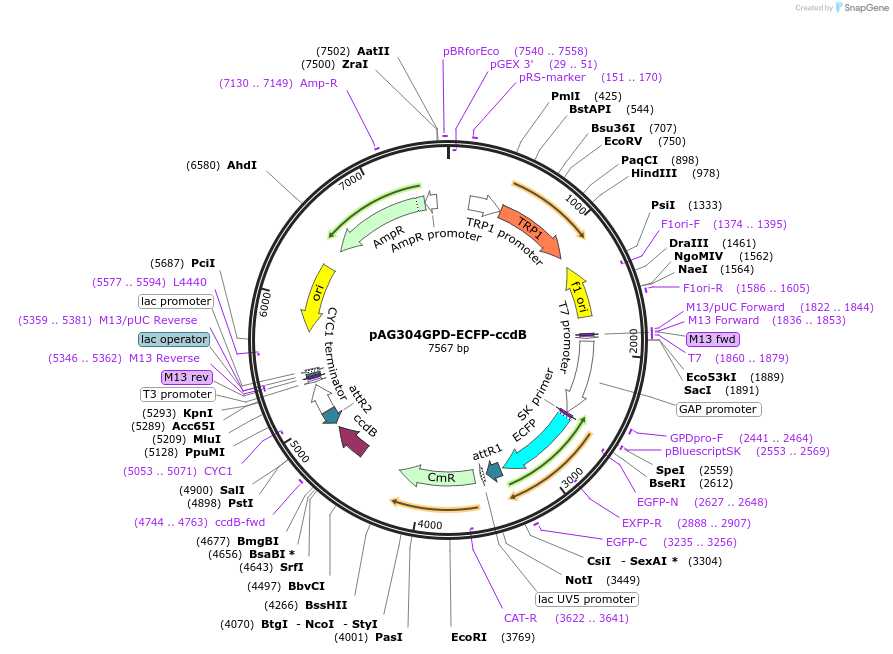
pAG304GPD-ECFP-ccdB
Plasmid#14280DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only