We narrowed to 23,042 results for: ESC
-
Plasmid#50835PurposeExpresses a tandem ddFP heterodimer in mammalian cells, construct 1 for a translocation-based green fluorescent caspase-3 biosensor, used together with plasmid 2 (BNLS).DepositorInsertddGFP A and ddRFP B
TagsNES sequence LALKLAGLDIGS and triplicated NLS seq…ExpressionMammalianPromoterCMVAvailable SinceJan. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
EYFP-p65
Plasmid#111192Purposefluorescent fusion proteinDepositorAvailable SinceJune 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
pAG104 FAST
Plasmid#130720PurposeExpresses FAST (also called YFAST) in mammalian cellsDepositorInsertFAST
ExpressionMammalianPromoterCMVAvailable SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
pAG87 FAST
Plasmid#130719PurposeExpresses His-tagged FAST (also called YFAST) in bacteriaDepositorInsertFAST
TagsHis-tagExpressionBacterialPromoterT7Available SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
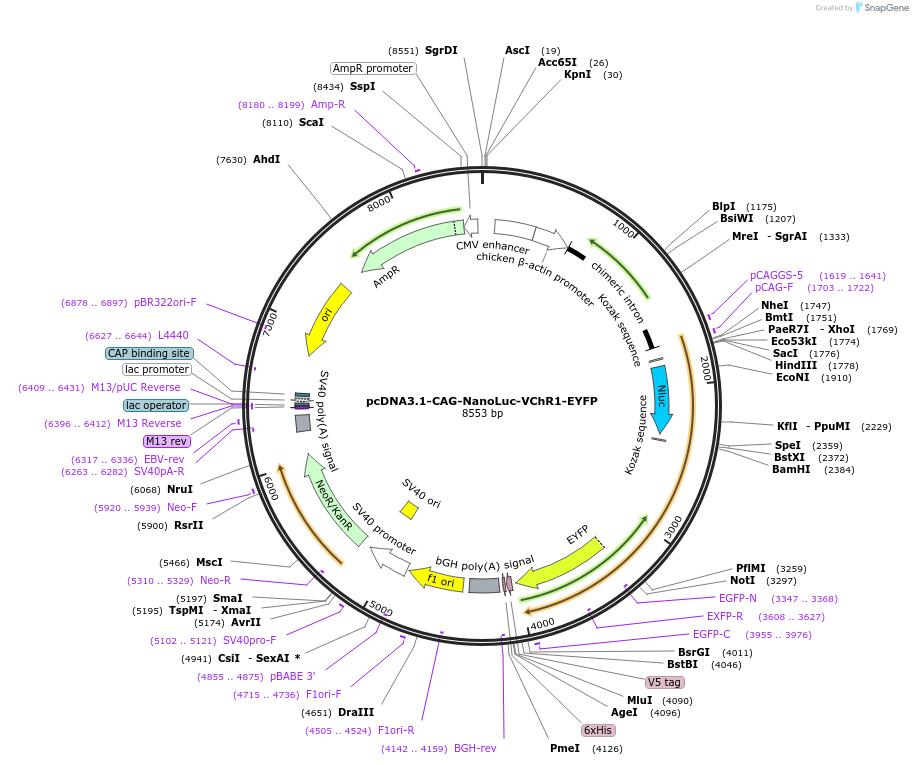
pcDNA3.1-CAG-NanoLuc-VChR1-EYFP
Plasmid#114112Purposefusion protein of NanoLuc, Volvox channelrhodopsin-1, and enhanced yellow fluorescent protein for bioluminescent optogeneticsDepositorInsertNanoLuc; Volvox channelrhodopsin 1; EYFP
ExpressionMammalianPromoterCAGAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
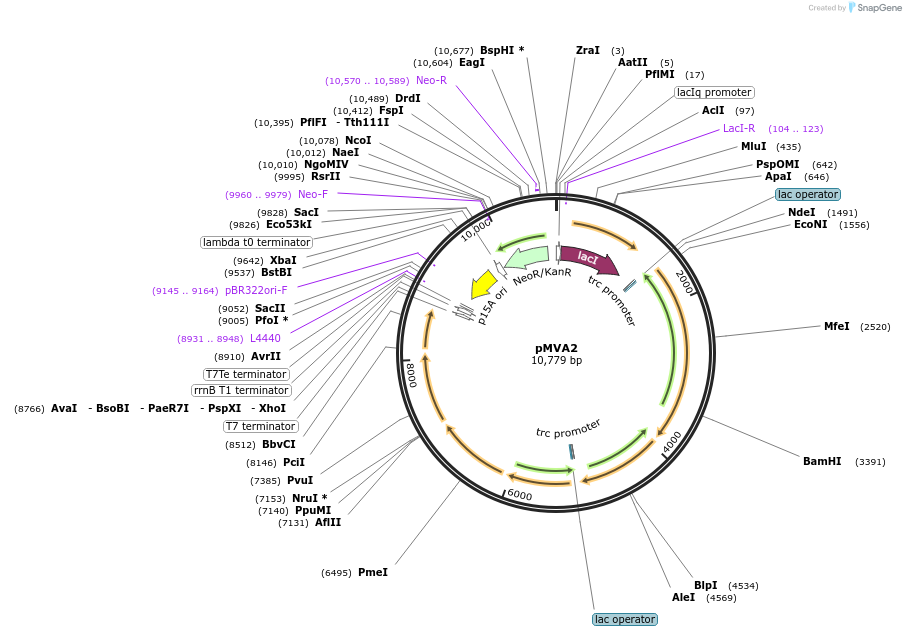
pMVA2
Plasmid#119951PurposeExpresses recombinant MVA pathway in Escherichia coliDepositorInsertsmvaE
mvaS
mvaK1
mvaK2
mvaD
idi
ExpressionBacterialMutationA110GAvailable SinceFeb. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
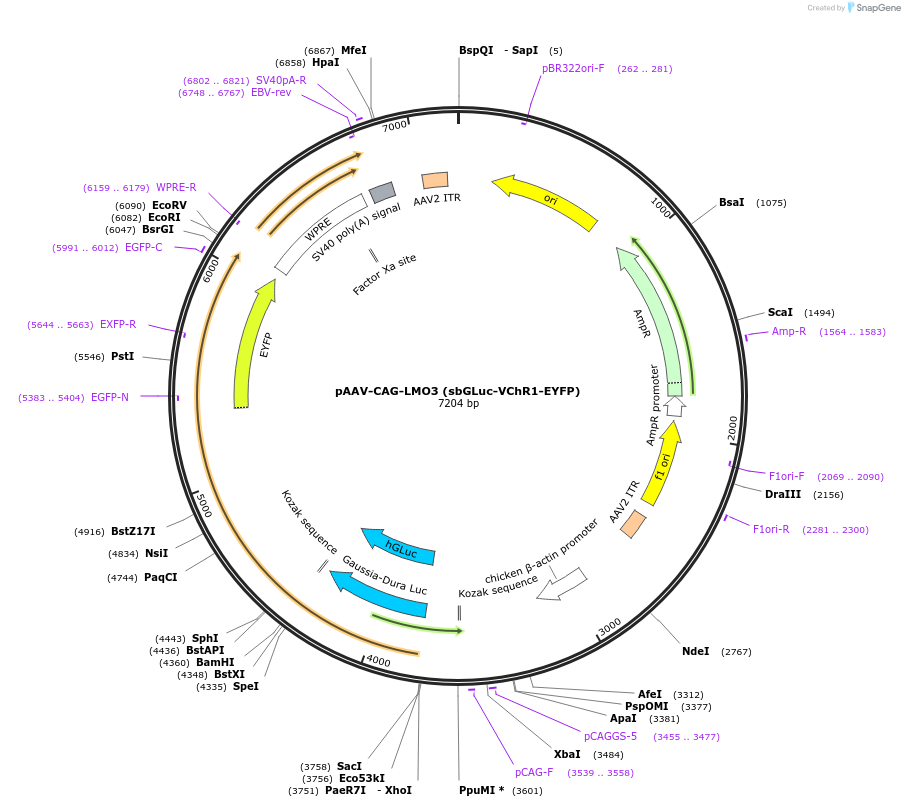
pAAV-CAG-LMO3 (sbGLuc-VChR1-EYFP)
Plasmid#114104Purposefusion protein of Gaussia luciferase variant slow burn, Volvox channelrhodopsin-1, and enhanced yellow fluorescent protein for bioluminescent optogeneticsDepositorInsertsbGLuc-VChR1-EYFP
UseAAVExpressionMammalianPromoterCAGAvailable SinceAug. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
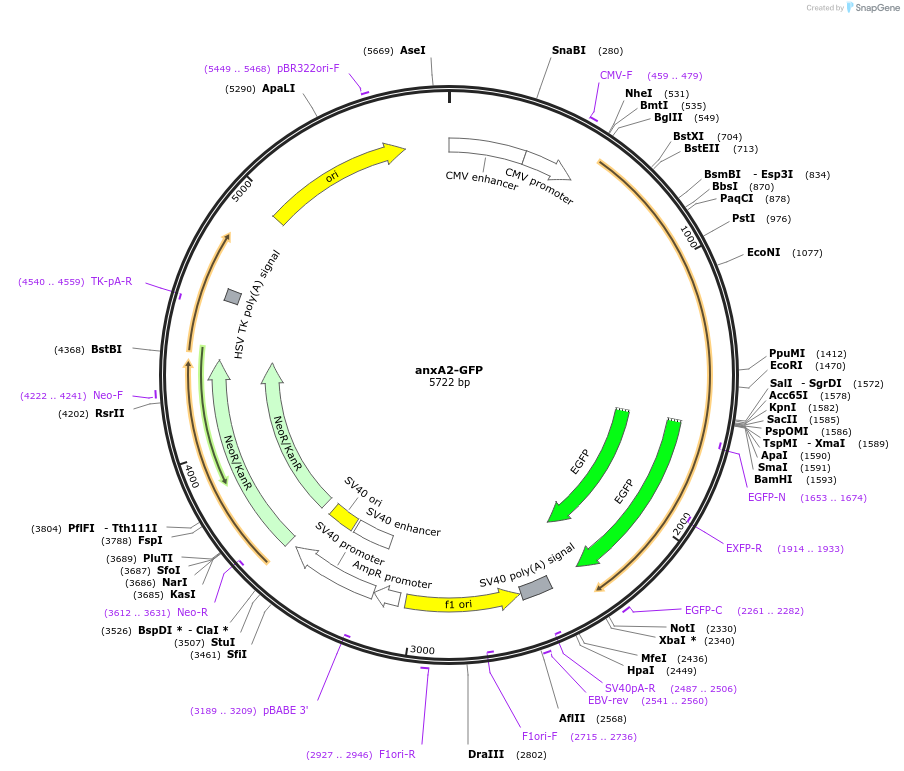
anxA2-GFP
Plasmid#107196PurposeExpresses human annexin A2-GFP fusion protein for live cell imagingDepositorInsertannexin A2 (ANXA2 Human)
TagsGreen Fluorescent ProteinExpressionMammalianMutationAla residue 65 in human sequence replaced by Glu …PromoterCMVAvailable SinceJune 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
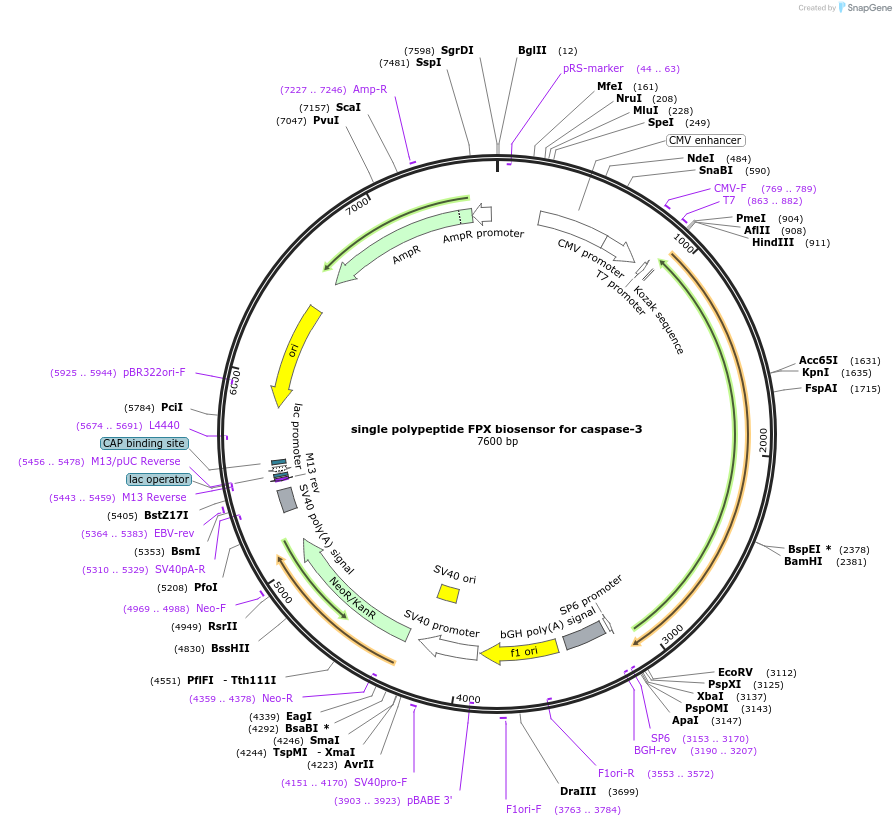
single polypeptide FPX biosensor for caspase-3
Plasmid#60883PurposeFPX biosensor, a new class of genetically encoded fluorescent biosensor based on reversible exchange of the heterodimeric partners of green and red dimerization dependent fluorescent proteins (ddFPs).DepositorInsertsingle polypeptide FPX biosensor for caspase-3
TagsHindIII site and NES sequence LALKLAGLDIGSExpressionMammalianPromoterCMVAvailable SinceJan. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pX037
Plasmid#167156PurposeMoClo-compatible Level2 vector for autonomous bioluminescence in plants encoding kanamycin resistance cassette, nnHispS, nnH3H, nnLuz and nnCPH. Does not contain NpgA.DepositorInsertcaffeic acid cycle genes
ExpressionPlantPromoterpNOS, p35sAvailable SinceJune 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
RANLS-IETD-BNES
Plasmid#60972PurposeExpresses a tandem ddFP heterodimer in mammalian cells, plasmid 1 for a translocation-based red fluorescent caspase-8 biosensor, used together with plasmid 2 (BNLS).DepositorInsertddRFP A and ddRFP B
TagsNES sequence LALKLAGLDIGS and triplicated NLS seq…ExpressionMammalianPromoterCMVAvailable SinceJan. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
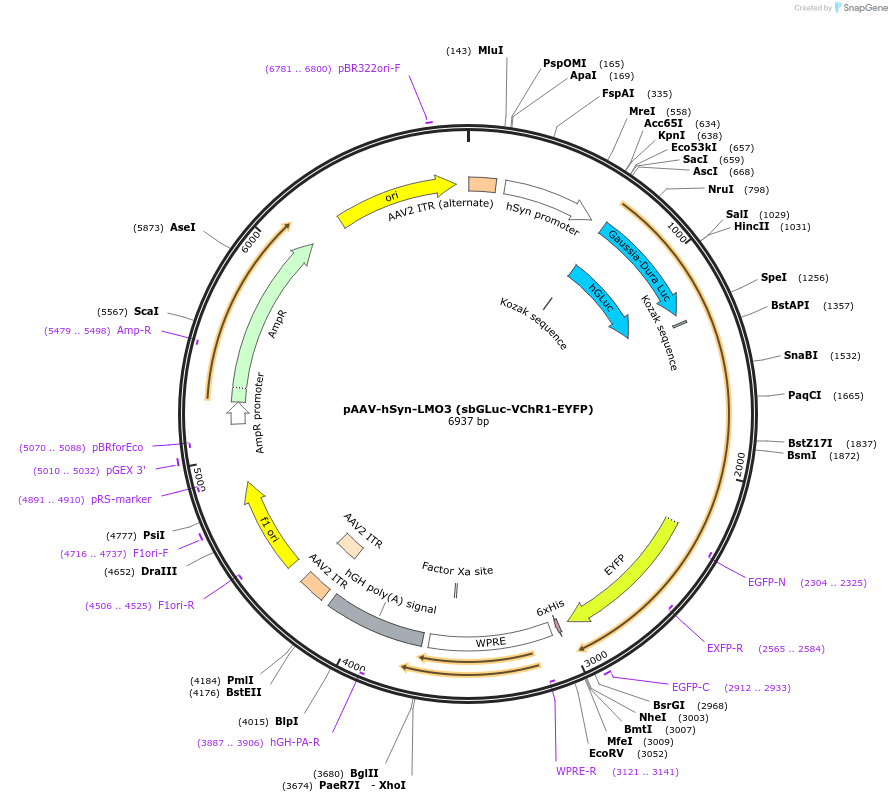
pAAV-hSyn-LMO3 (sbGLuc-VChR1-EYFP)
Plasmid#114099Purposefusion protein of Gaussia luciferase variant slow burn, Volvox channelrhodopsin-1, and enhanced yellow fluorescent protein for bioluminescent optogeneticsDepositorInsertGaussia luciferase, slow burn; Volvox Channelrhodopsin 1; EYFP
UseAAVExpressionMammalianPromoterhSynAvailable SinceAug. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
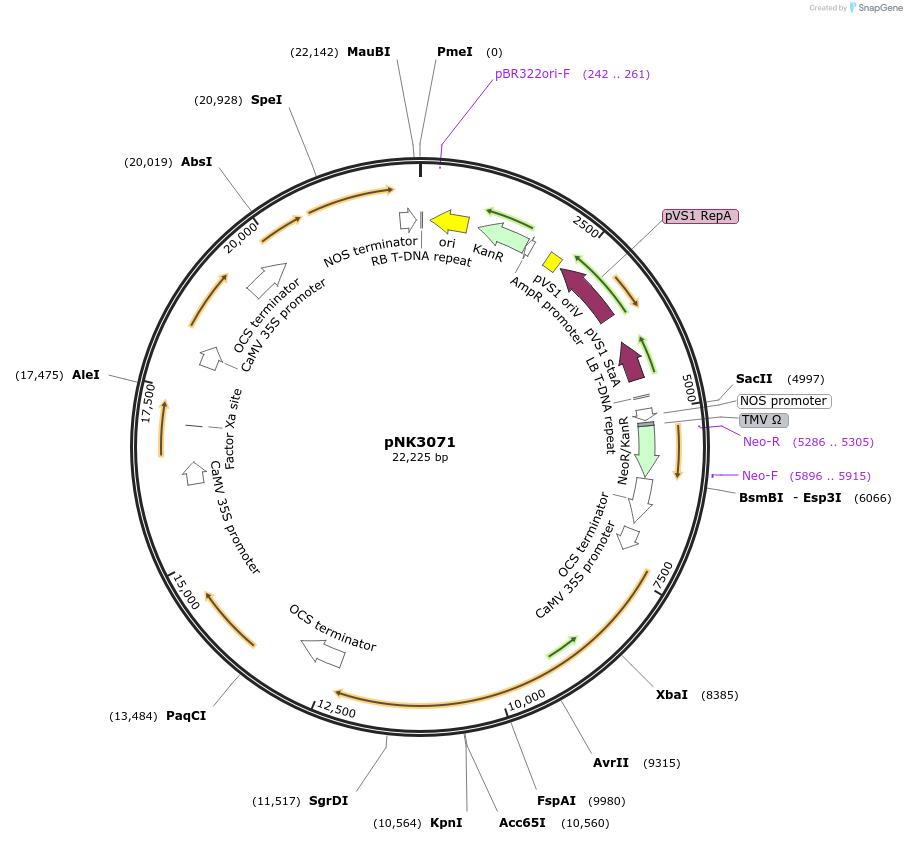
pNK3071
Plasmid#219755PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, mcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertmcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceOct. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
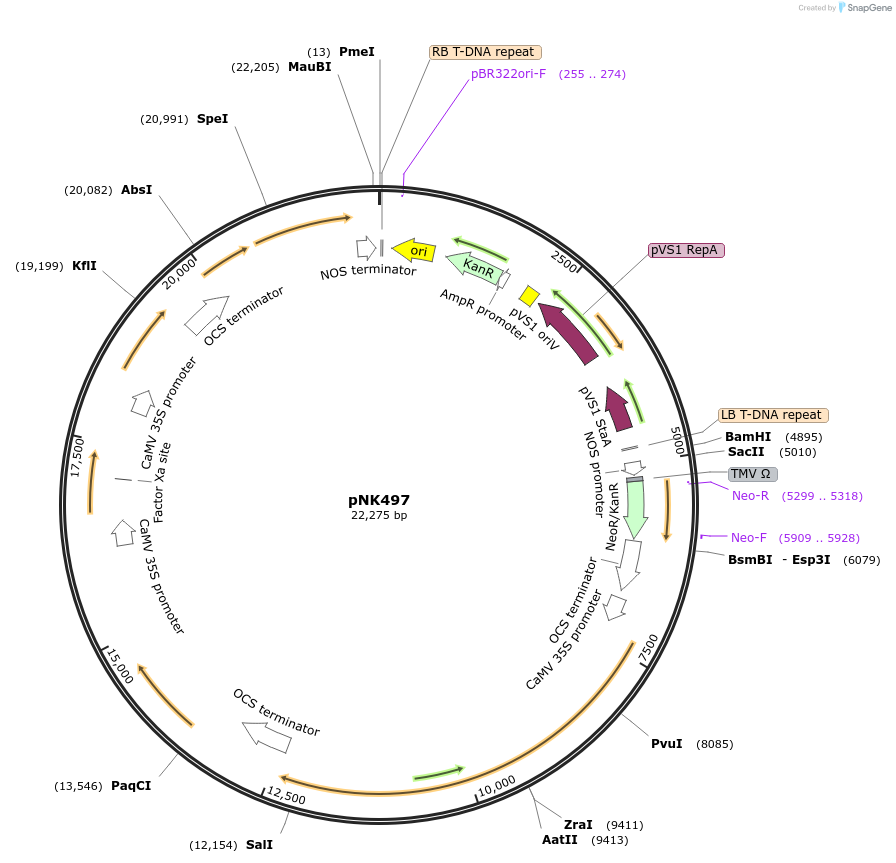
pNK497
Plasmid#219754PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, nnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertnnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceJan. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
RANLS-LEHD-BNES
Plasmid#60973PurposeExpresses a tandem ddFP heterodimer in mammalian cells, plasmid 1 for a translocation-based red fluorescent caspase-9 biosensor, used together with plasmid 2 (BNLS).DepositorInsertddRFP A and ddRFP B
TagsCaspase9 substrate sequence (LEHD) is after NLS s…ExpressionMammalianPromoterCMVAvailable SinceJan. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
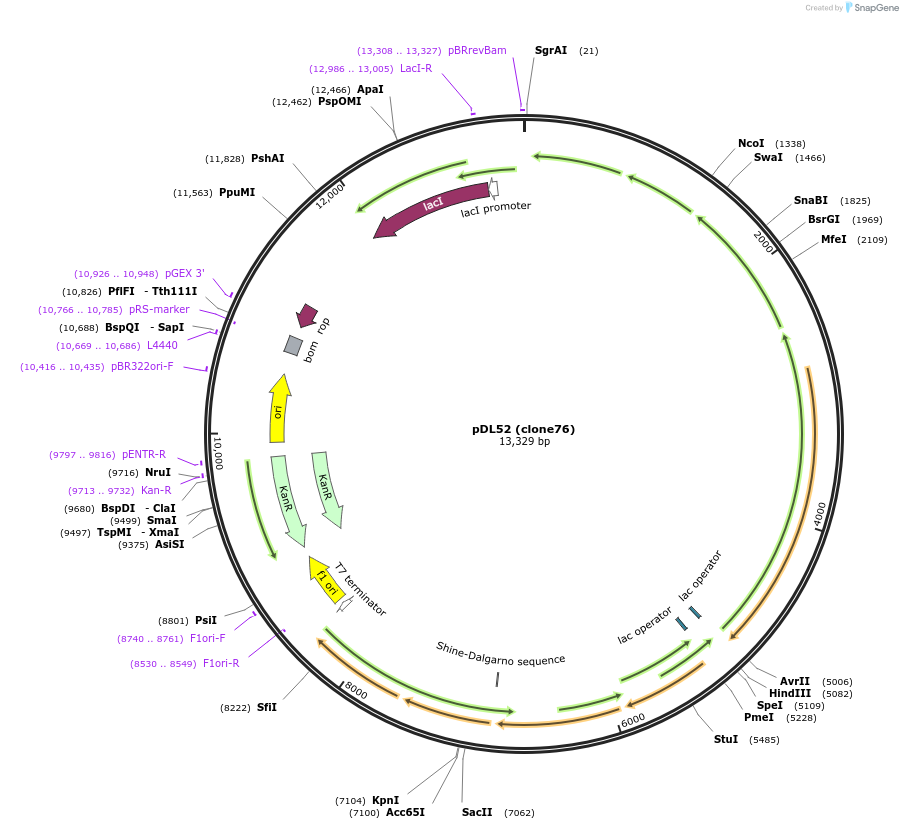
pDL52 (clone76)
Plasmid#188579PurposeHigh-yield production of coenzyme F420 in E. coliDepositorInsertsribA
ribD
yigB
fbiC
cofC
cofD
cofI
cofE
ExpressionBacterialMutationCodon optimization, synthetic ribosome binding si…PromoterT7 variantsAvailable SinceSept. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
CMV Marina-T2A-nls-mCherry
Plasmid#74216Purposegenetically-encoded fluorescent voltage indicatorDepositorInsertfluorescent protein voltage sensor
TagsmCherryExpressionMammalianMutationCi-VSP contains R217Q mutation; super ecliptic pH…PromoterCMVAvailable SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
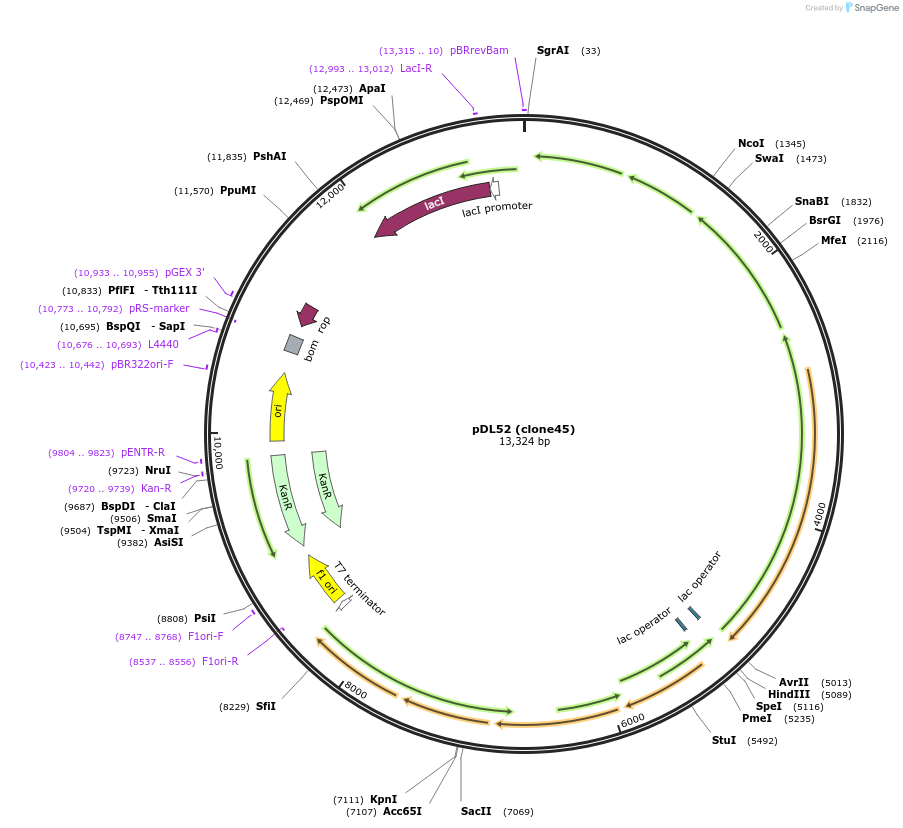
pDL52 (clone45)
Plasmid#188578PurposeHigh-yield production of coenzyme F420 in E. coliDepositorInsertsribA
ribD
yigB
fbiC
cofC
cofD
cofI
cofE
ExpressionBacterialMutationCodon optimization, synthetic ribosome binding si…PromoterT7 variantsAvailable SinceSept. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
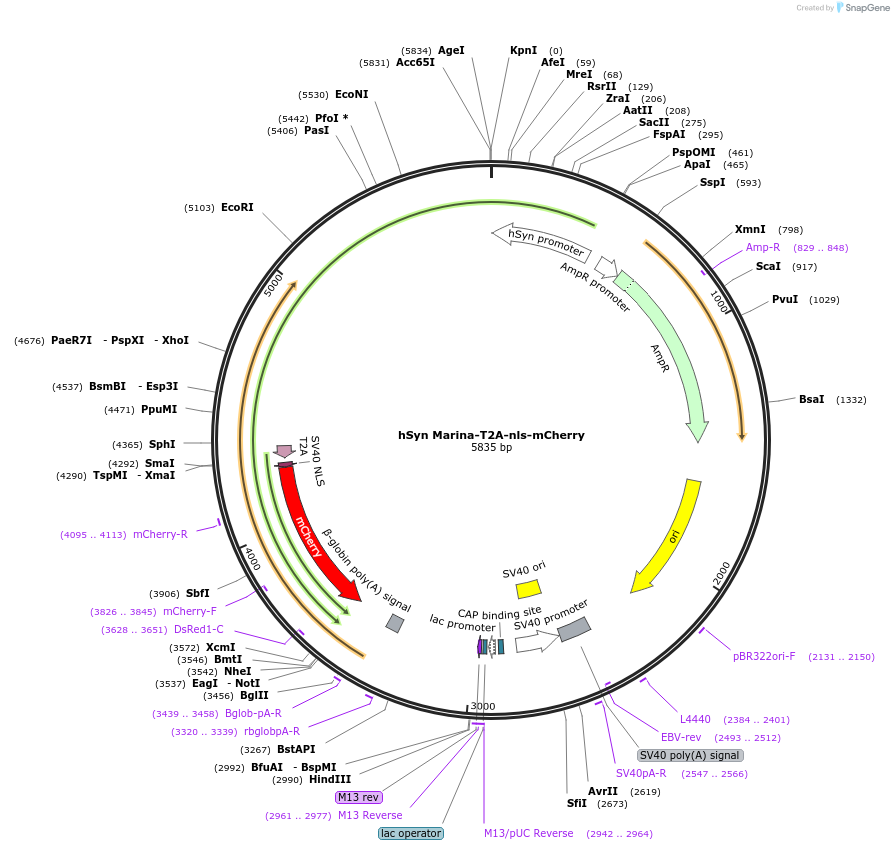
hSyn Marina-T2A-nls-mCherry
Plasmid#85843Purposegenetically-encoded fluorescent voltage indicatorDepositorInsertGEVI Marina
TagsmCherryExpressionMammalianMutationCi-VSP contains R217Q mutation; super ecliptic pH…PromoterhSyn1Available SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
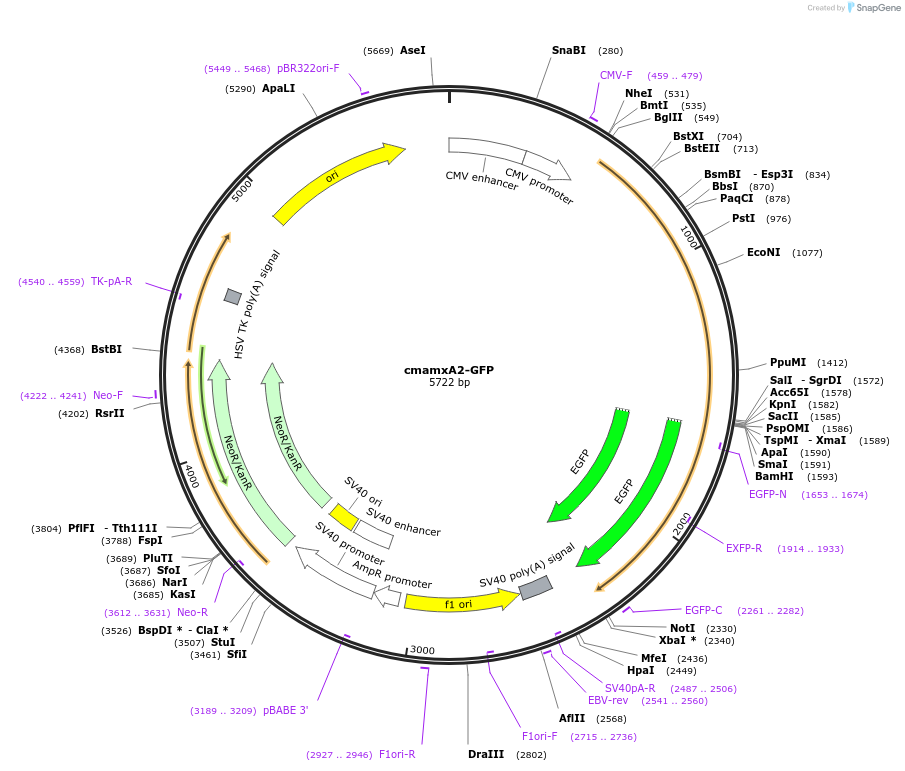
cmamxA2-GFP
Plasmid#107197PurposeExpresses human annexin A2-GFP fusion protein for live cell imaging, all type II Ca2+ binding sites deletedDepositorInsertcmannexin A2 (ANXA2 Human)
TagsGreen Fluorescent ProteinExpressionMammalianMutationall type II Ca2+ binding sites deleted ( D161A, E…PromoterCMVAvailable SinceJune 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
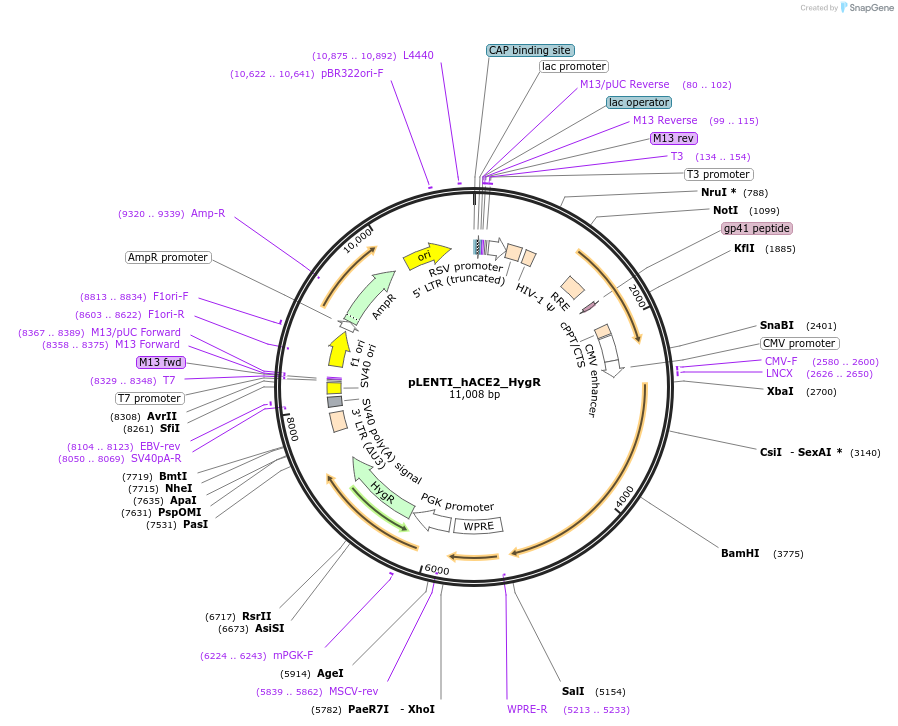
pLENTI_hACE2_HygR
Plasmid#155296PurposeLentiviral vector to generate hACE2 stable expressing cell line, Hygromycin resistanceDepositorAvailable SinceAug. 5, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
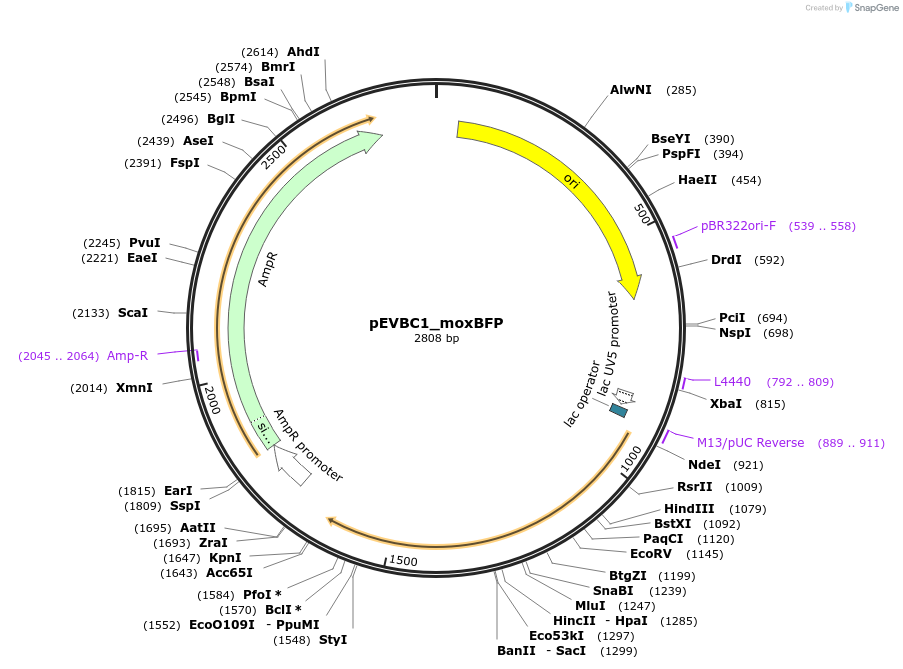
pEVBC1_moxBFP
Plasmid#248345PurposeBlue fluorescent protein control for Fluorescent Protein LibraryDepositorInsertmoxBFP
UseSynthetic BiologyExpressionBacterialPromoterP_LacUV5Available SinceNov. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
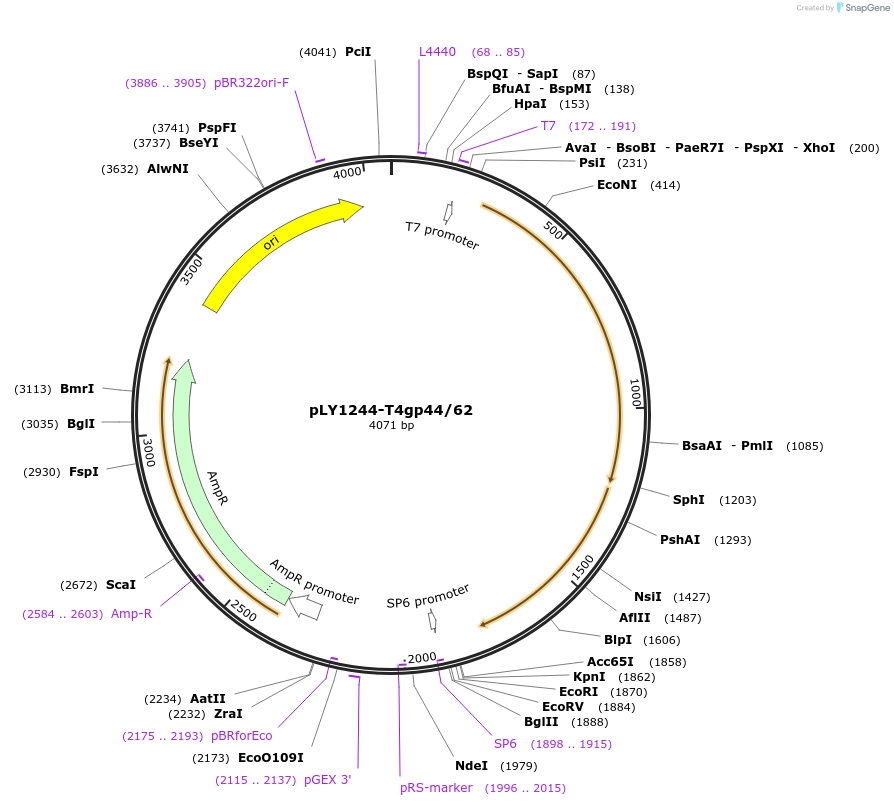
pLY1244-T4gp44/62
Plasmid#228205PurposeExpression in BL21(DE3) of WT T4 clamp loader consisting of gp44 and gp62DepositorInsert44/62
ExpressionBacterialPromoterT7Available SinceMay 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
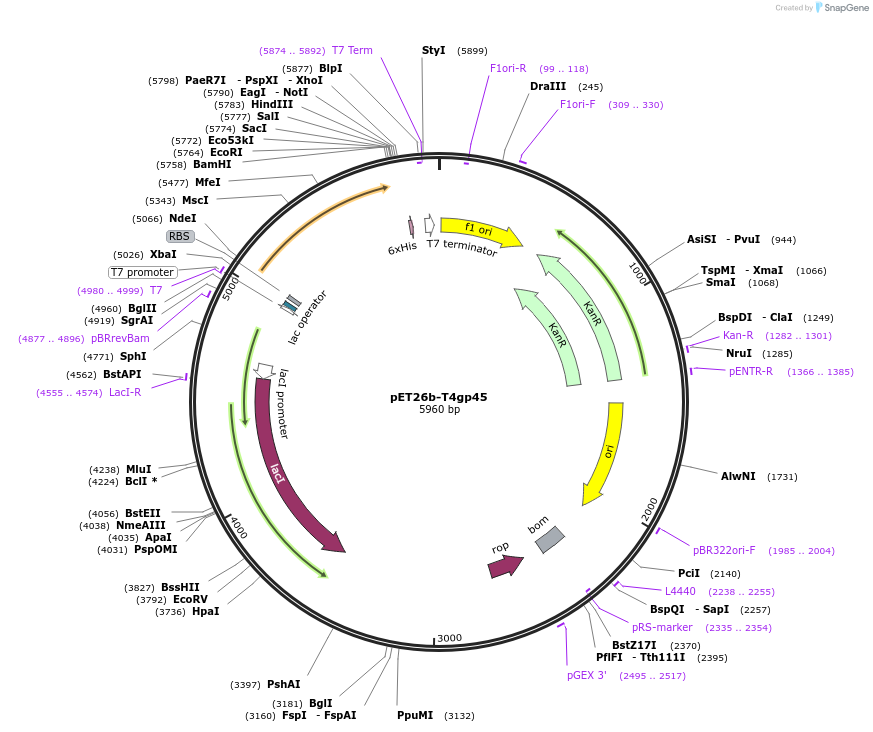
pET26b-T4gp45
Plasmid#228204PurposeExpression in BL21(DE3) of WT T4 clampDepositorAvailable SinceNov. 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
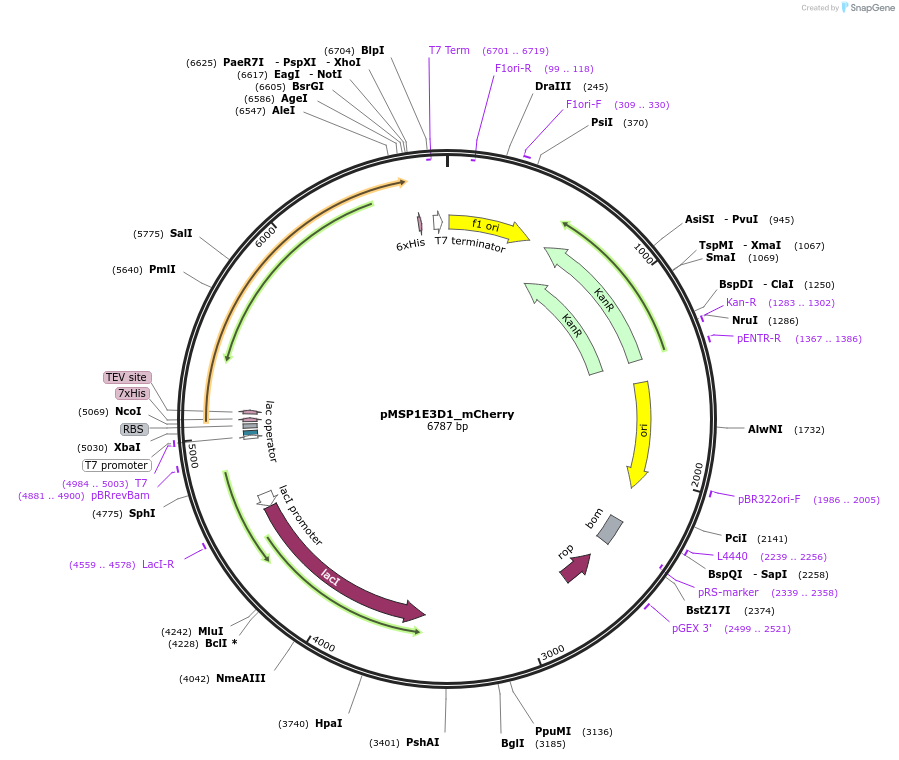
pMSP1E3D1_mCherry
Plasmid#248929PurposeBacterial expression of fluorescent membrane scaffold protein MSP1E3D1 fused to mCherry for nanodisc formationDepositorInsertMSP1E3D1_mCherry
TagsHisExpressionBacterialPromoterT7 PromoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
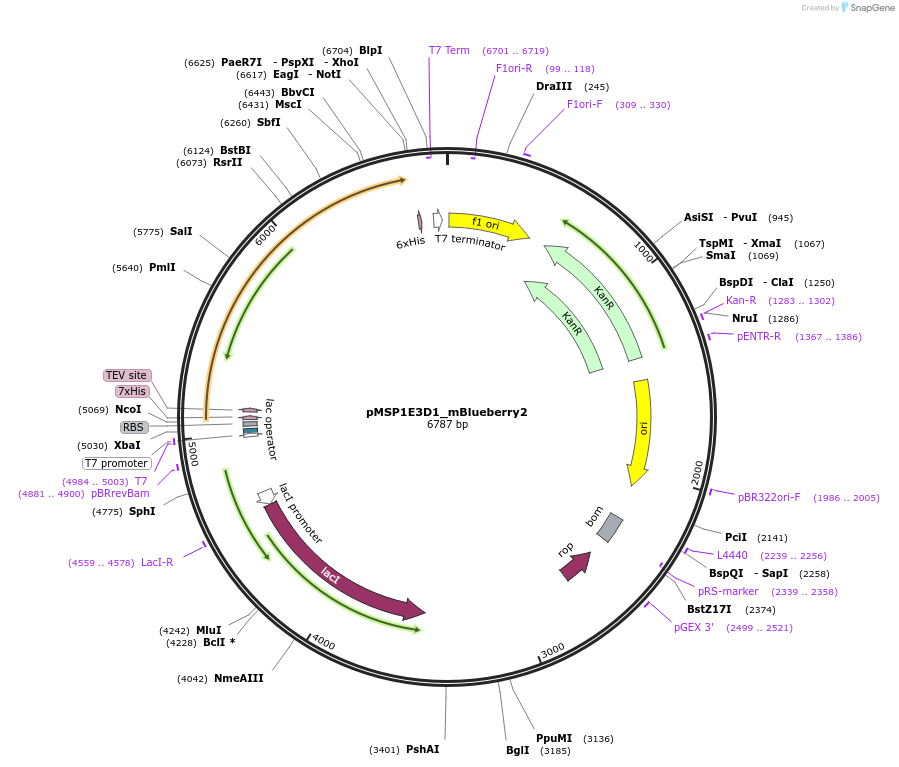
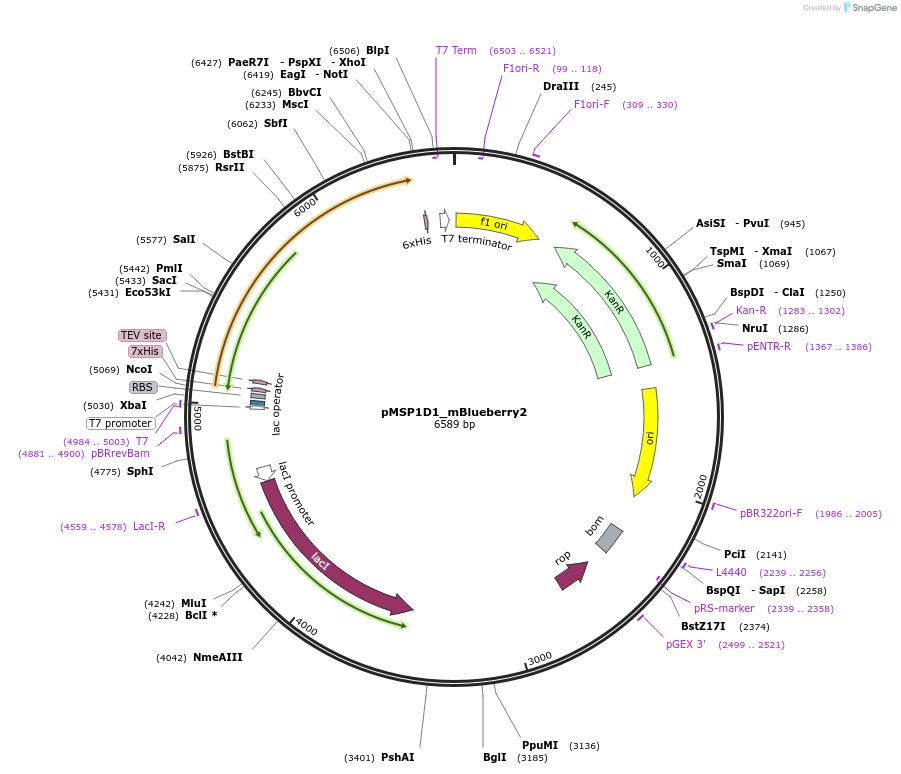
pMSP1E3D1_mBlueberry2
Plasmid#248930PurposeBacterial expression of fluorescent membrane scaffold protein MSP1E3D1 fused to mBlueberry 2 for nanodisc formationDepositorInsertMSP1E3D1_mBlueberry2
TagsHisExpressionBacterialPromoterT7 PromoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
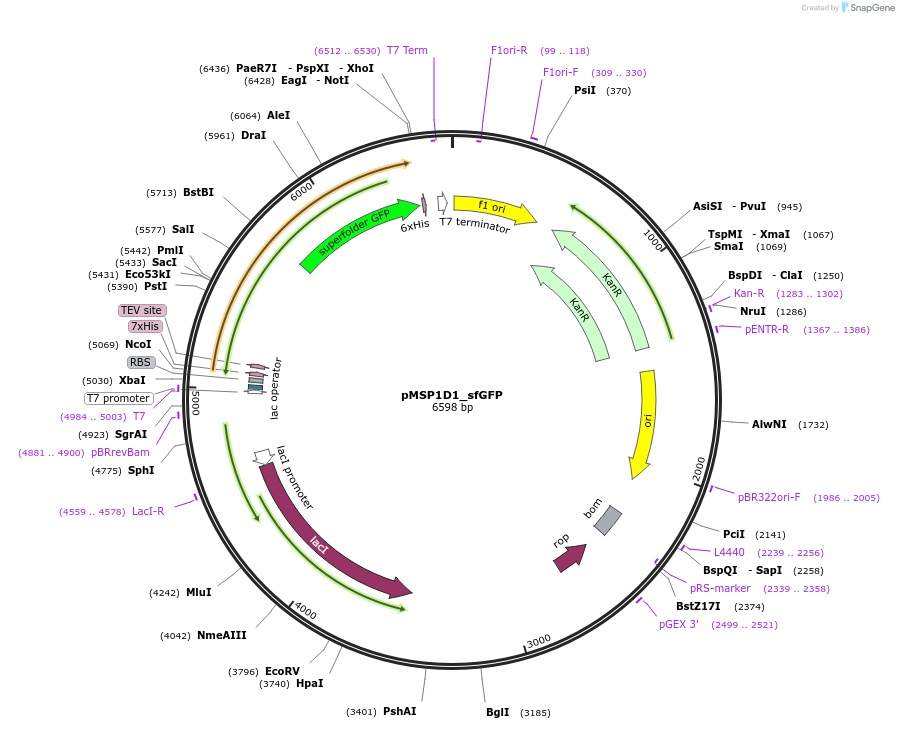
pMSP1D1_sfGFP
Plasmid#248920PurposeBacterial expression of fluorescent membrane scaffold protein MSP1D1 fused to sfGFP for nanodisc formationDepositorInsertMSP1D1_sfGFP
TagsHisExpressionBacterialPromoterT7 PromoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
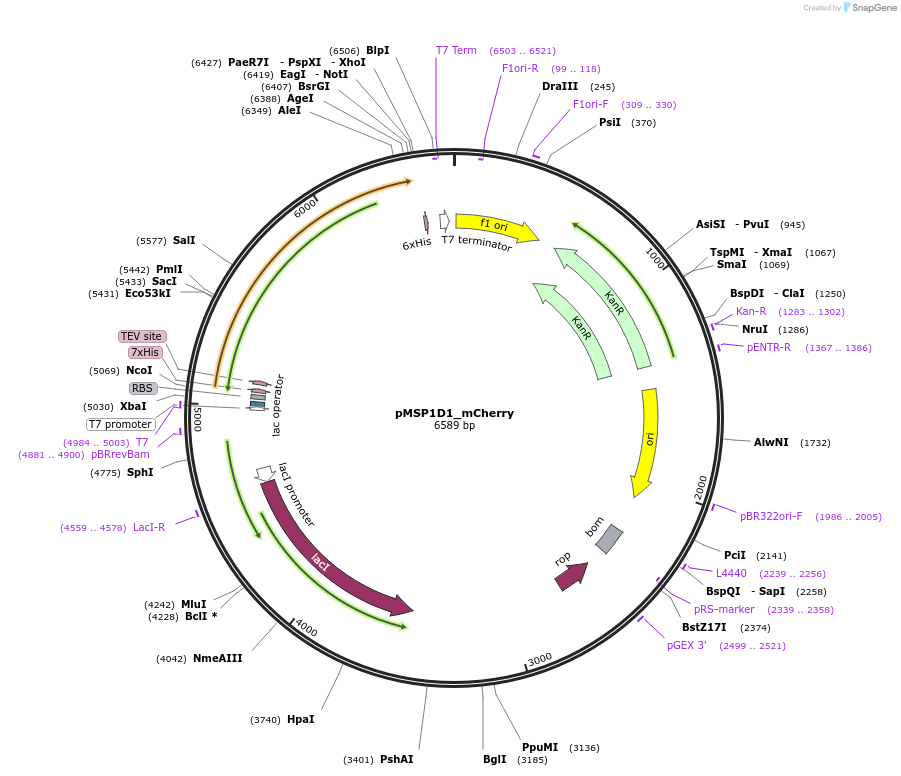
pMSP1D1_mCherry
Plasmid#248922PurposeBacterial expression of fluorescent membrane scaffold protein MSP1D1 fused to mCherry for nanodisc formationDepositorInsertMSP1D1_mCherry
TagsHisExpressionBacterialPromoterT7 PromoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
pMSP1D1_mBlueberry2
Plasmid#248925PurposeBacterial expression of fluorescent membrane scaffold protein MSP1D1 fused to mBlueberry2 for nanodisc formationDepositorInsertMSP1D1_mBlueberry2
TagsHisExpressionBacterialPromoterT7 PromoterAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
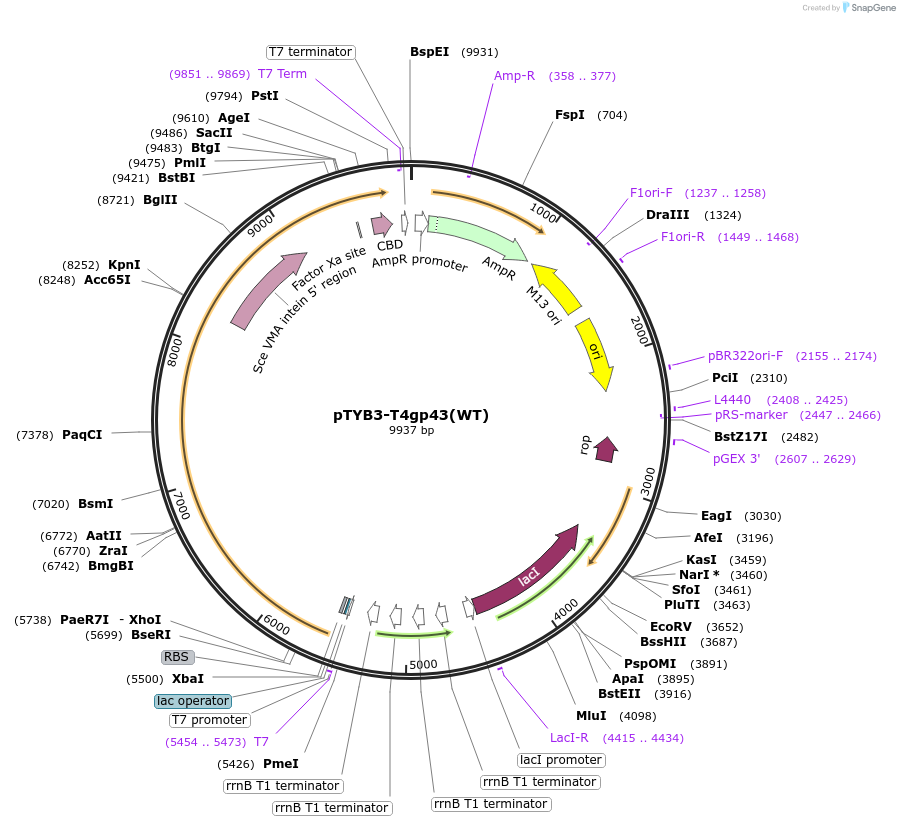
pTYB3-T4gp43(WT)
Plasmid#228203PurposeExpression in BL21(DE3) of WT T4 DNA polymeraseDepositorInsert43
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceMay 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
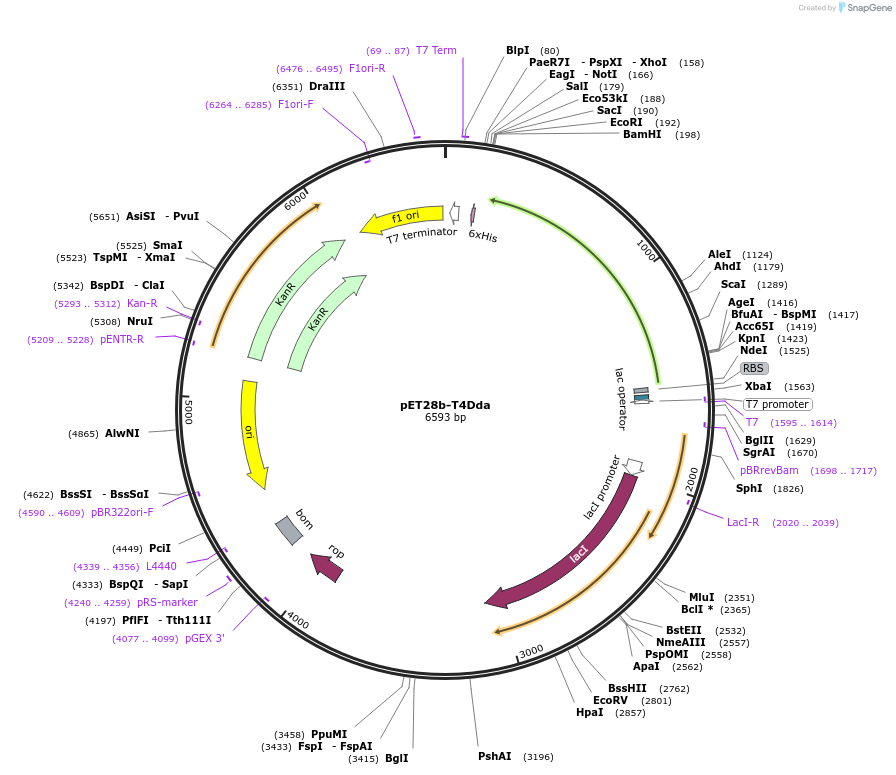
pET28b-T4Dda
Plasmid#228215PurposeExpression in BL21(DE3) of T4 Dda-like helicaseDepositorAvailable SinceMarch 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
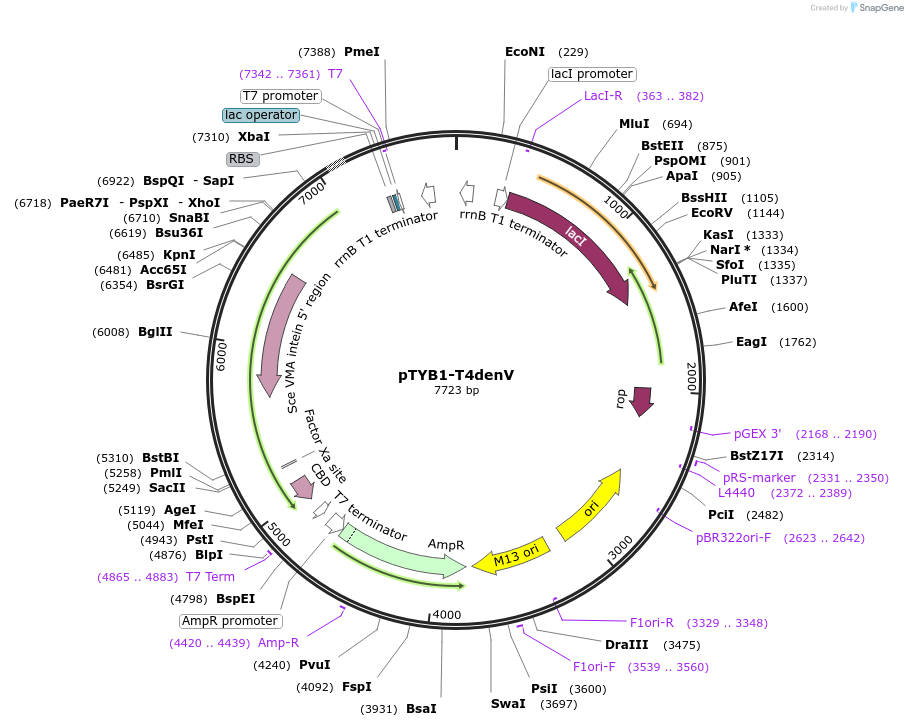
pTYB1-T4denV
Plasmid#228216PurposeExpression in BL21(DE3) of T4 endonuclease V N-glycosylase UV repair enzymeDepositorInsertdenV
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
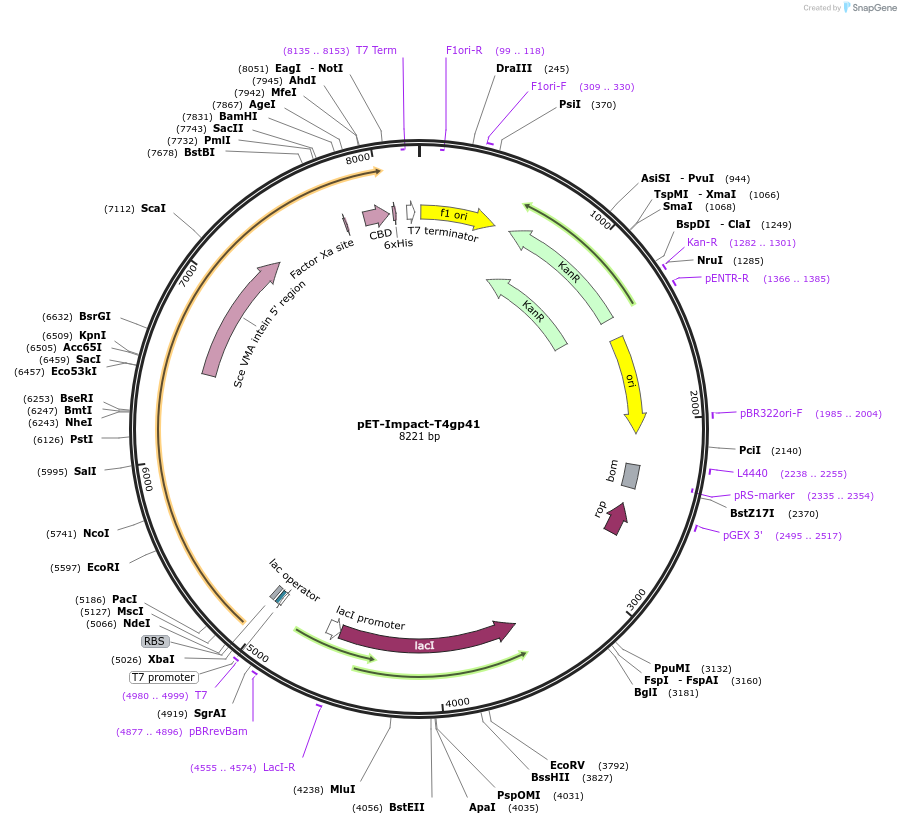
pET-Impact-T4gp41
Plasmid#228207PurposeExpression in BL21(DE3) of T4 helicaseDepositorInsert41 (41 Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceMarch 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
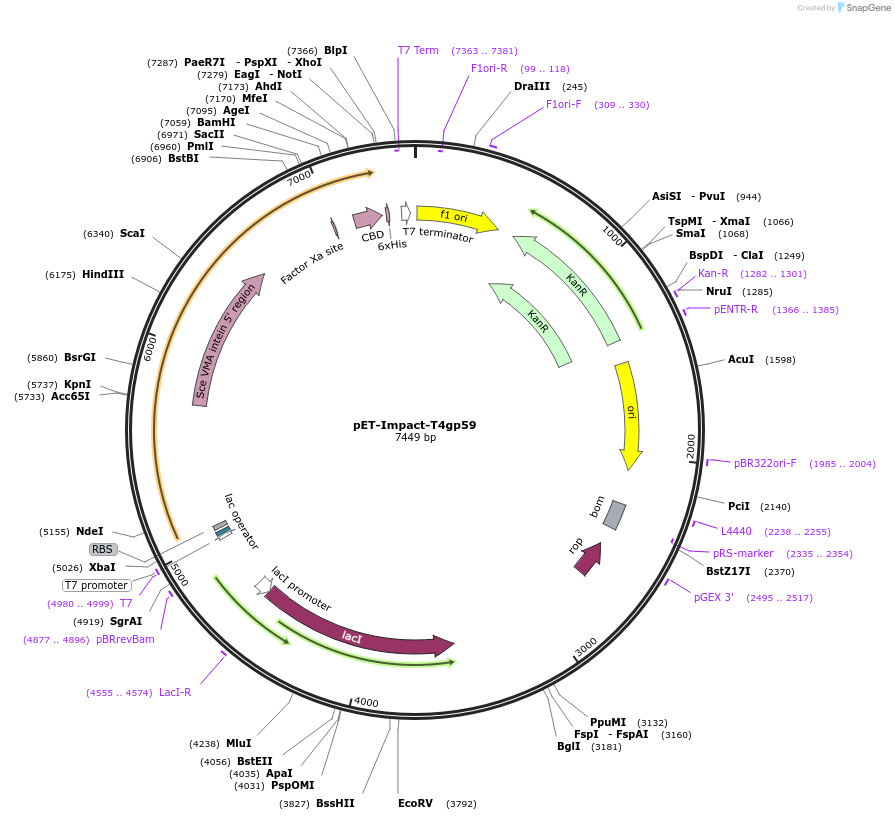
pET-Impact-T4gp59
Plasmid#228206PurposeExpression in BL21(DE3) of WT T4 helicase loaderDepositorInsert59 (59 Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceMarch 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
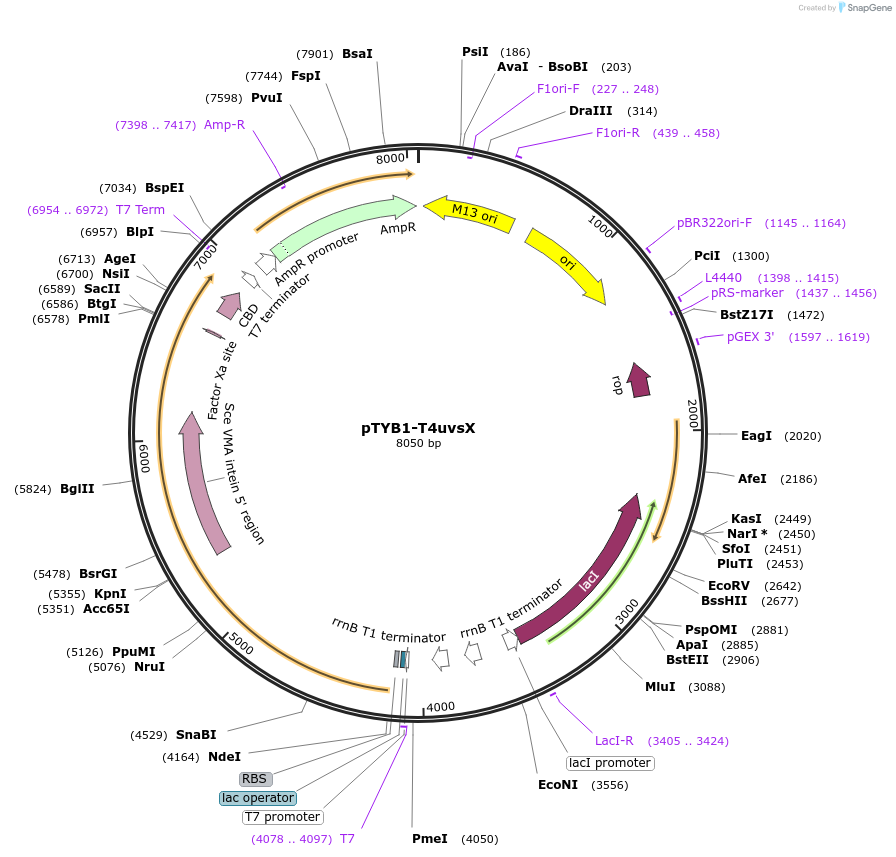
pTYB1-T4uvsX
Plasmid#228213PurposeExpression in BL21(DE3) of T4 recombinaseDepositorInsertuvsX (uvsX Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
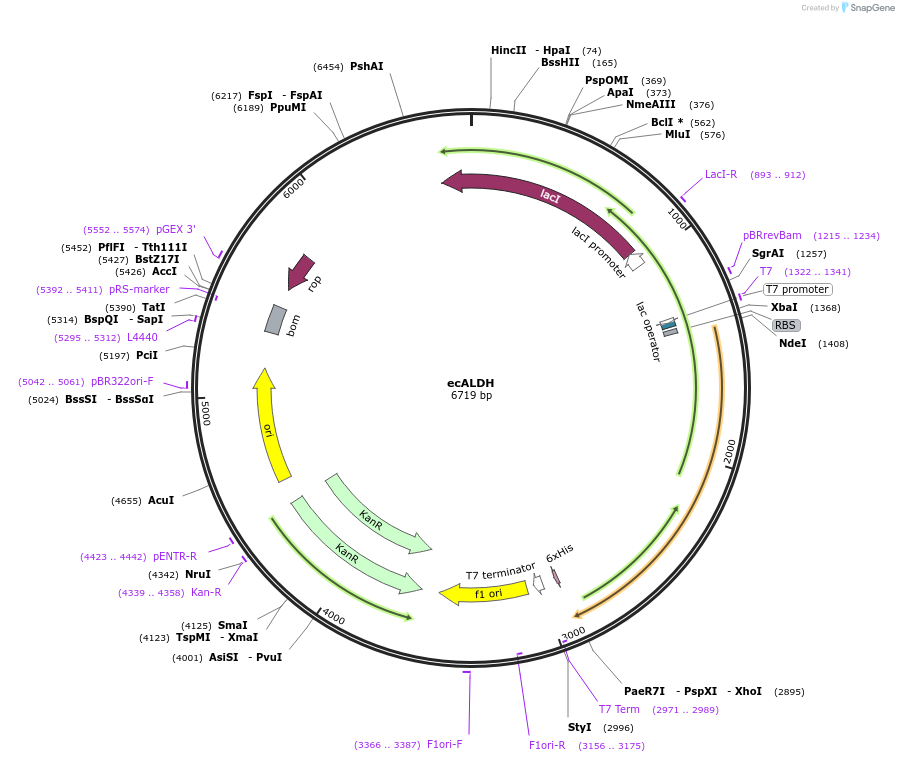
ecALDH
Plasmid#67921PurposeExpresses aldehyde dehydrogenase from Escherichia coli, codon optimized for expression in E. coliDepositorInsertEscherichia coli aldehyde dehydrogenase (puuC Escherichia coli)
Tags6x-HisExpressionBacterialPromoterT7Available SinceSept. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
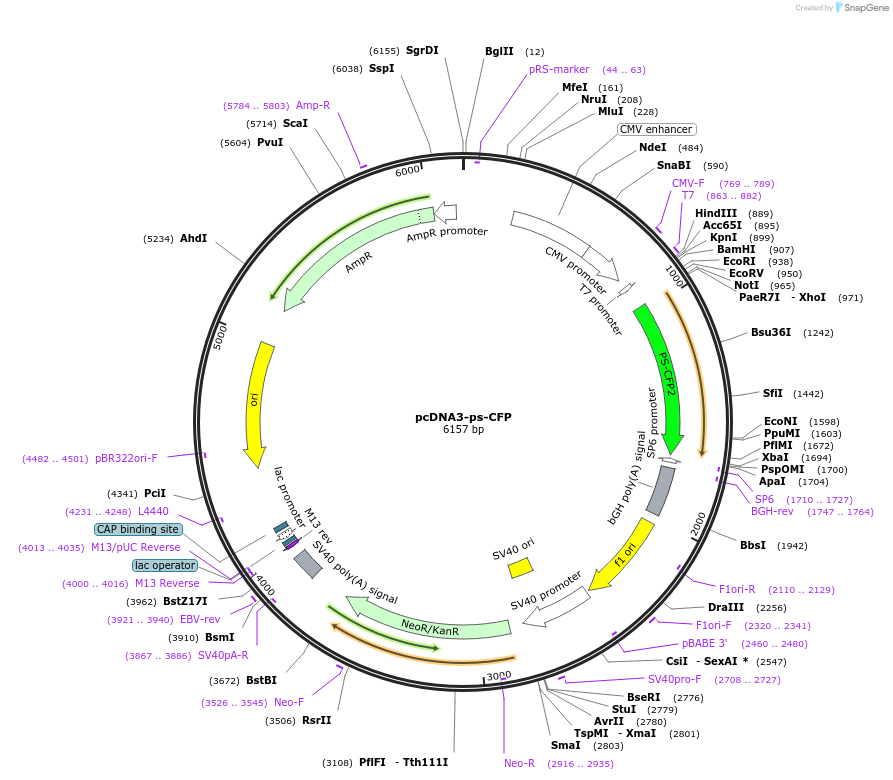
pcDNA3-ps-CFP
Plasmid#13034PurposeExpresses photo-switchable CFP in mammalian cells. Can undergo irreversible photoconversion from cyan to green fluorescenceDepositorInsertphoto-switchable cyan fluorescent protein
ExpressionMammalianPromoterCMVAvailable SinceOct. 2, 2013AvailabilityAcademic Institutions and Nonprofits only -
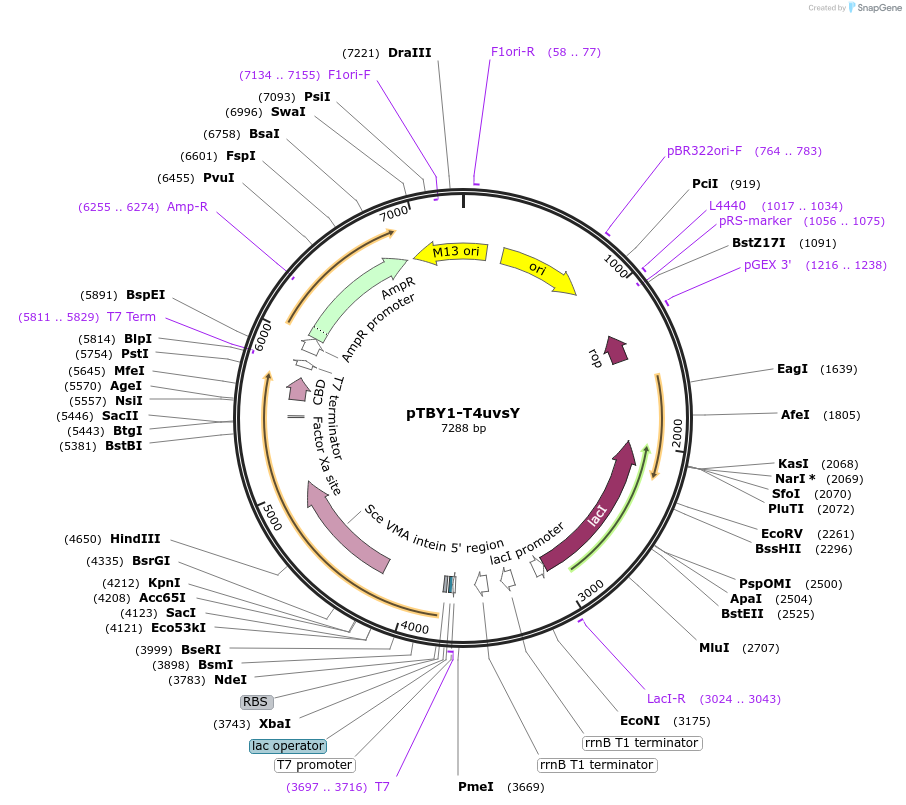
pTBY1-T4uvsY
Plasmid#228214PurposeExpression in BL21(DE3) of T4 UvsY-like recombination mediatorDepositorInsertuvsY (uvsY Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
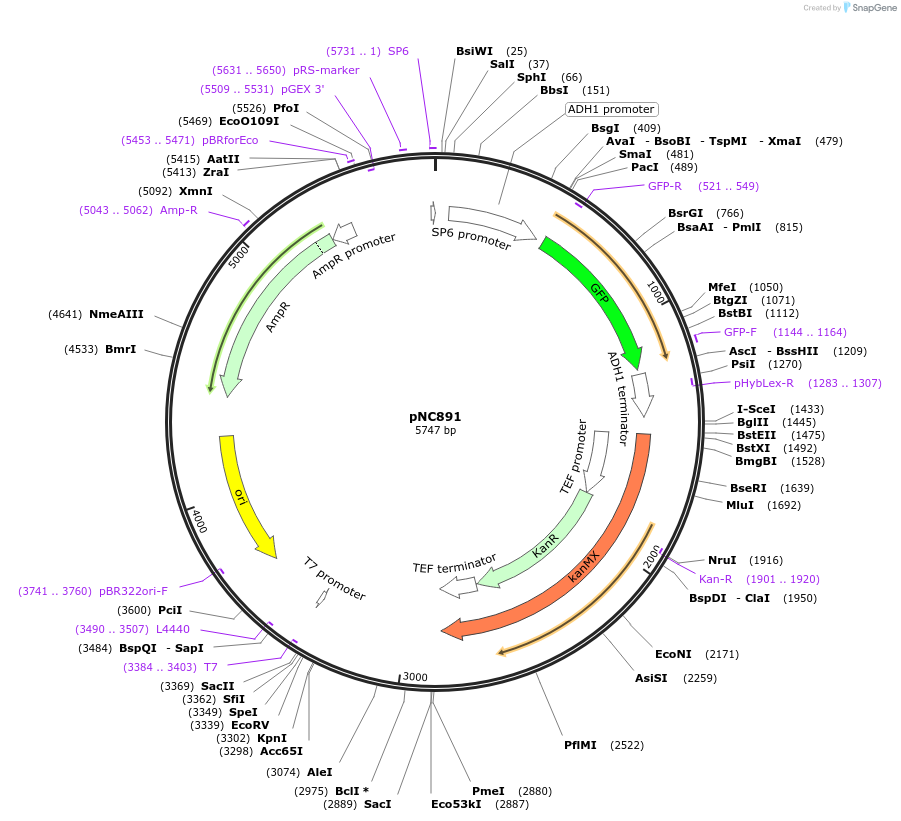
pNC891
Plasmid#20397DepositorInsertYFP-reporter
ExpressionYeastMutationX=M (see schematic)Available SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
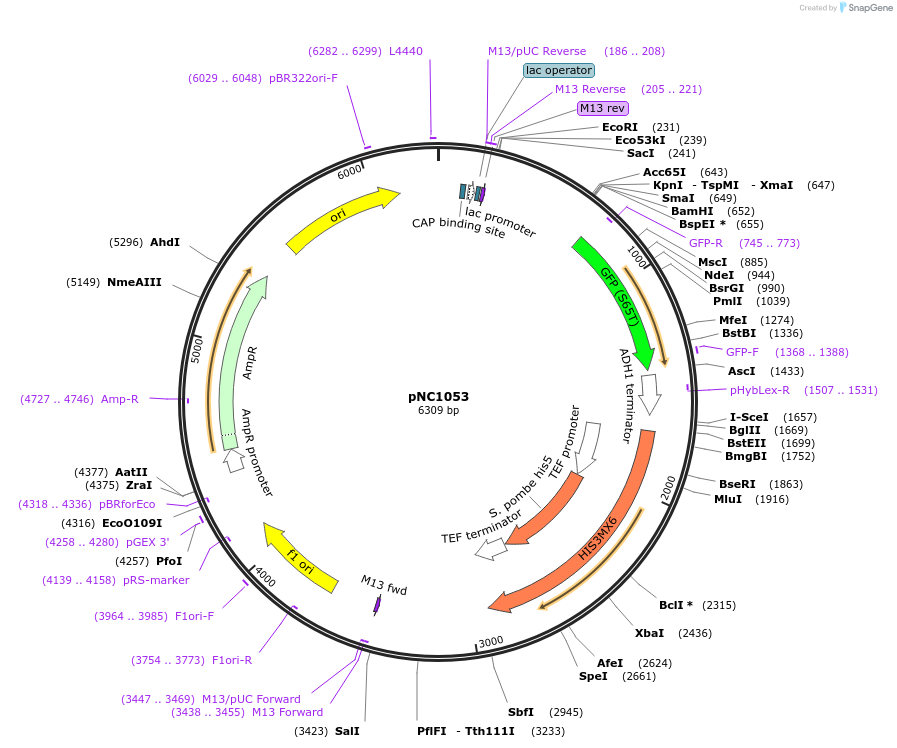
pNC1053
Plasmid#20406DepositorInsertCFP
ExpressionYeastMutationlinker: delta kAvailable SinceMarch 6, 2009AvailabilityAcademic Institutions and Nonprofits only