We narrowed to 387 results for: amh
-
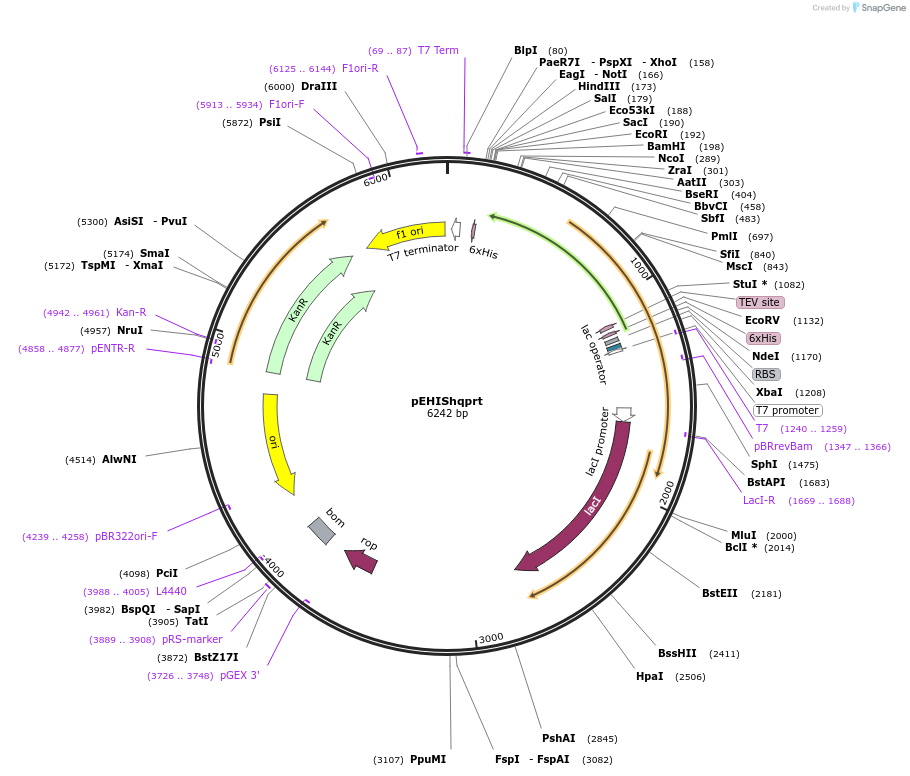
Plasmid#92166PurposePlasmid pEHIShqprt expressing the full-length of human quinolinate phosphoribosyltransferase (hQPRTase) was constrcucted by inserting BspHI/BamHI-fragment into NcoI/BamHI digested pEHISTEV vectorDepositorInsertQuinolinate Phosphoribosyltransferase (QPRT Human)
TagsTEV protease -cleavable N-terminal 6HisExpressionBacterialMutationchanged Asp 2 to Asn to create BspHI sitePromoterT7Available SinceAug. 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
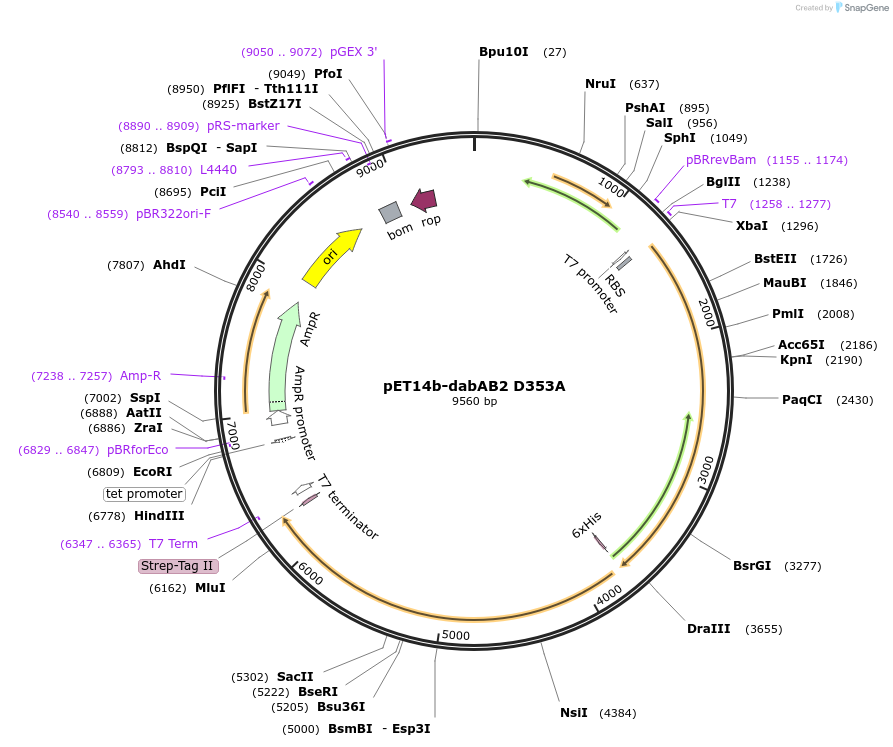
pET14b-dabAB2
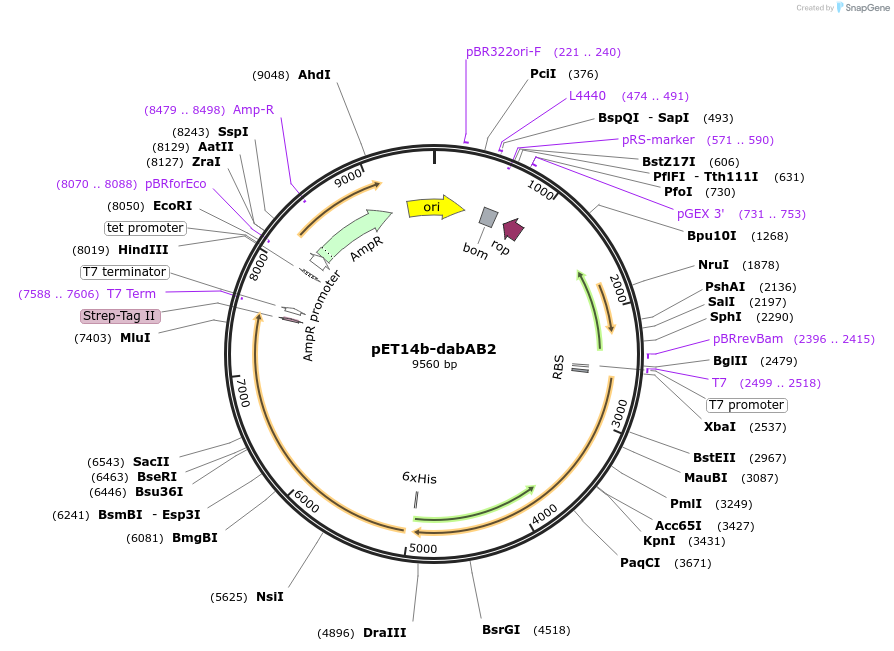
Plasmid#133013PurposepET14b carrying the dabA2 gene (Uniprot: D0KWS7) with a c-terminal strep tag fusion and the dabB2 gene (Uniprot: D0KWS8) with a c-terminal sfGFP V206K fusion and 6xHis tagDepositorInsertsdabB2
dabA2
Tags6xHis, StrepII tag, and sfGFPExpressionBacterialPromotert7Available SinceNov. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
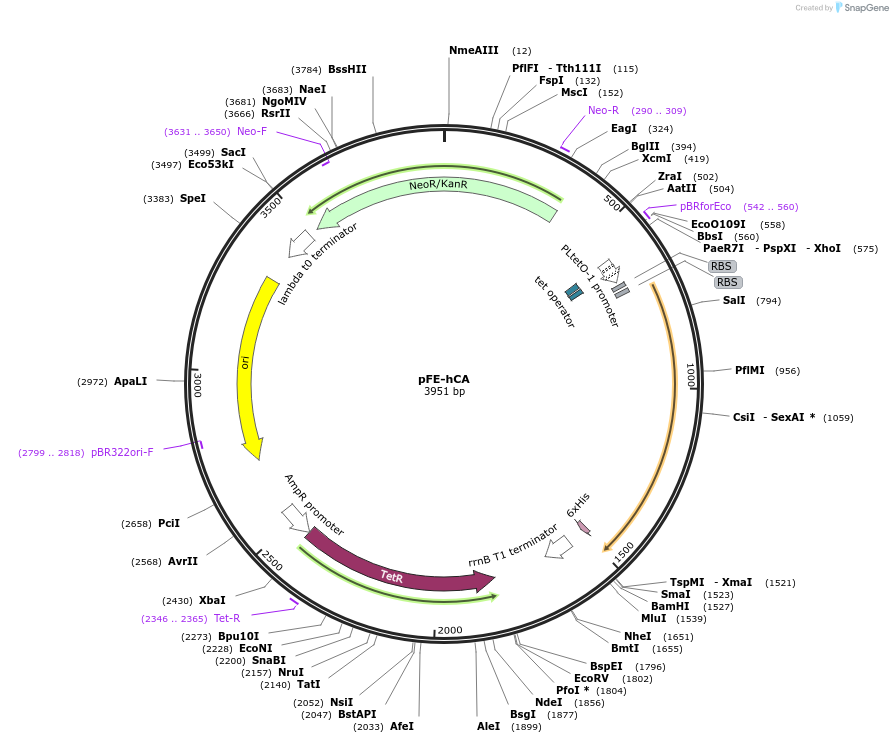
pFE-hCA
Plasmid#132999PurposepFE carrying a human carbonic anhydrase II gene (Uniprot: P00918)DepositorAvailable SinceNov. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
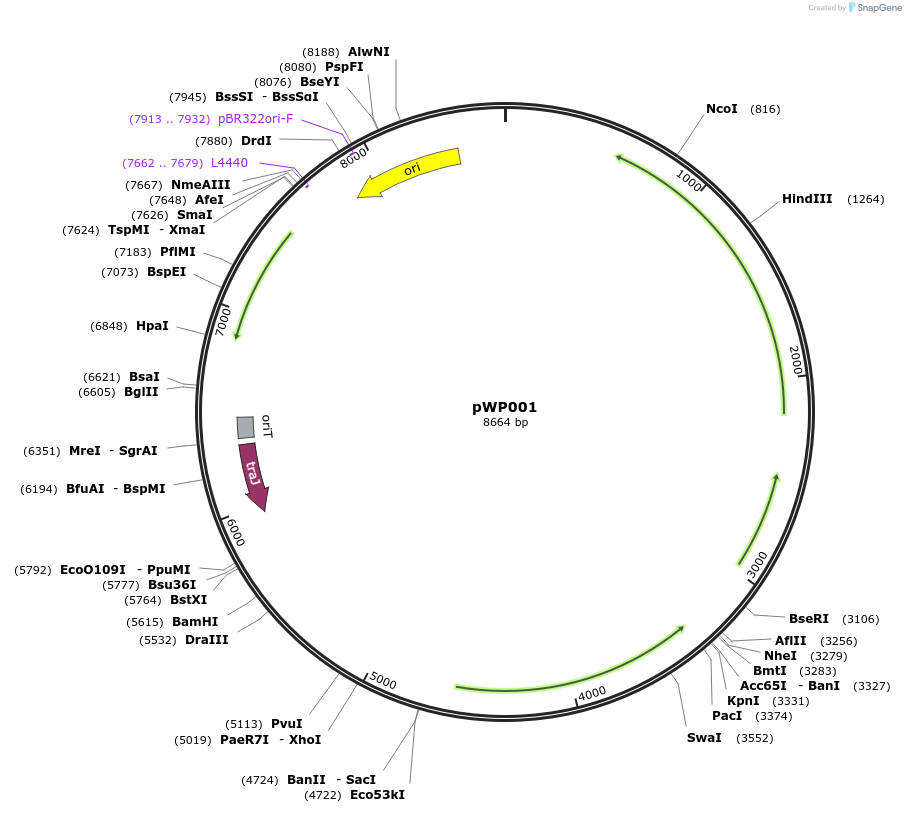
pWP001
Plasmid#178596PurposepAF256 derived plasmid encoding a xylose inducible CD16/50L CBD-SmBiT and LgBiTDepositorInsertCBD-SmBiT/LgBiT
ExpressionBacterialPromoterPxylAvailable SinceApril 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
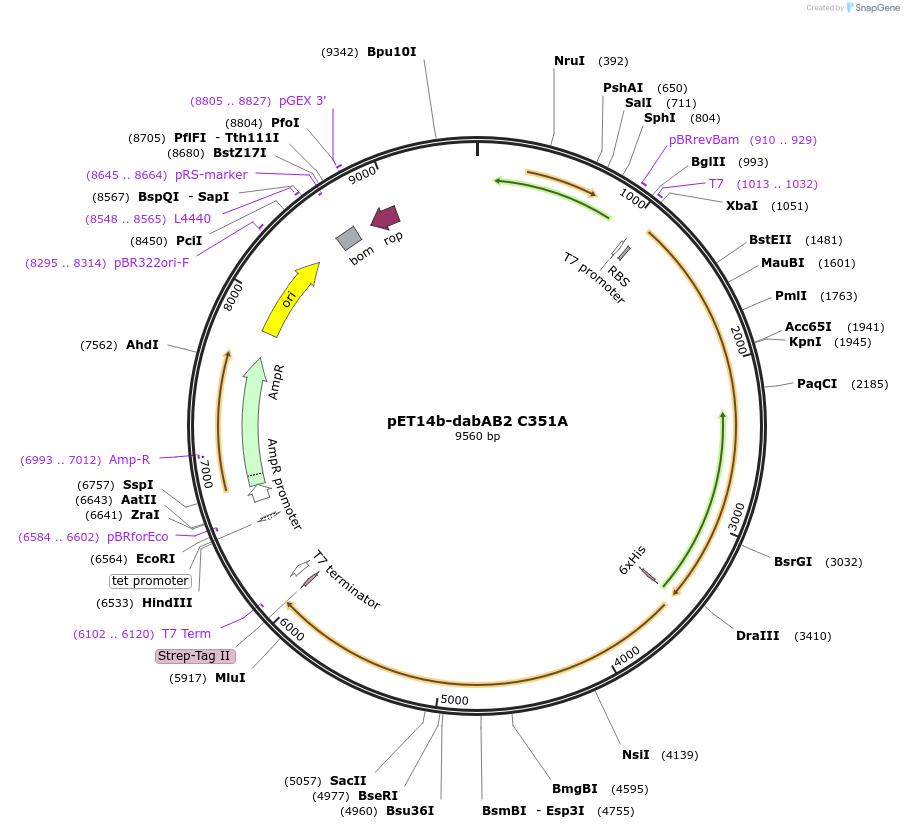
pET14b-dabAB2 D353A
Plasmid#133016PurposepET14b carrying the dabA2 gene (Uniprot: D0KWS7) with a D353A mutation fused to a c-terminal strep tag and the dabB2 gene (Uniprot: D0KWS8) with a c-terminal sfGFP V206K fusion and 6xHis tagDepositorInsertsdabB2
dabA2
Tags6xHis, StrepII tag, and sfGFPExpressionBacterialMutationD353APromotert7Available SinceNov. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET14b-dabAB2 C351A
Plasmid#133014PurposepET14b carrying the dabA2 gene (Uniprot: D0KWS7) with a C351A mutation fused to a c-terminal strep tag and the dabB2 gene (Uniprot: D0KWS8) with a c-terminal sfGFP V206K fusion and 6xHis tagDepositorInsertsdabB2
dabA2
Tags6xHis, StrepII tag, and sfGFPExpressionBacterialMutationC351APromotert7Available SinceNov. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
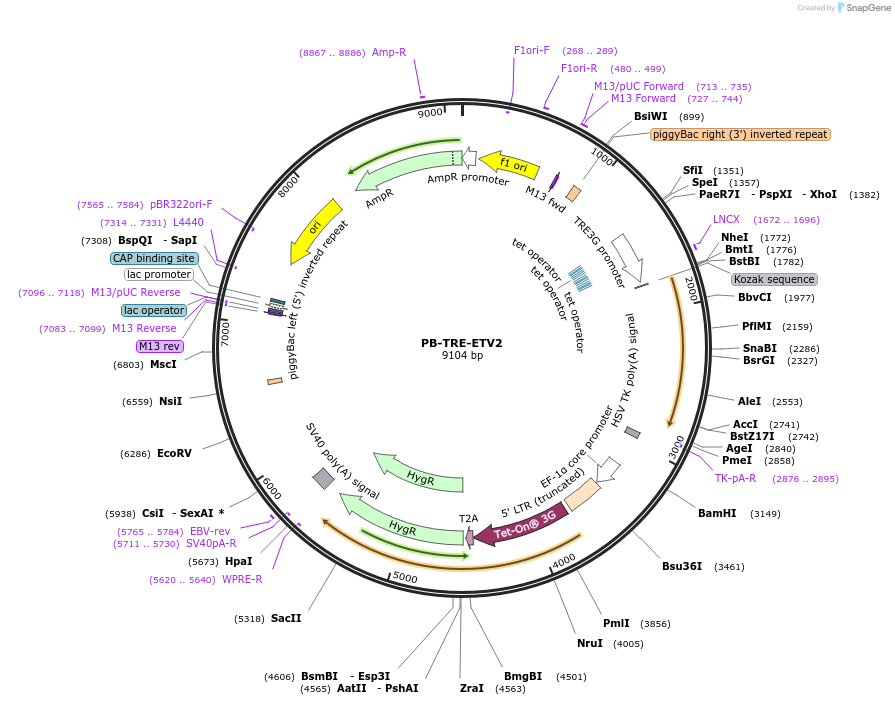
PB-TRE-ETV2
Plasmid#225051PurposePiggyBac vector encoding doxycycline-inducible codon-optimized human ETV2DepositorInsertETS Variant Transcription Factor 2 (ETV2 Human)
ExpressionMammalianMutationCodon OptimizedPromoterTRE3GAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
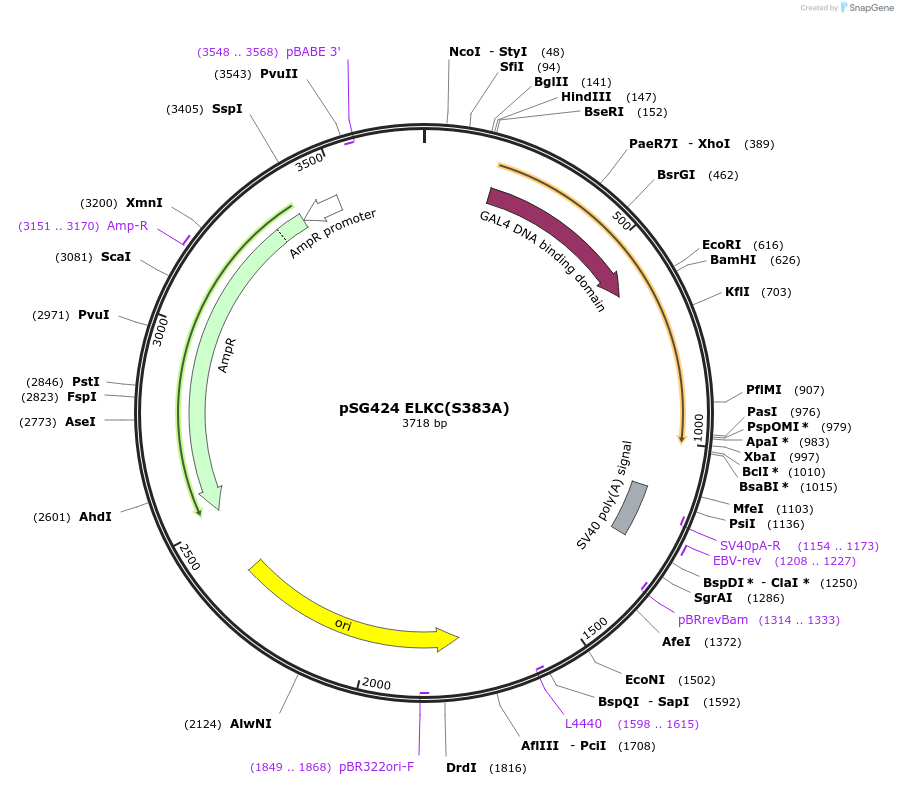
pSG424 ELKC(S383A)
Plasmid#89334PurposeElk1 cDNA encoding aa 307-428, aa383 mutated from ser-Ala, The PCR product was cut with BamH1 and Xba 1(365bp). pSG424 was cut with Bam H1 and Xba1. The Elk1 fragment was ligated into the BamH1DepositorInsertELK1 (ELK1 Human)
TagsGal 4ExpressionMammalianMutationencodes aa 307-428, changed serine 383 to alanine…Available SinceApril 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
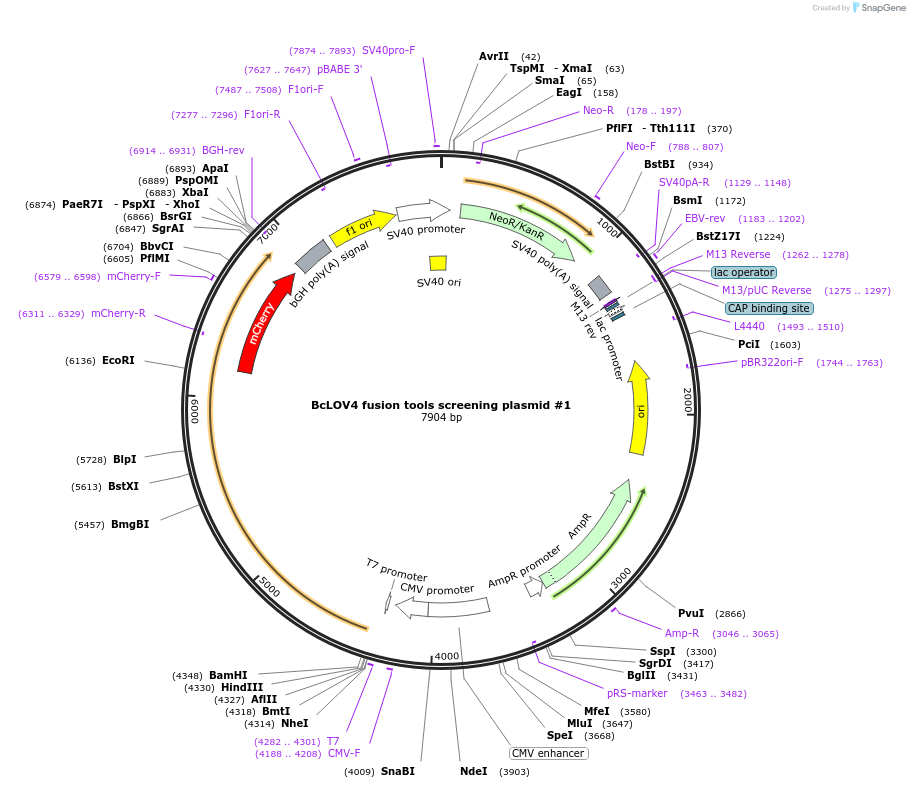
BcLOV4 fusion tools screening plasmid #1
Plasmid#174511PurposeCloning plasmid for creating BcLOV4 optogenetic tool fusions. Expresses [BamHI]-BcLOV4-[EcoRI]-GGGSx2-mCherry-[XhoI]-STOP in a pcDNA3.1 backbone.DepositorInsert[BamHI]-BcLOV4-[EcoRI]-GGGSx2-mCherry-[XhoI]-STOP
TagsmCherryExpressionMammalianPromoterCMVAvailable SinceDec. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
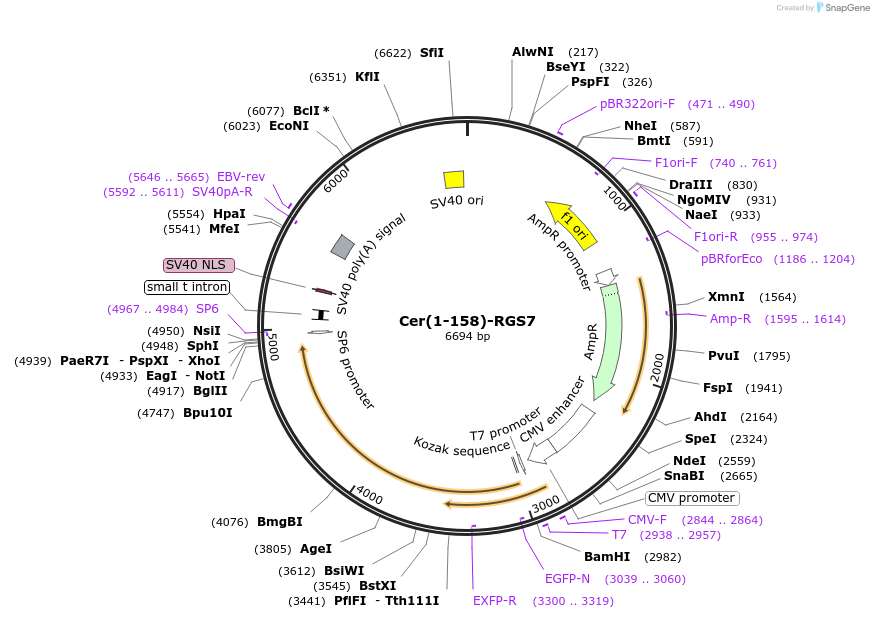
Cer(1-158)-RGS7
Plasmid#55760PurposeAn amino-terminal mCerulean fragment was fused to RGS7. When co-expressed with a carboxyl terminal fragment of CFP fused to Gbeta-5, a fluorescent signal is produced.DepositorInsertmCer(1-158)-RGS7 (RGS7 Aequorea victoria, Human)
TagsCer(1-158) was fused to the amino terminus of RGS…ExpressionMammalianMutationRGS7 was amplified via PCR, which added an N-term…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
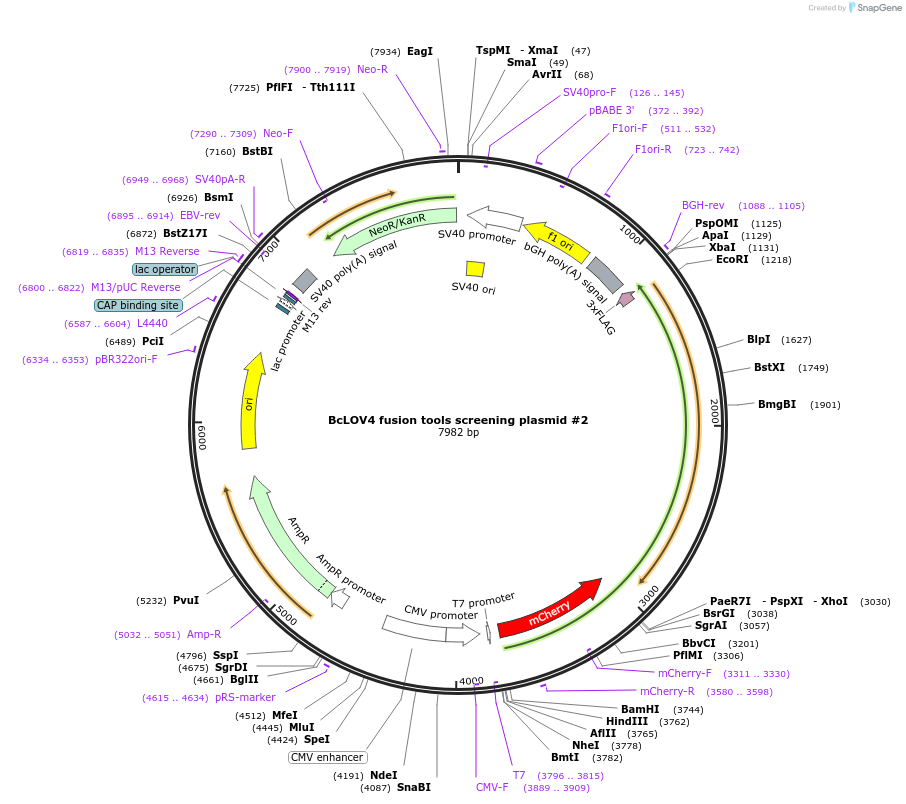
BcLOV4 fusion tools screening plasmid #2
Plasmid#174512PurposeCloning plasmid for creating BcLOV4 optogenetic tool fusions. Expresses [BamHI]-mCherry-[XhoI]-GGGSx2-BcLOV4-[EcoRI]-GGGS-3xFLAG-STOP in a pcDNA3.1 backbone.DepositorInsertplasmid B [BamHI]-mCherry-[XhoI]-GGGSx2-BcLOV4-[EcoRI]-GGGS-3xFLAG-STOP
Tags3xFLAG and mCherryExpressionMammalianPromoterCMVAvailable SinceDec. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
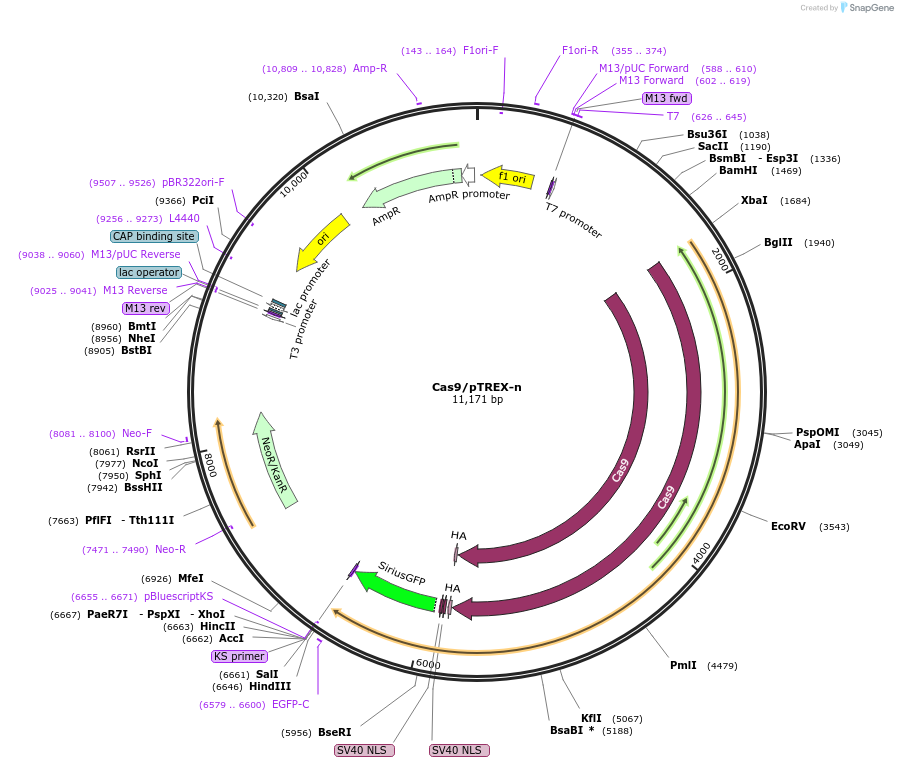
Cas9/pTREX-n
Plasmid#68708PurposeExpresses the fusion gene CAS9-HA-2xNLS-GFP in the pTREX-n backbone. This vector is used for cloning a specific sgRNA by BamHI, to be co-expressed with Cas9 for genome editing in Trypanosoma cruzi.DepositorInsertFusion gene Cas9-HA-2xNLS-GFP from vector pMJ920 (Addgene) .
UseCRISPRTagsFusion gene Cas9-HA-2xNLS-GFP from vector pMJ920 …MutationC4239T mutation that eliminates a BamHI restricti…Available SinceSept. 14, 2015AvailabilityAcademic Institutions and Nonprofits only -
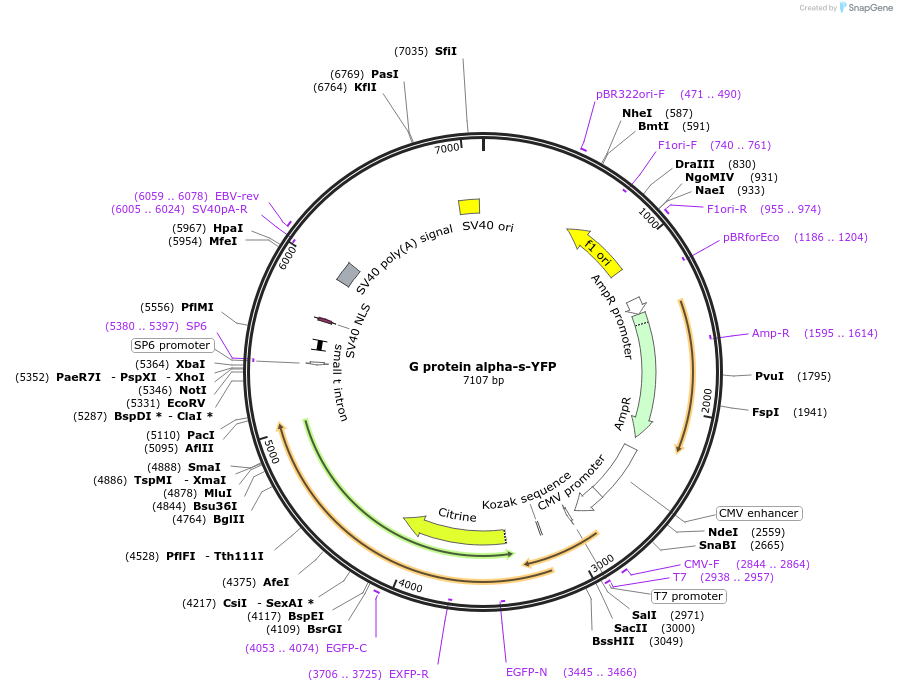
G protein alpha-s-YFP
Plasmid#55781PurposeThis G protein alpha-s construct contains internal insertions of YFP and the EE epitope.DepositorInsertalpha-s-EE YFP (Gnas Rat)
TagsEYFP flanked by SGGGS on each side was inserted i…ExpressionMammalianMutationMet was substituted for Gln69 in EYFP (Clontech).…PromoterCMVAvailable SinceAug. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
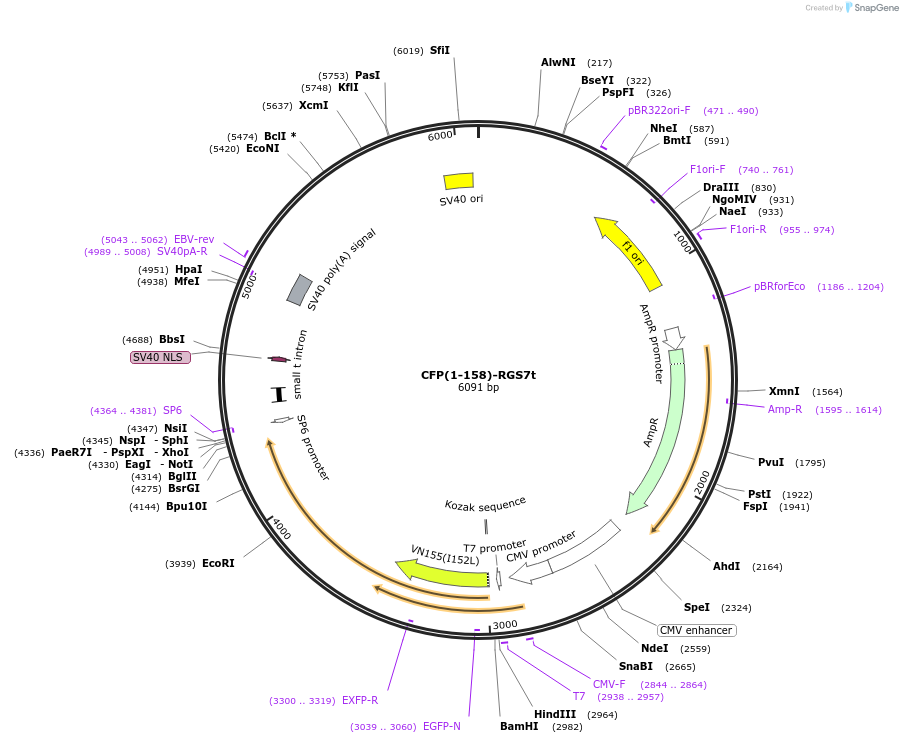
CFP(1-158)-RGS7t
Plasmid#55761PurposeAn amino-terminal CFP Fragment was fused to residues 202-477 of RGS7. When co-expressed with a carboxyl terminal fragment of CFP fused to Gbeta-5, a fluorescent signal is produced.DepositorInsertRGS7(202-477)-CFP(1-158) (RGS7 Aequorea victoria, Human)
TagsCFP(1-158) was fused to the amino terminus of RGS…ExpressionMammalianMutationThe RGS sequence amino terminal to the GGL domain…PromoterCMVAvailable SinceAug. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
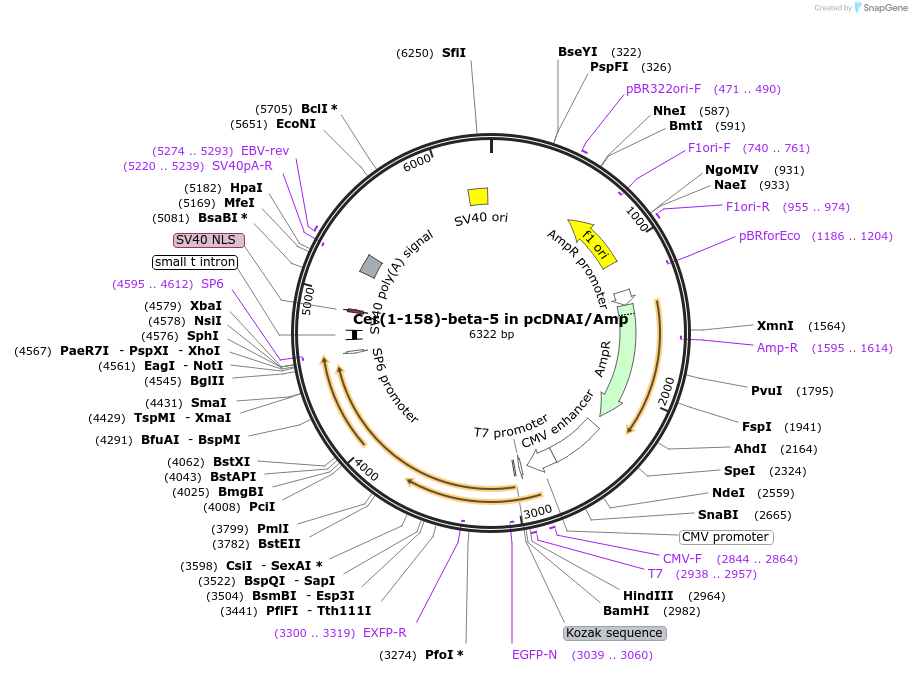
Cer(1-158)-beta-5 in pcDNAI/Amp
Plasmid#55708PurposeAn amino-terminal fragment of mCerulean was fused to Gbeta5. When co-expressed with a carboxyl terminal CFP fragment fused to a Ggamma subunit, a fluorescent signal is produced.DepositorInsertmCer(1-158)-beta5 (GNB5 Aequorea victoria, Human)
TagsCer(1-158) was fused to the amino terminus of Gbe…ExpressionMammalianMutationGBeta-5 was amplified via PCR, which added an N-t…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
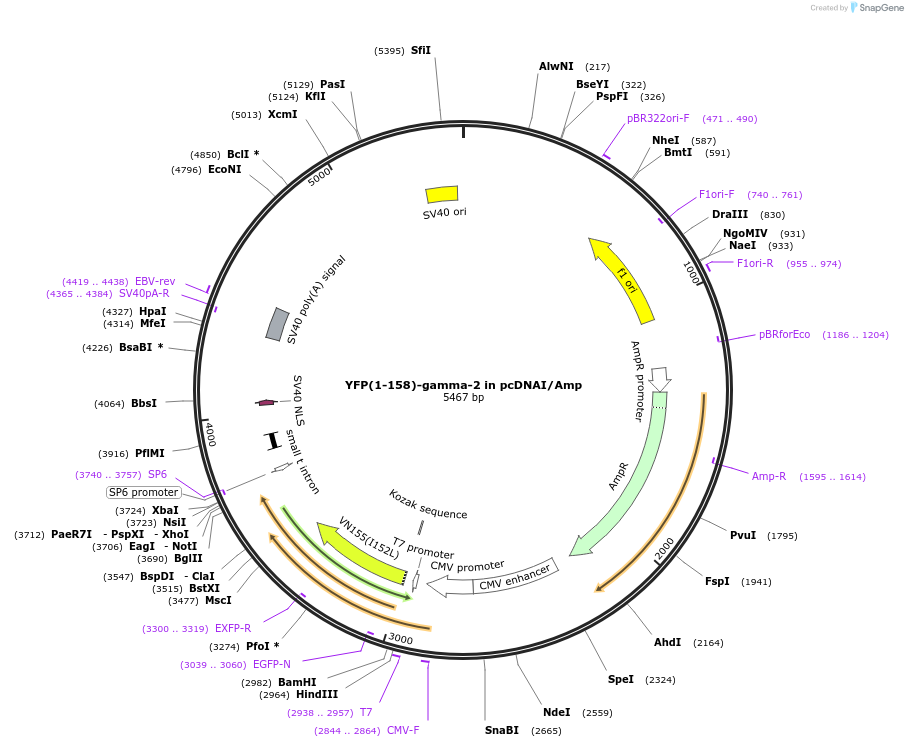
YFP(1-158)-gamma-2 in pcDNAI/Amp
Plasmid#55762PurposeAn amino-terminal YFP fragment was fused to Ggamma2. When co-expressed with a carboxyl terminal YFP or CFP fragment fused to a Gbeta subunit with which it interacts, a fluorescent signal is produced.DepositorInsertYFP(1-158)-gamma-2 (GNG2 Aequorea victoria, Human)
TagsYFP(1-158) was fused to the amino terminus of Gga…ExpressionMammalianMutationYFP (1-158) includes a substitution of Met for Gl…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
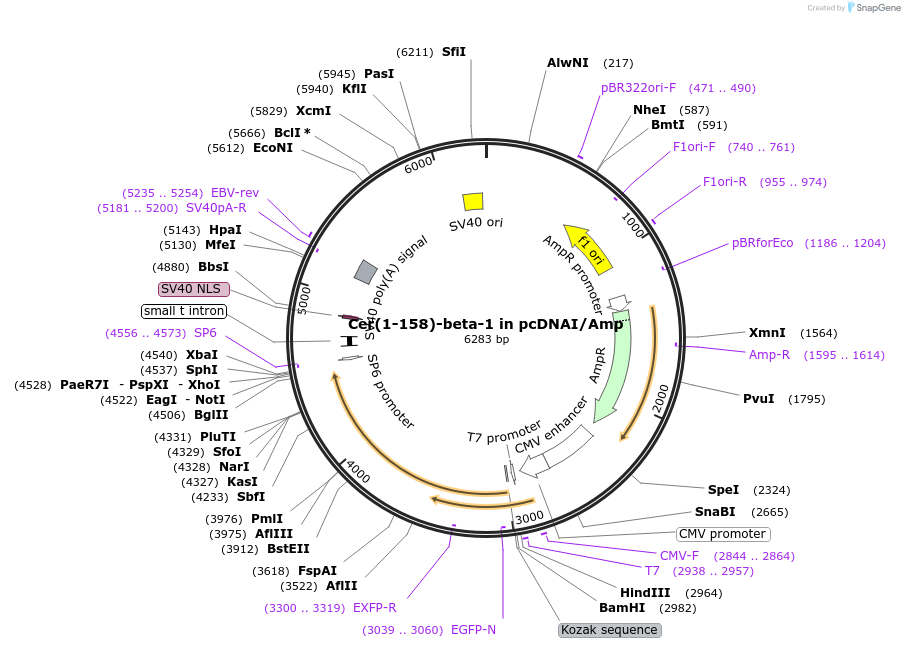
Cer(1-158)-beta-1 in pcDNAI/Amp
Plasmid#55707PurposeAn amino-terminal fragment of mCerulean was fused to Gbeta1. When co-expressed with a carboxyl terminal CFP fragment fused to a Ggamma subunit, a fluorescent signal s produced.DepositorInsertmCer(1-158)-beta 1 (GNB1 Aequorea victoria, Human)
TagsCer(1-158) was fused to the amino terminus of Gbe…ExpressionMammalianMutationGBeta-1 was amplified via PCR, which removed an i…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
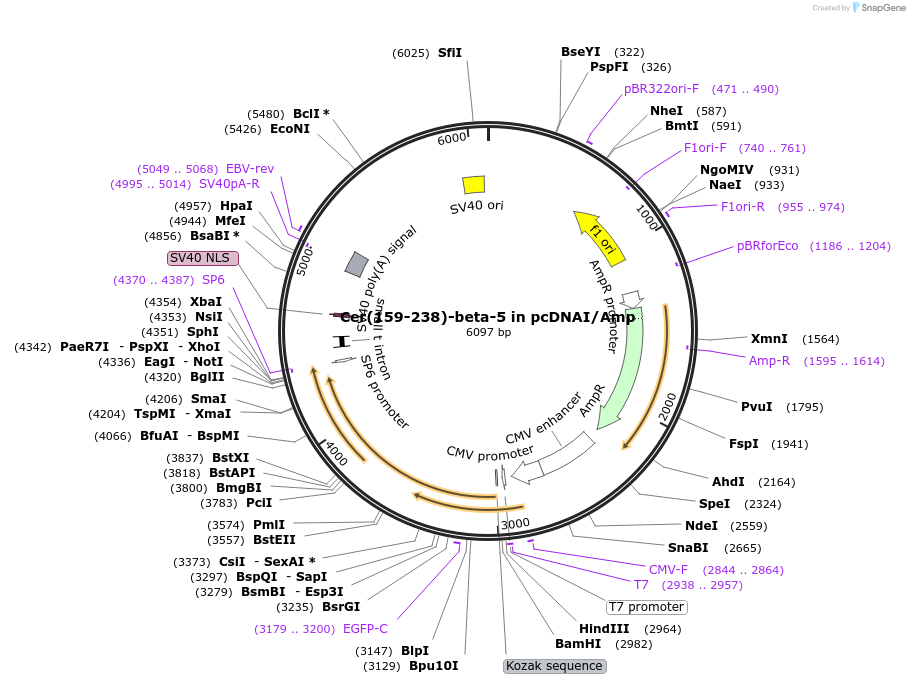
Cer(159-238)-beta-5 in pcDNAI/Amp
Plasmid#55778PurposeA carboxyl-terminal mCerulean fragment was fused to Gbeta-5. When co-expressed with an amino terminal mCerulean fragment fused to a Ggamma subunit, a fluorescent signal is produced.DepositorInsertCer(159-238)-beta-5 (GNB5 Aequorea victoria, Human)
TagsmCer(159-238) was fused to the amino terminus of …ExpressionMammalianMutationGBeta-5 was amplified via PCR, which added an N-t…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
G protein alpha-s-GFP
Plasmid#66083PurposeG protein alpha-s internally tagged with EGFP and EE epitopeDepositorInsertG-alpha-s-EE-EGFP (Gnas Rat)
TagsEGFP flanked by SGGGS on each side was inserted i…ExpressionMammalianMutationBamHI sites in the alpha-s cDNA were removed and …PromoterCMVAvailable SinceAug. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
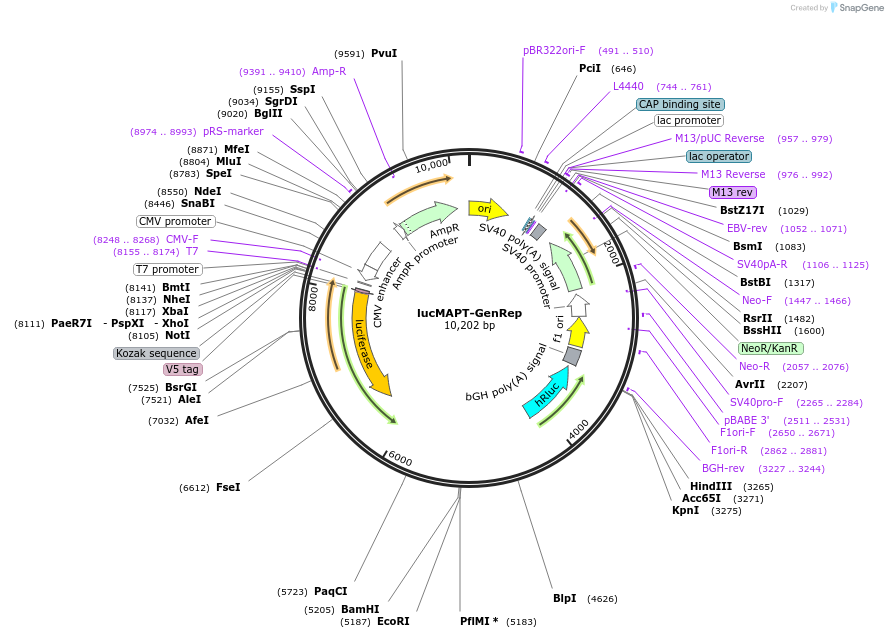
lucMAPT-GenRep
Plasmid#214674PurposeMAPT Exon 10 reporter, with Exon 10 and 100bp flanking intronic regions replaced with a BamHI/EcoRI cloning site for testing different exonsDepositorInsertMAPT (MAPT Human)
TagsFirefly Luciferase, Renilla Luciferase, and V5ExpressionMammalianMutationContains Exon 9 and Exon 11, with a BamHI EcoRI c…PromoterCMVAvailable SinceMarch 15, 2024AvailabilityAcademic Institutions and Nonprofits only