We narrowed to 10,377 results for: plasmids 101
-
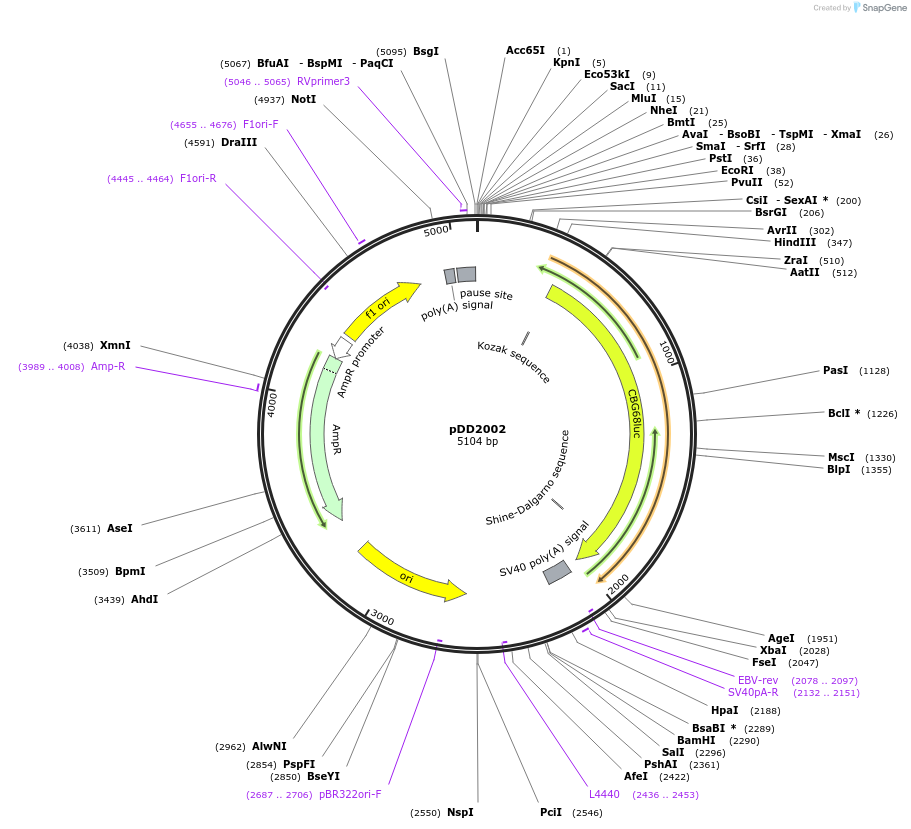
Plasmid#101198PurposeName: pCBG68-LANApi. LANApi driving green luciferase reporter for CBG68 in pCBG68 backbone.DepositorInsertLANApi driving green luciferase reporter for CBG68 in pCBG68 backbone.
UseLuciferaseAvailable SinceApril 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
pDD2005
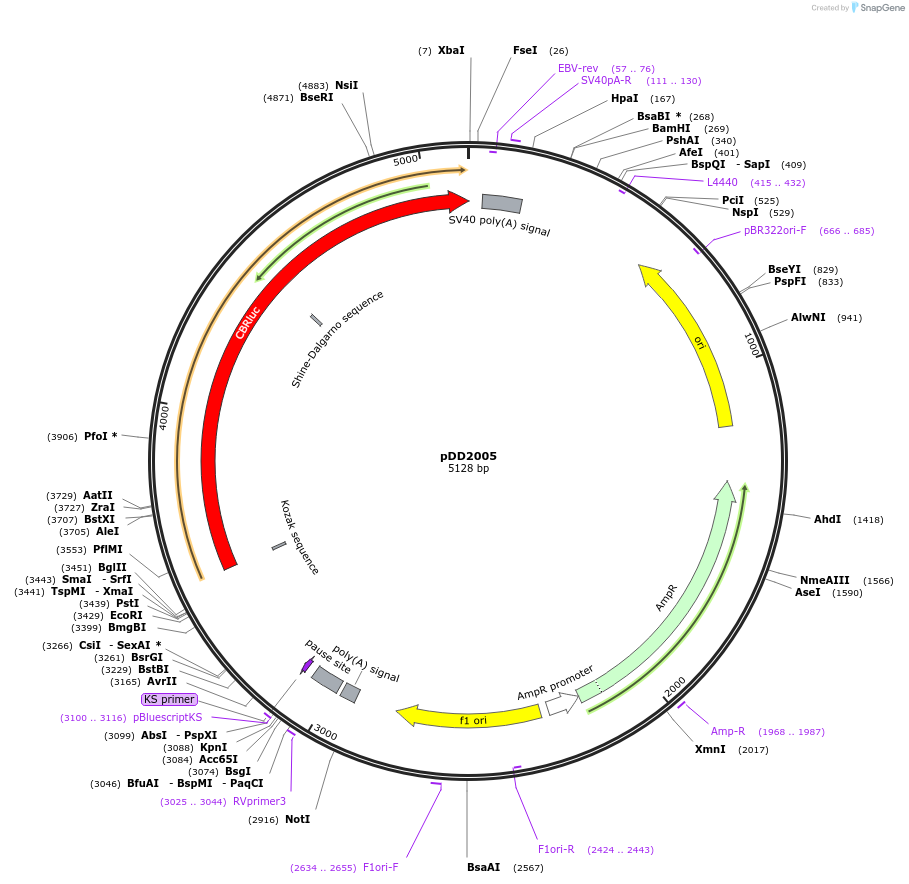
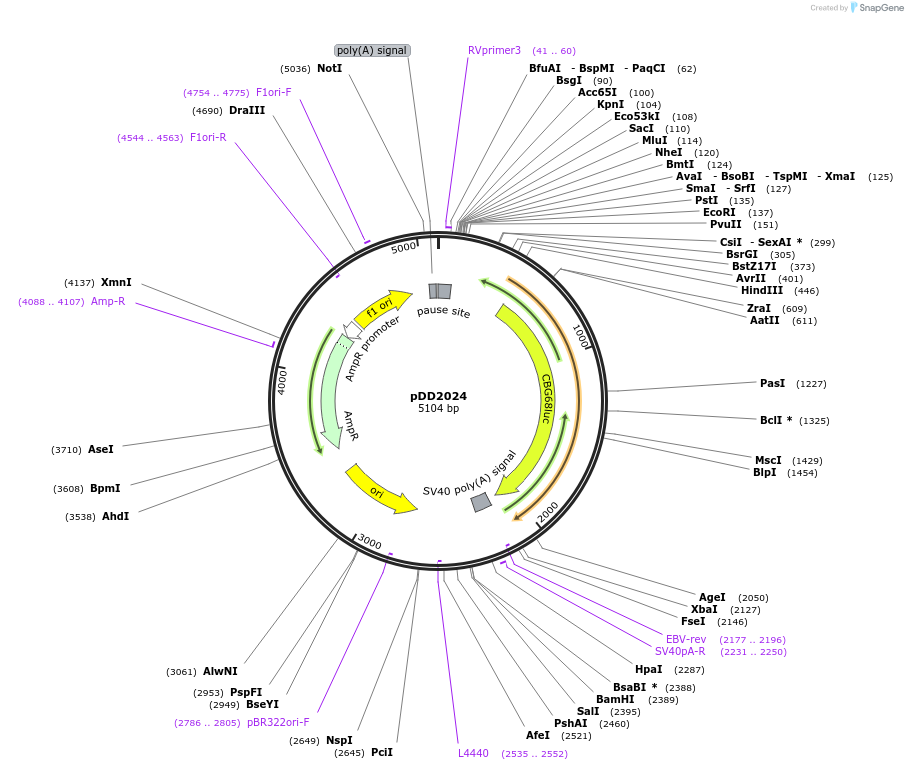
Plasmid#101219PurposeName: pCBR-K14. K14 driving red luciferase reporter for CBR in pCBR backbone.DepositorInsertK14 driving red luciferase reporter for CBR in pCBR backbone.
UseLuciferaseAvailable SinceApril 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
pDD2024
Plasmid#101368PurposeName: pCBG68-RBPjkΔA. RBPjk binding site A mutation in pCBG68-LANApi (pDD2002).DepositorInsertRBPjk binding site A mutation in pCBG68-LANApi (pDD2002).
UseLuciferaseMutationRBPjk binding site A mutation in pCBG68-LANApi (p…Available SinceDec. 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
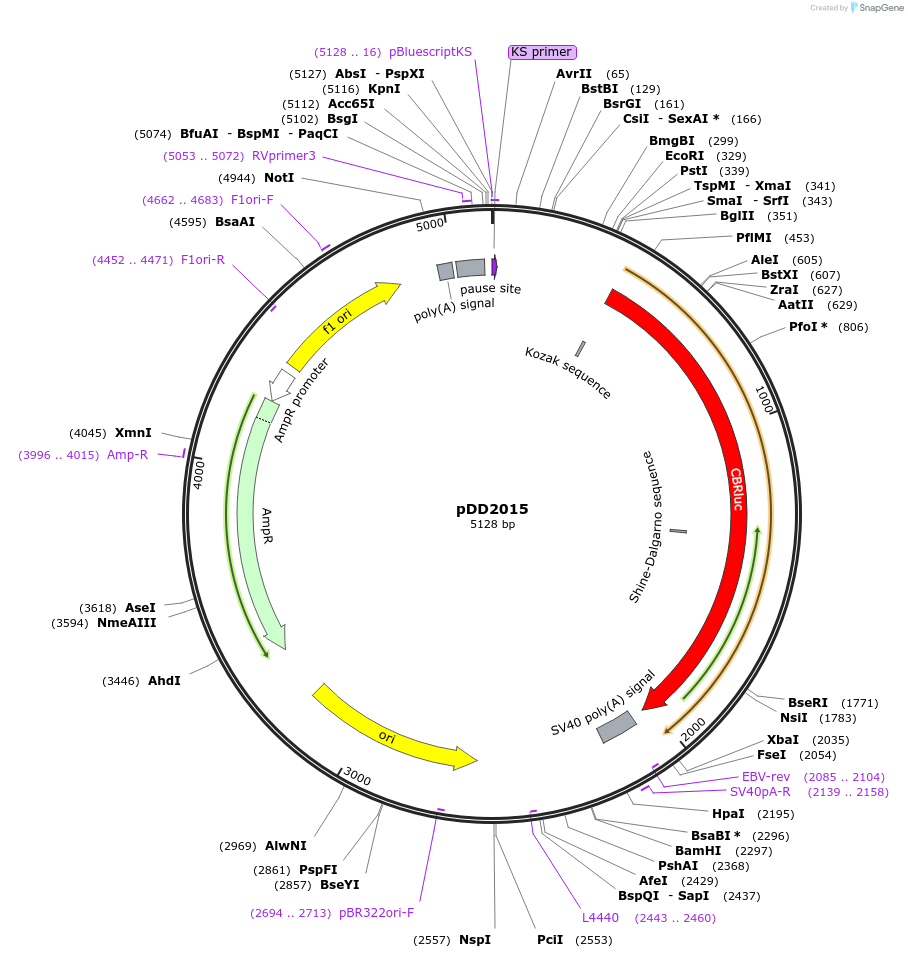
pDD2015
Plasmid#101365PurposeName: pCBR-K14ΔTATA. K14 TATA box mutation in pCBR-K14 (pDD2005).DepositorInsertK14 TATA box mutation in pCBR-K14 (pDD2005)
UseLuciferaseMutationK14 TATA box mutation in pCBR-K14 (pDD2005)Available SinceOct. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
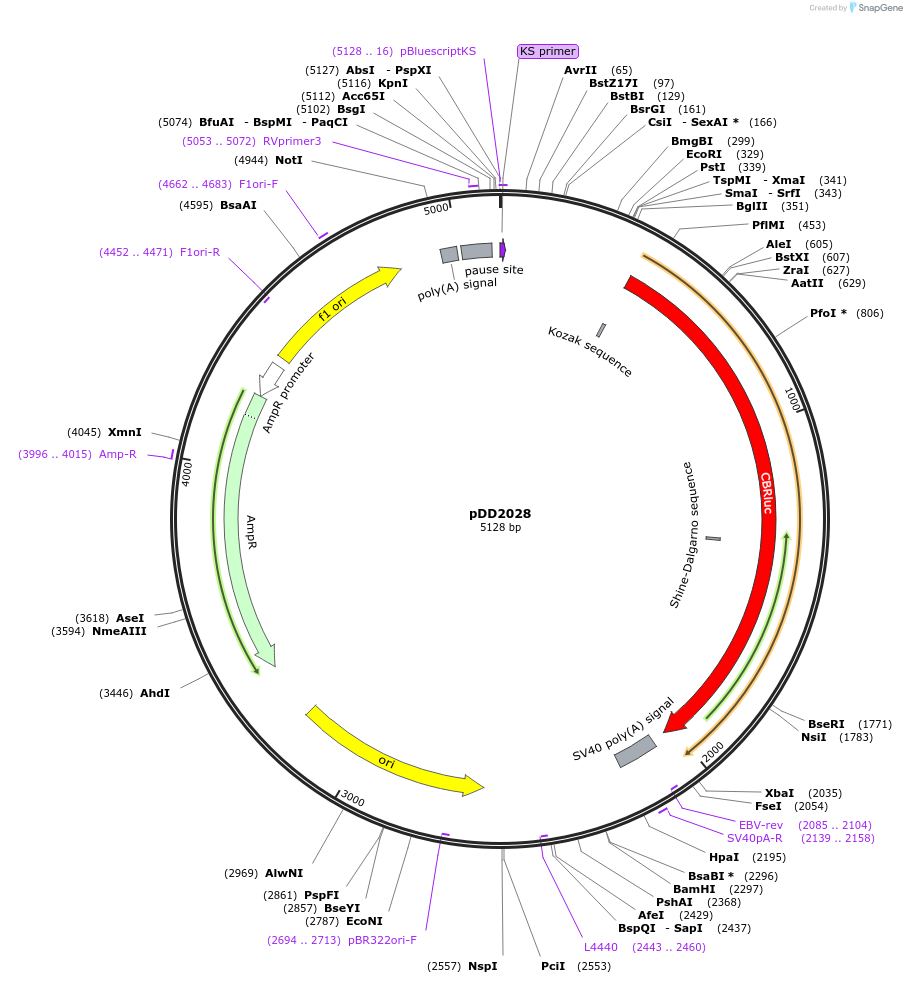
pDD2028
Plasmid#101371PurposeName: pCBR-RBPjkΔA. RBPjk binding site A mutation in pCBR-K14 (pDD2005).DepositorInsertRBPjk binding site A mutation in pCBR-K14 (pDD2005).
UseLuciferaseMutationRBPjk binding site A mutation in pCBR-K14 (pDD200…Available SinceOct. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
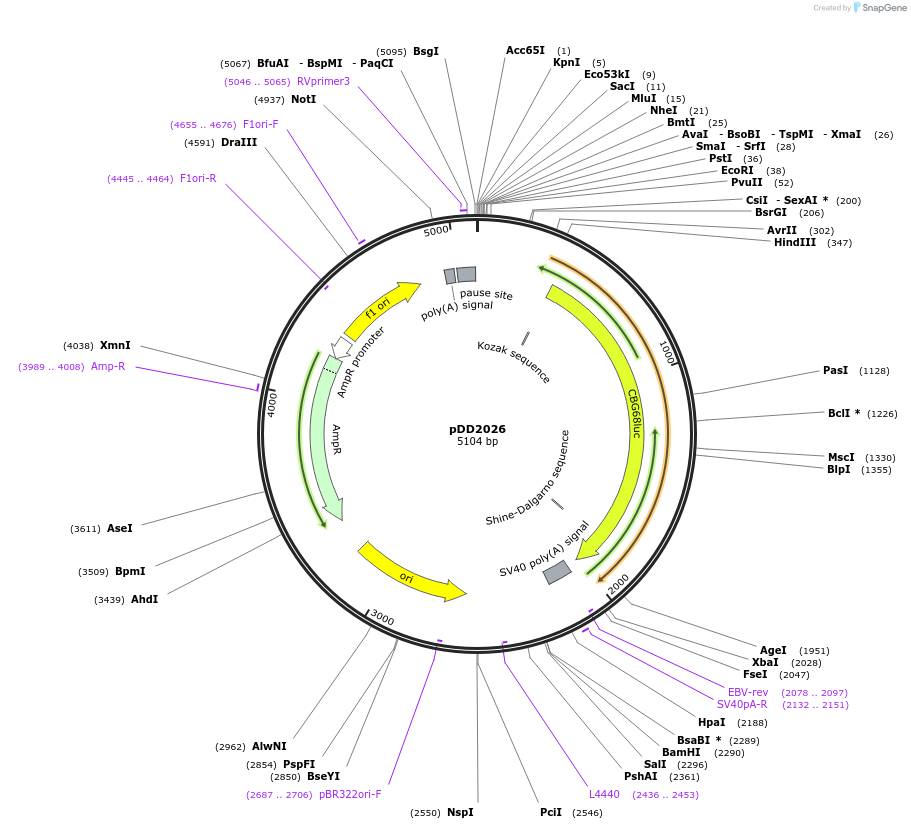
pDD2026
Plasmid#101369PurposeName: pCBG68-LANApiΔTATA. LANApi TATA box mutation in pCBG68-LANApi (pDD2002).DepositorInsertLANApi TATA box mutation in pCBG68-LANApi (pDD2002).
UseLuciferaseMutationLANApi TATA box mutation in pCBG68-LANApi (pDD200…AvailabilityAcademic Institutions and Nonprofits only -
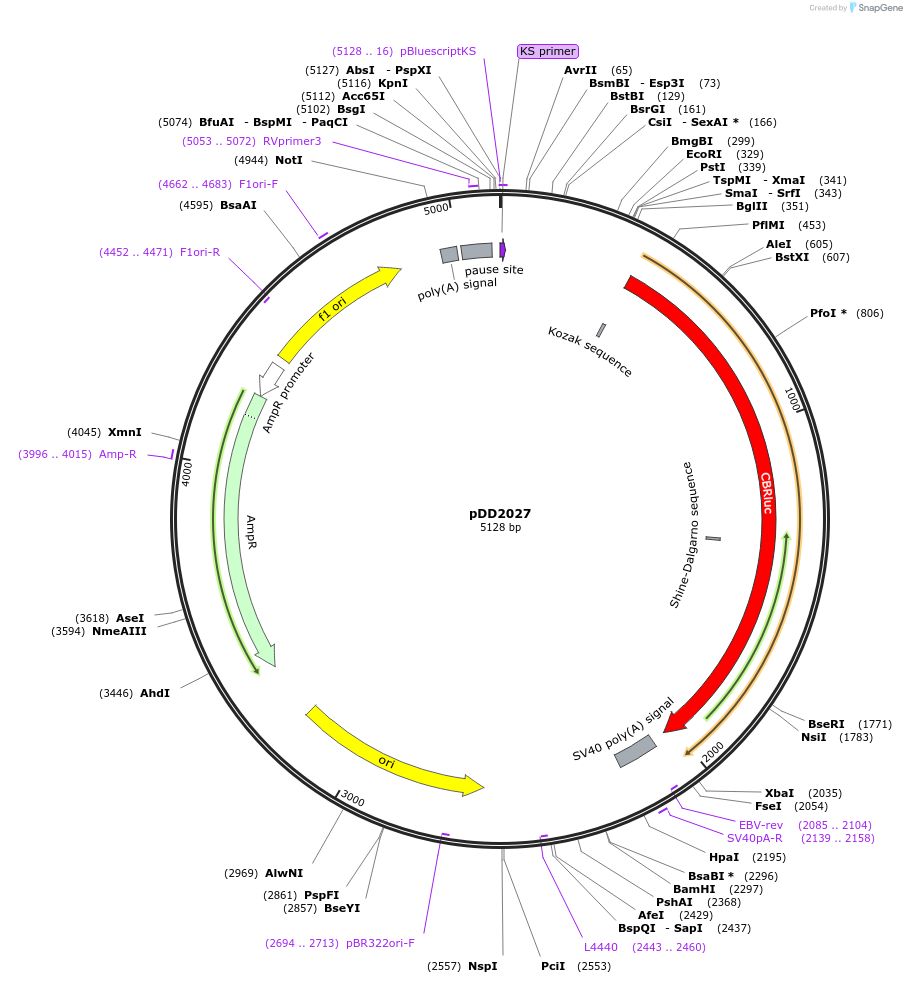
pDD2027
Plasmid#101370PurposeName: pCBR-LANApiΔTATA. LANApi TATA box mutation in pCBR-K14 (pDD2005).DepositorInsertLANApi TATA box mutation in pCBR-K14 (pDD2005).
UseLuciferaseMutationLANApi TATA box mutation in pCBR-K14 (pDD2005).AvailabilityAcademic Institutions and Nonprofits only -
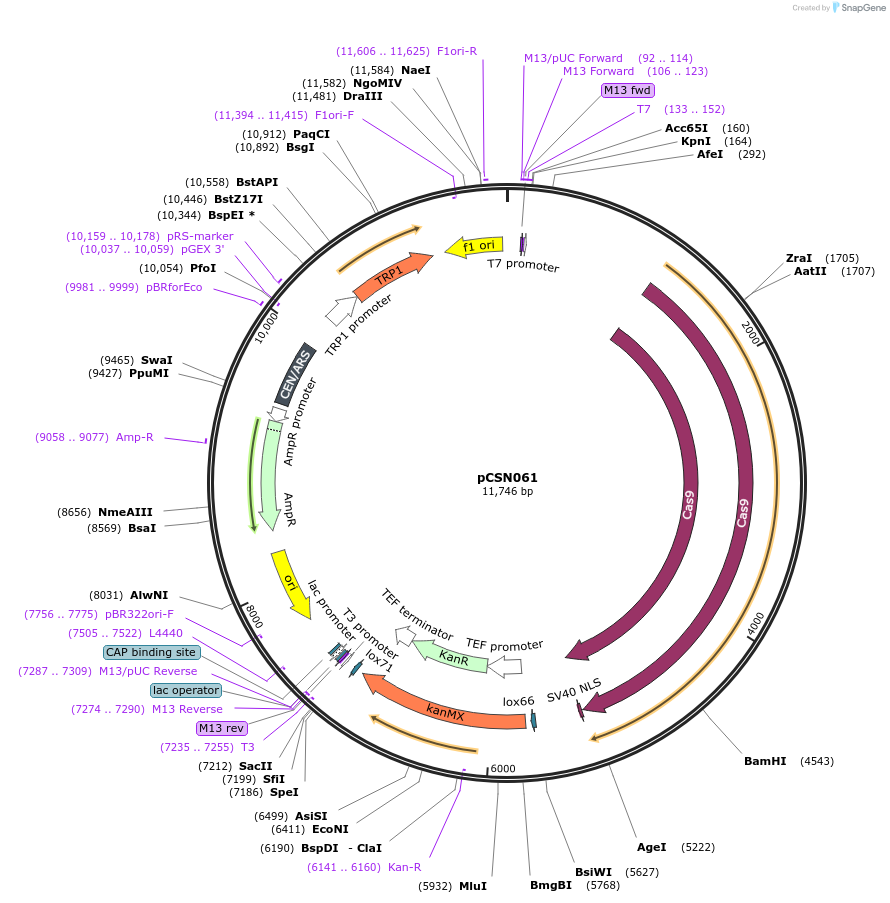
pCSN061
Plasmid#101725PurposeSingle copy yeast plasmid expressing Cas9 from Streptococcus pyogenes (SpCas9), codon optimized for expression in Saccharomyces cerevisiae.DepositorInsertsS. pyogenes Cas9 codon optimized for expression in S. cerevisiae. (NEWENTRY )
KanMX marker expression cassette.
UseCRISPRTagsSV40 NLSExpressionYeastPromoterHeterologous TEF1 promoter from A. gossypii. and …Available SinceDec. 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
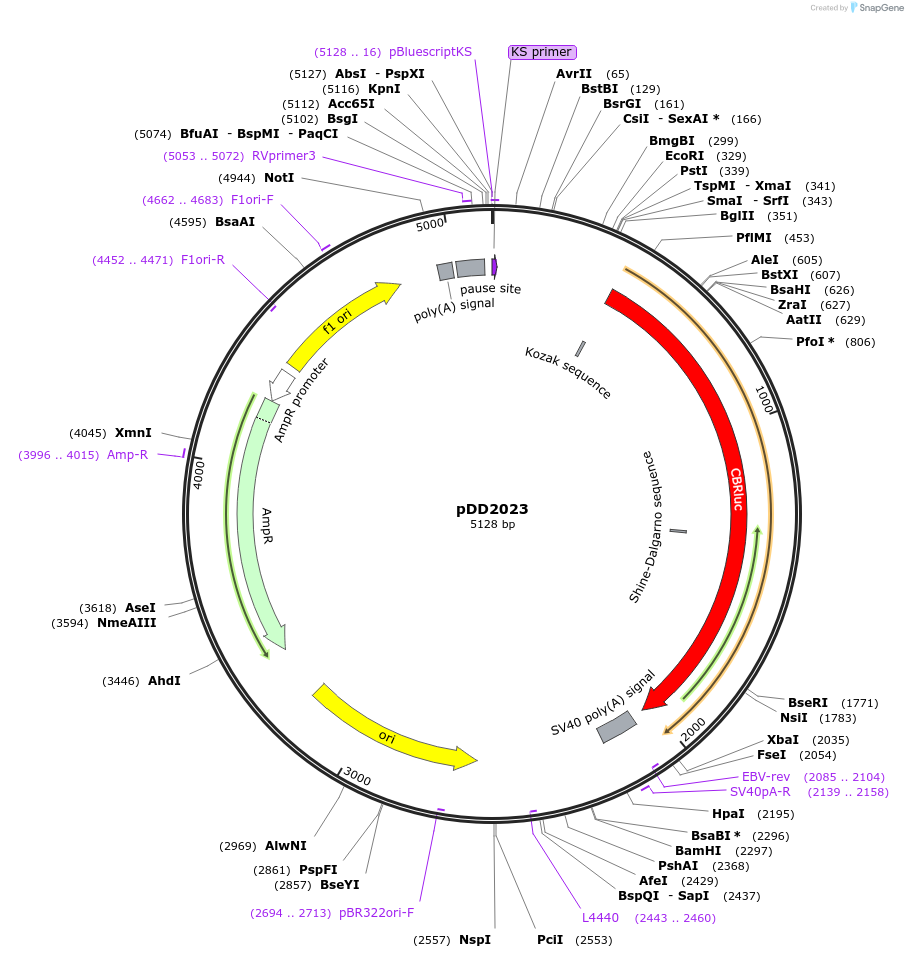
pDD2023
Plasmid#101367PurposeName: pCBR-RBPjkΔB. RBPjk binding site B mutation in pCBR-K14 (pDD2005).DepositorInsertRBPjk binding site B mutation in pCBR-K14 (pDD2005).
UseLuciferaseMutationRBPjk binding site B mutation in pCBR-K14 (pDD200…Available SinceOct. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
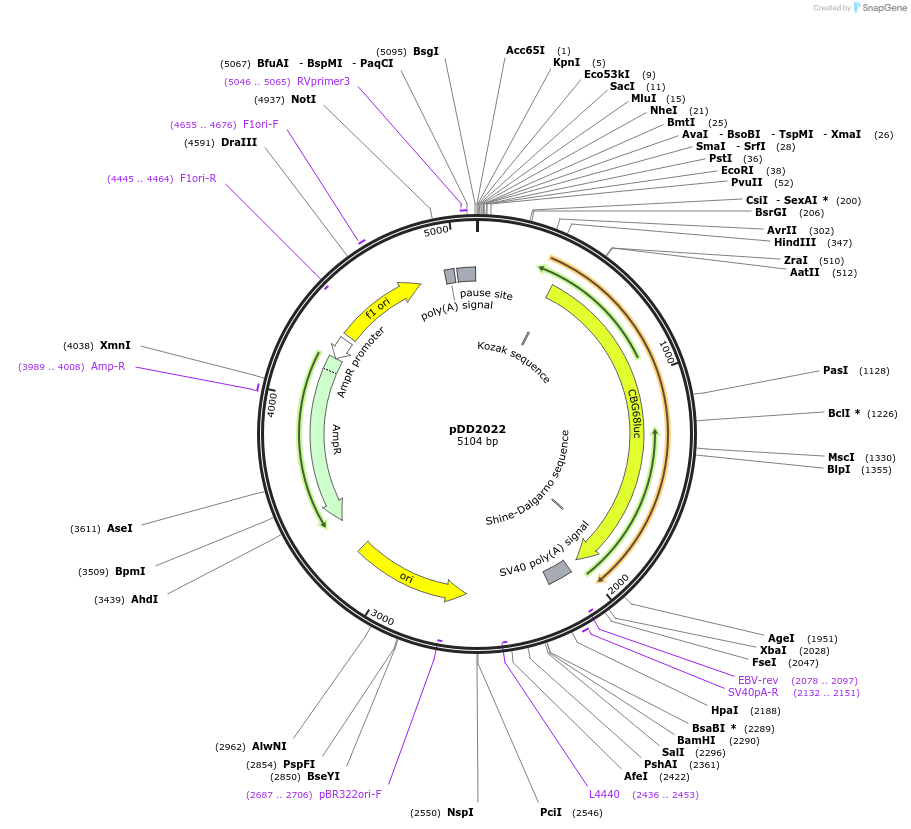
pDD2022
Plasmid#101366PurposeName: pCBG68-RBPjkΔB. RBPjk binding site B mutation in pCBG68-LANApi (pDD2002).DepositorInsertRBPjk binding site B mutation in pCBG68-LANApi (pDD2002).
UseLuciferaseMutationRBPjk binding site B mutation in pCBG68-LANApi (p…AvailabilityAcademic Institutions and Nonprofits only -
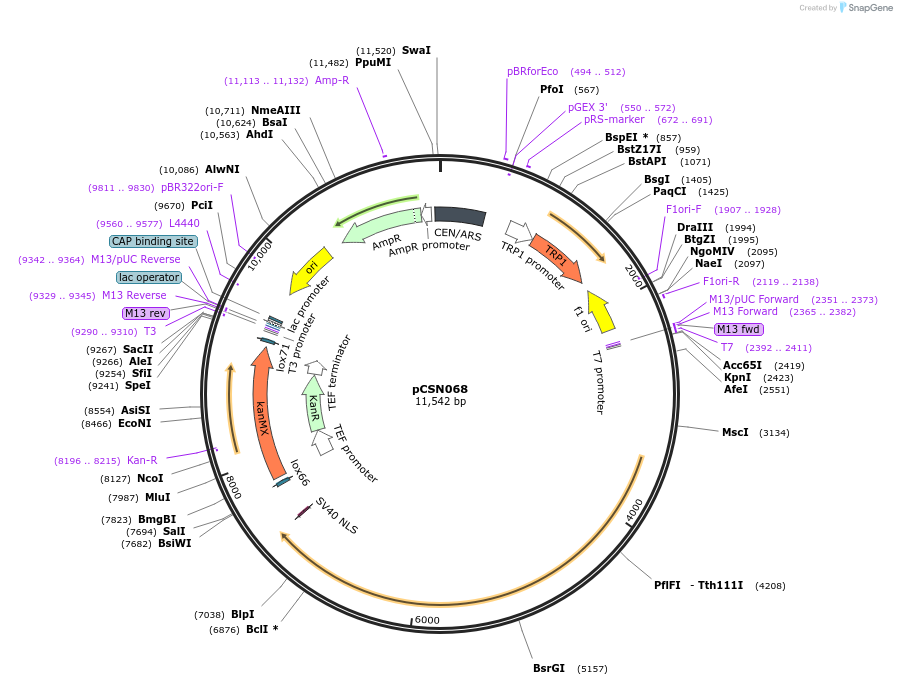
pCSN068
Plasmid#101749PurposeSingle copy yeast plasmid expressing Cpf1 from Francisella novicida U112 (FnCpf1), codon optimized for expression in Saccharomyces cerevisiae.DepositorInsertsCpf1 from Francisella novicida U112 (FnCpf1) codon optimized for expression in S. cerevisiae.
KanMX marker expression cassette.
UseCRISPRTagsSV40 NLSExpressionYeastPromoterHeterologous TEF1 promoter from A. gossypii. and …Available SinceJan. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
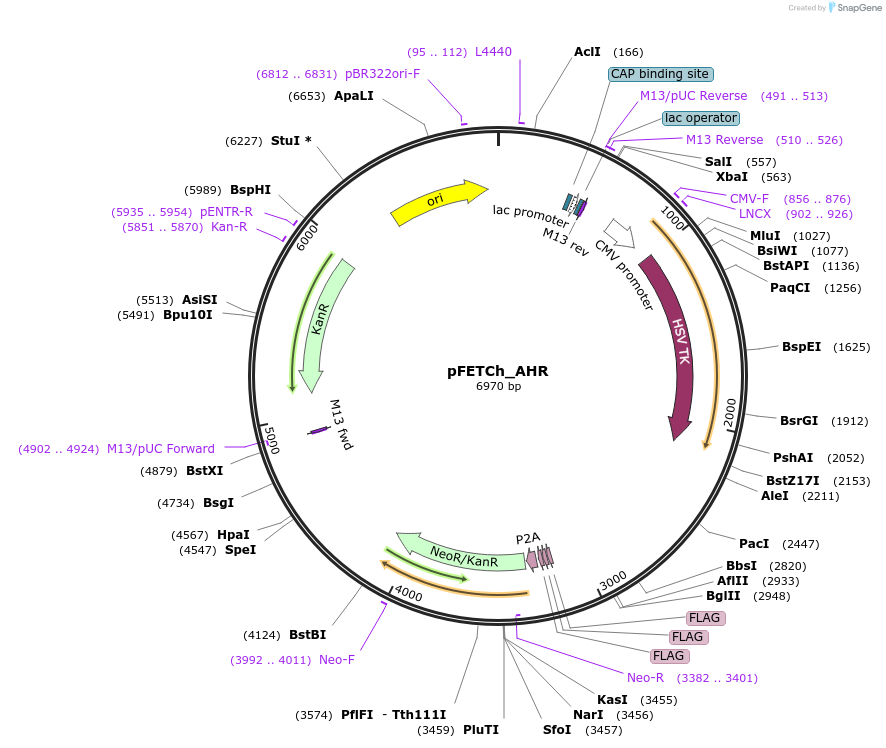
pFETCh_AHR
Plasmid#101071PurposeDonor vector for 3' FLAG tag of human AHRDepositorAvailable SinceSept. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
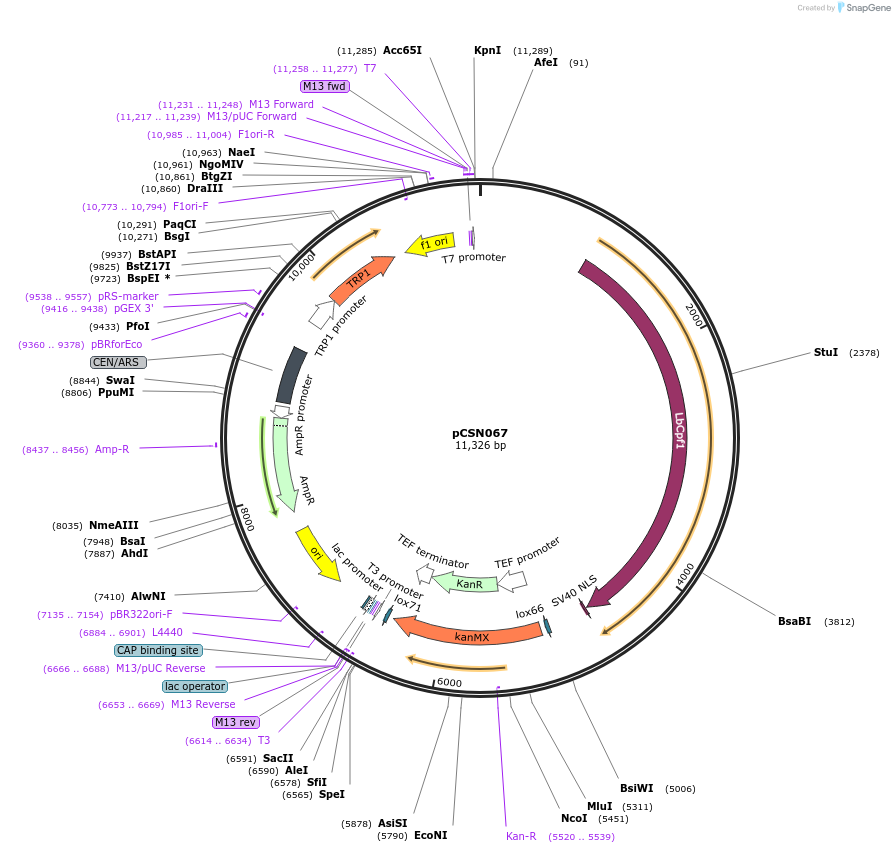
pCSN067
Plasmid#101748PurposeSingle copy yeast plasmid expressing Cpf1 from Lachnospiraceae bacterium ND2006 (LbCpf1), codon optimized for expression in Saccharomyces cerevisiae.DepositorInsertsCpf1 from Lachnospiraceae bacterium ND2006 (LbCpf1) codon optimized for expression in S. cerevisiae.
KanMX marker expression cassette.
UseCRISPRTagsSV40 NLSExpressionYeastPromoterHeterologous TEF1 promoter from A. gossypii. and …Available SinceDec. 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
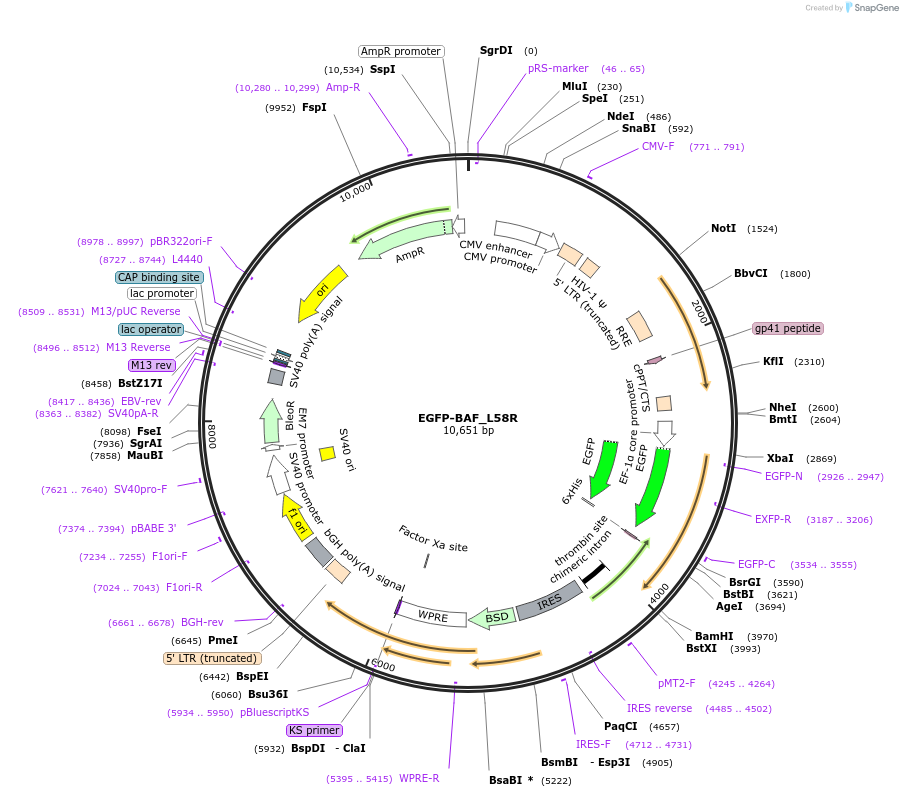
EGFP-BAF_L58R
Plasmid#101776PurposeExpresses EGFP tagged mutant BAF (L58R) in human cellsDepositorInsertBAF (BANF1) (BANF1 Human)
UseLentiviralTagsEGFPExpressionMammalianMutationL58R mutationPromoterEF1aAvailable SinceJan. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
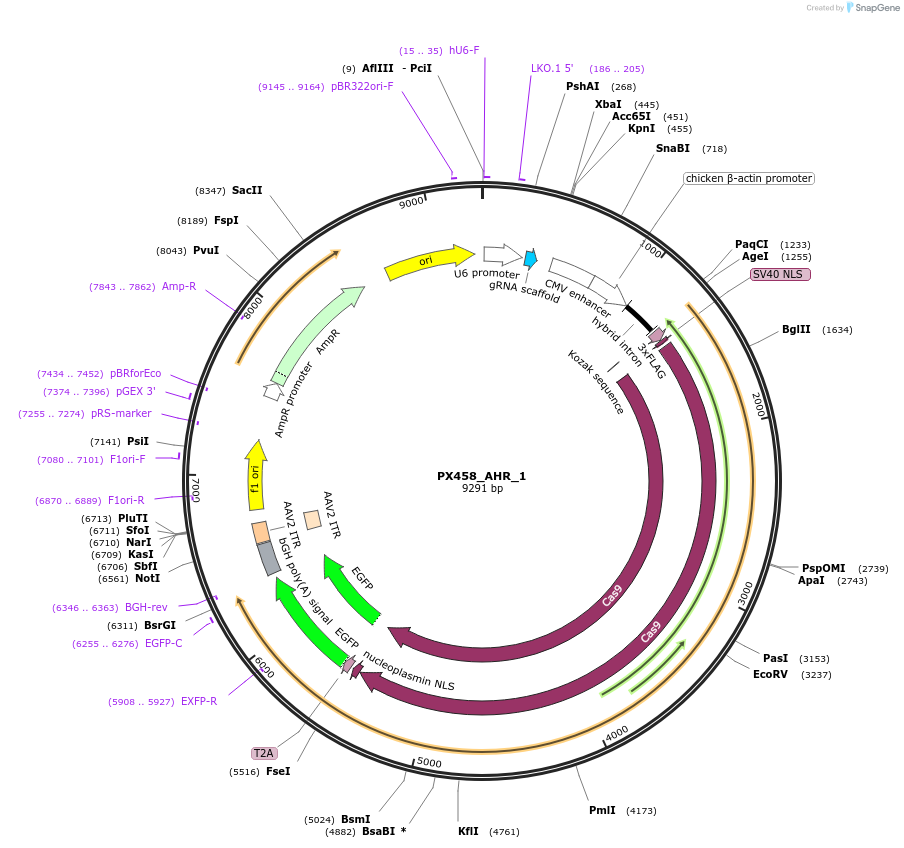
PX458_AHR_1
Plasmid#101076PurposeEncodes gRNA for 3' target of human AHRDepositorInsertgRNA against AHR (AHR Human)
UseCRISPRAvailable SinceOct. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
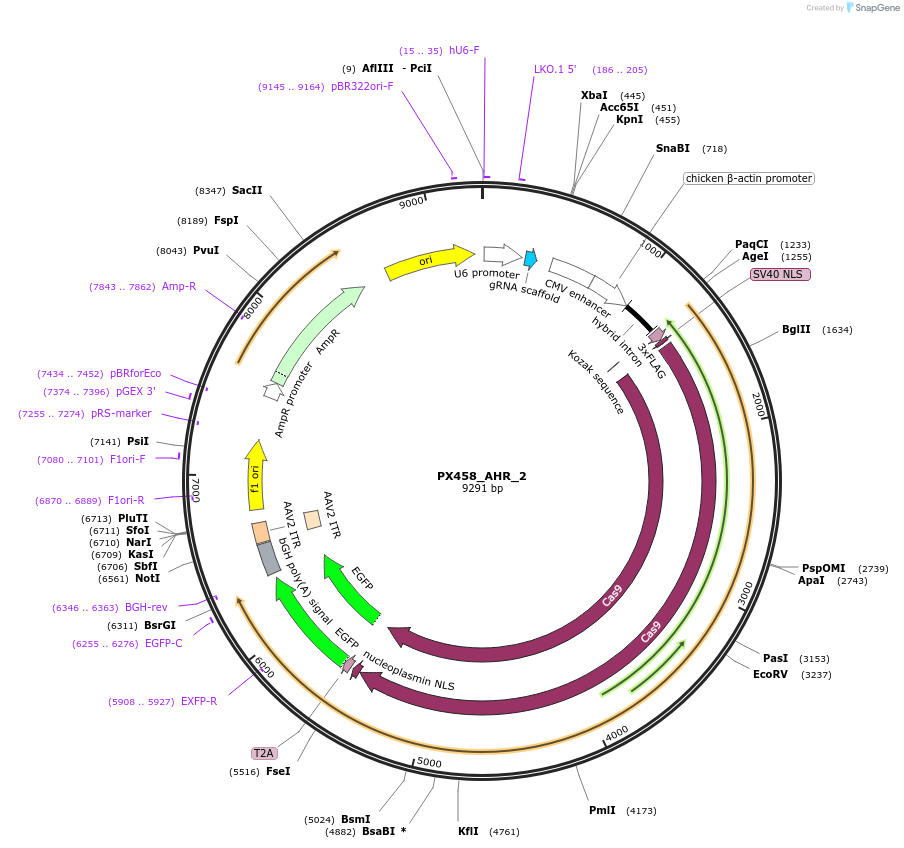
PX458_AHR_2
Plasmid#101077PurposeEncodes gRNA for 3' target of human AHRDepositorInsertgRNA against AHR (AHR Human)
UseCRISPRAvailable SinceOct. 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
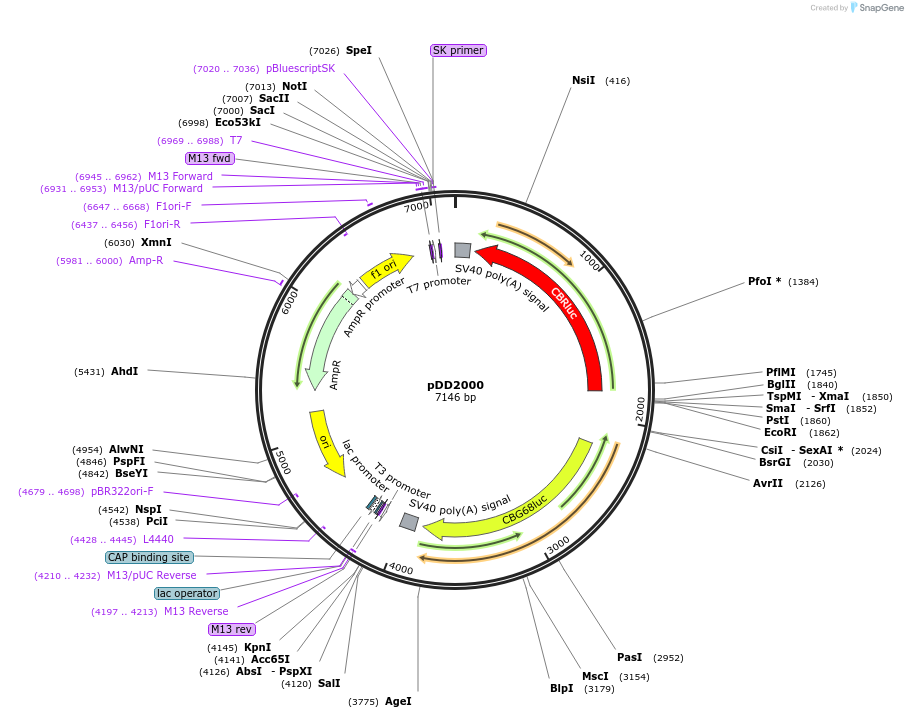
pDD2000
Plasmid#101197PurposeName: pBluescript-LANApi,K14. Bidirectional reporter driving LANApi side [green luciferase isoform from pCBG68] and K14 side [red luciferase isoform from pCBR] in pBluescript backbone.DepositorInsertLANApi side [green luciferase isoform of pCBG68], K14 side [red luciferase isoform of pCBR]
UseLuciferaseAvailabilityAcademic Institutions and Nonprofits only -
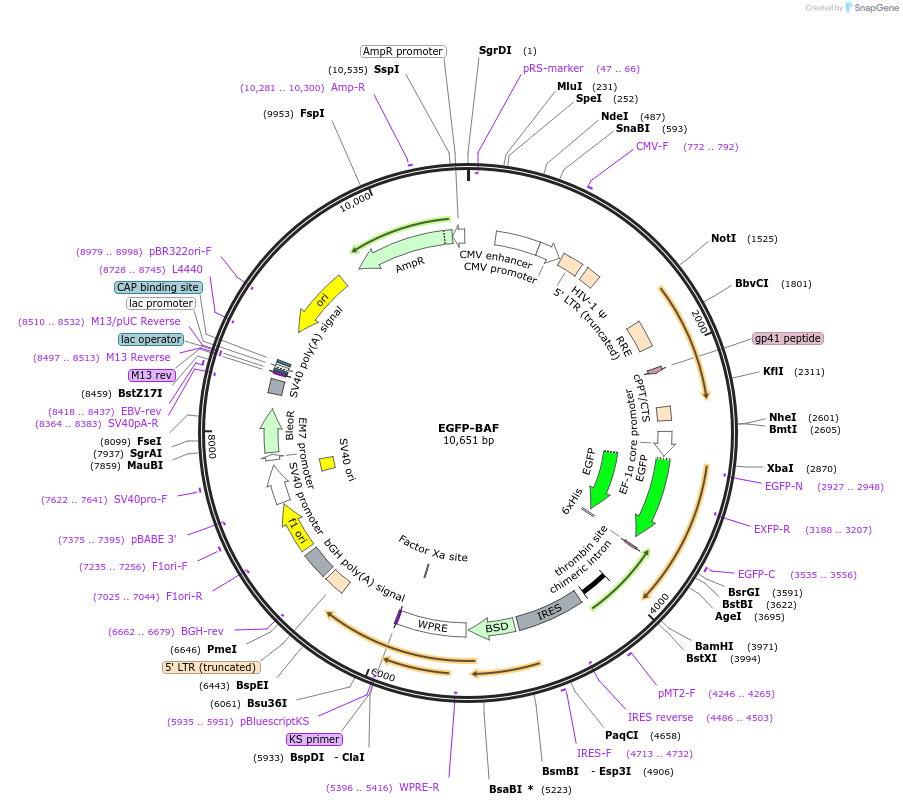
EGFP-BAF
Plasmid#101772PurposeExpresses EGFP tagged BAF in human cellsDepositorAvailable SinceJan. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
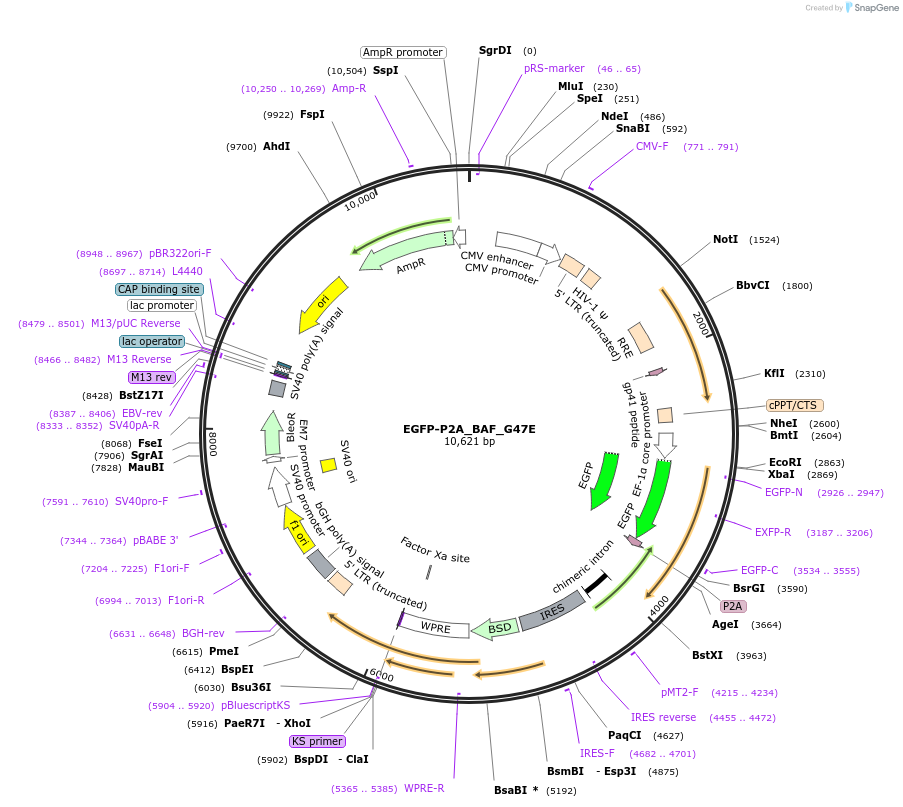
EGFP-P2A_BAF_G47E
Plasmid#101774PurposeExpresses mutant BAF (G47E) in human cells (with EGFP produced from the same transcript as expression control)DepositorInsertBAF (BANF1) (BANF1 Human)
UseLentiviralTagsEGFP-P2AExpressionMammalianMutationG47E mutationPromoterEF1aAvailable SinceDec. 12, 2017AvailabilityAcademic Institutions and Nonprofits only -
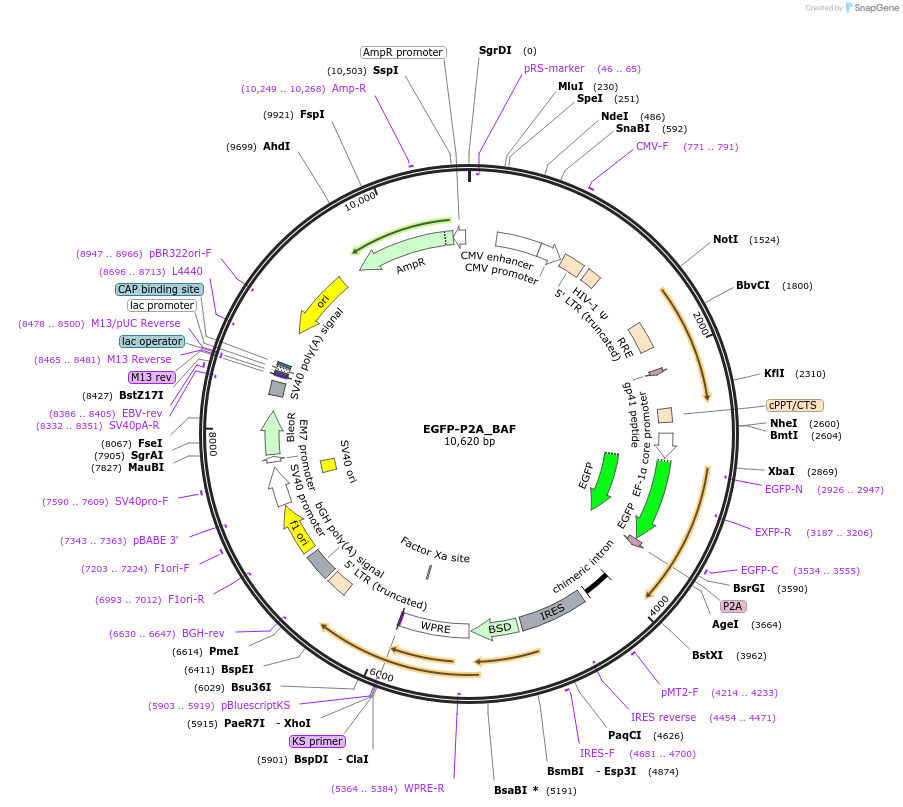
EGFP-P2A_BAF
Plasmid#101773PurposeExpresses BAF in human cells (with EGFP produced from the same transcript as expression control)DepositorAvailable SinceMarch 5, 2018AvailabilityAcademic Institutions and Nonprofits only