We narrowed to 10,278 results for: CAD;
-
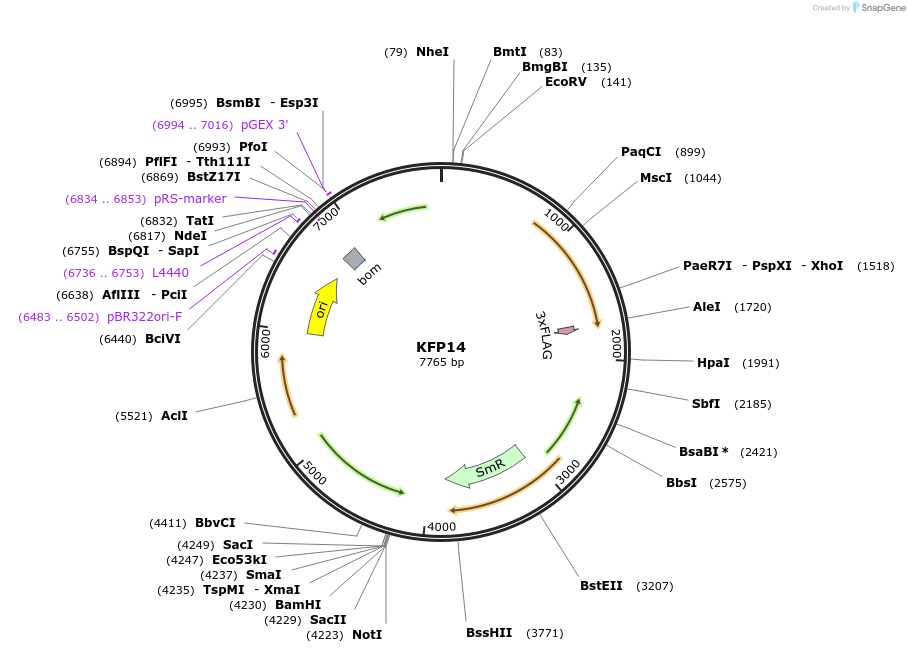
Plasmid#119796PurposeRpoD6 epitope tagged construct for integration at neutral site 1DepositorInsertSynpcc7942_1557 (Synpcc7942_1557 )
ExpressionBacterialAvailable SinceMarch 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
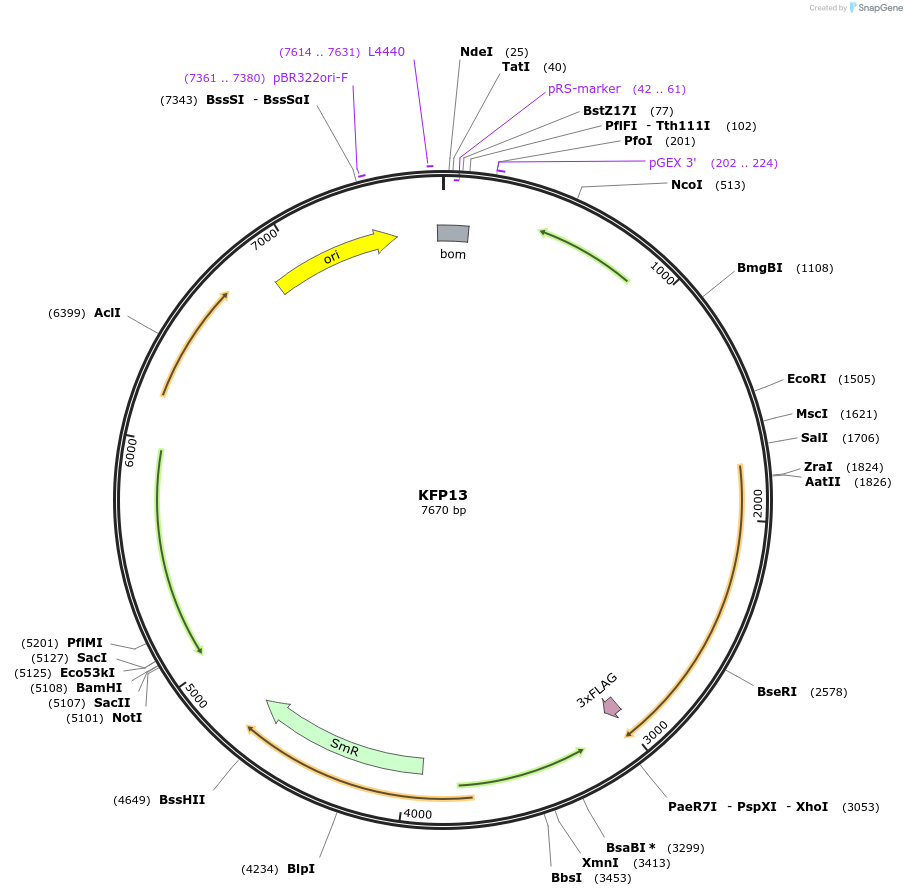
KFP13
Plasmid#119795PurposeRpoD5 epitope tagged construct for integration at neutral site 1DepositorInsertSynpcc7942_1849 (Synpcc7942_1849 )
ExpressionBacterialAvailable SinceJan. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
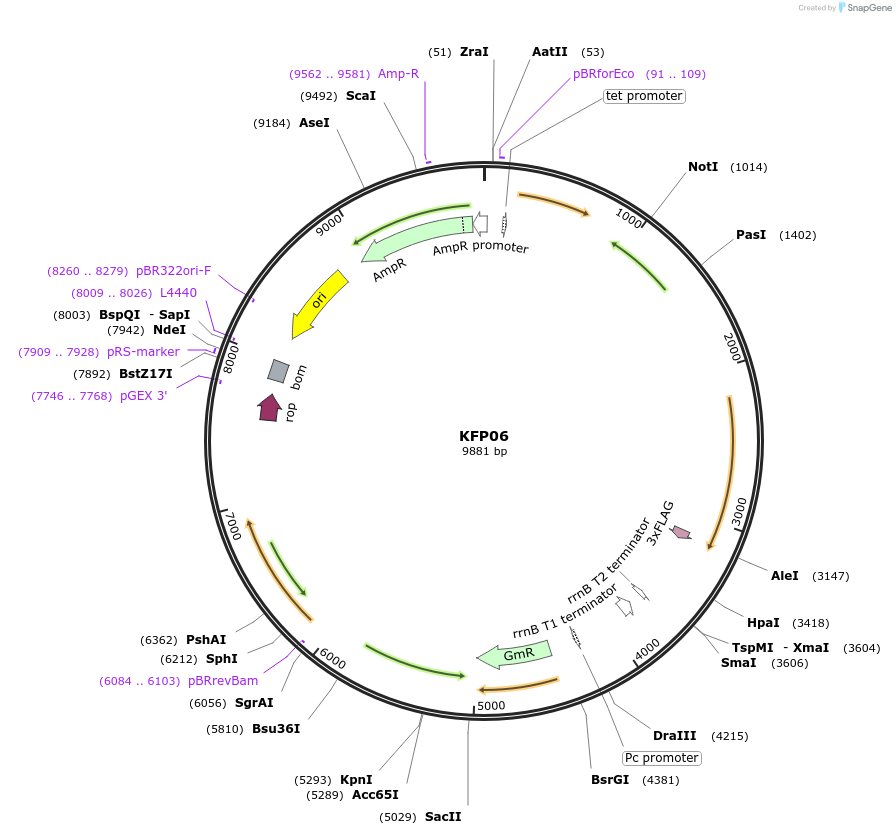
KFP06
Plasmid#119799PurposeRpoD6 epitope tagged construct for integration at neutral site 2.2DepositorInsertSynpcc7942_1557 (Synpcc7942_1557 )
ExpressionBacterialAvailable SinceDec. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
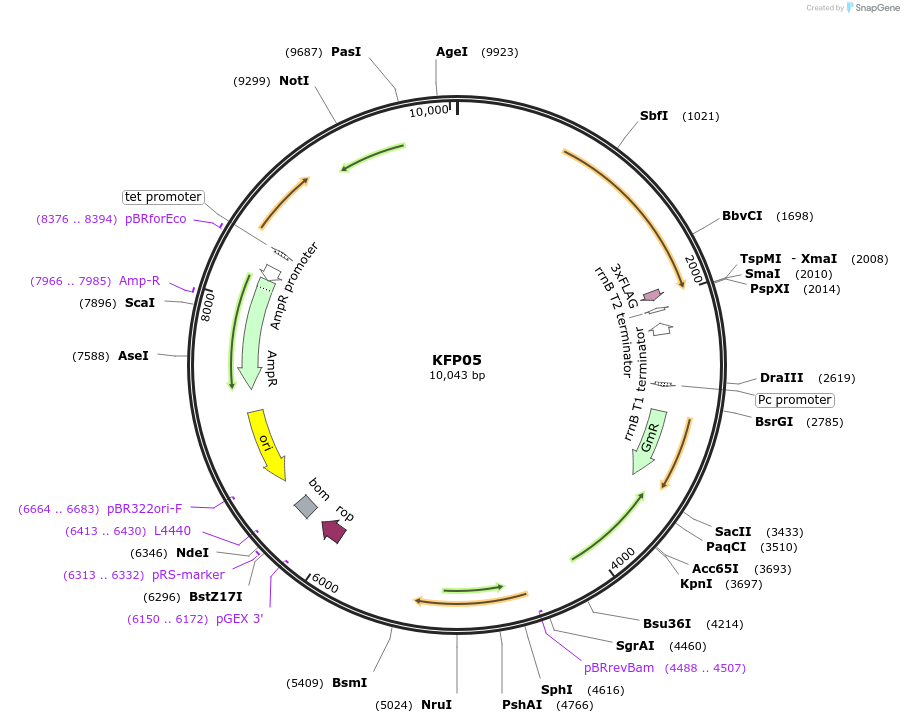
KFP05
Plasmid#119800PurposeRpoD5 epitope tagged construct for integration at neutral site 2.2DepositorInsertSynpcc7942_1849 (Synpcc7942_1849 )
ExpressionBacterialAvailable SinceDec. 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
pHL1801
Plasmid#53039PurposePLlacO-1:RBS(st7):gfpDepositorInsertGFP
UseSynthetic BiologyTagsRBS(st7)ExpressionBacterialPromoterpLlac0-1Available SinceAug. 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL1506
Plasmid#53026PurposePLlacO-1:RBS(st7):csrADepositorInsertcsrA
UseSynthetic BiologyTagsRBS(st7)ExpressionBacterialPromoterpLlac0-1Available SinceMay 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
KaiC CII cat-/EE (E318Q; S431E; T432E)
Plasmid#42498DepositorInsertKaiC CII cat-/EE (E318Q; S431E; T432E)
ExpressionBacterialMutationE318Q; S431E; T432EPromotert7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
KaiC CI cat- (E77Q;E78Q)
Plasmid#41886DepositorInsertKaiC CI cat -(E77Q;E78Q)
ExpressionBacterialMutationchanged glutamic acid 77 and 78 to glutaminePromotert7 promoterAvailable SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
KaiC CII cat- (E318Q)
Plasmid#41887DepositorInsertKai C CII cat (E318Q)
UseBl de3 21ExpressionBacterialMutationchanged amino acid 318 E to QPromotert7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
pHL1318
Plasmid#37637DepositorInsertsTetR
CsrA
UseSynthetic BiologyExpressionBacterialPromoterpConNoHindAvailable SinceJuly 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
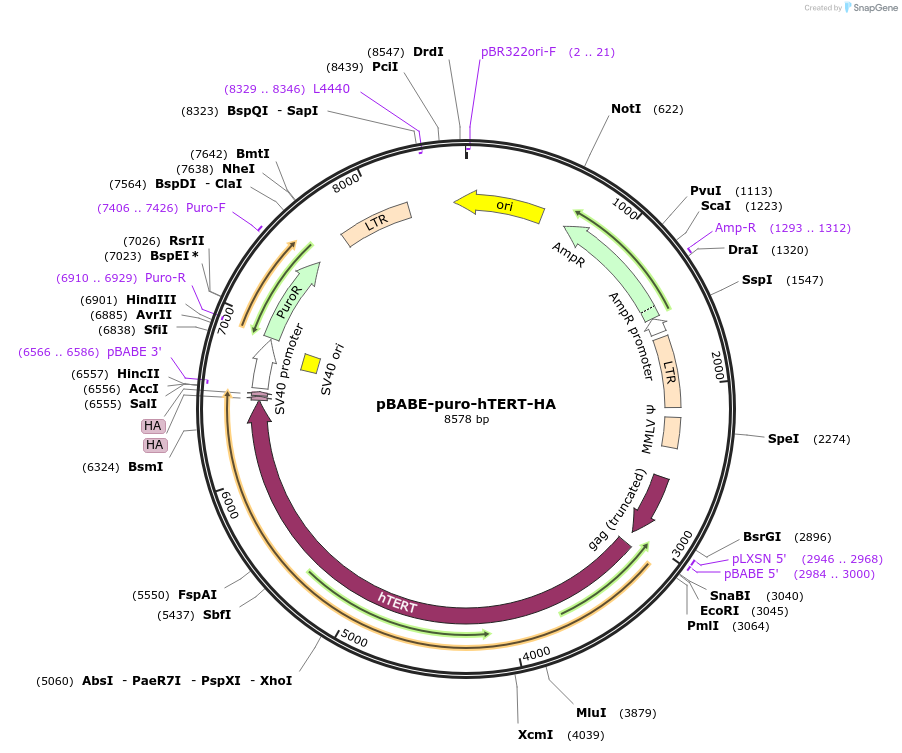
pBABE-puro-hTERT-HA
Plasmid#1772DepositorAvailable SinceJune 3, 2005AvailabilityAcademic Institutions and Nonprofits only -
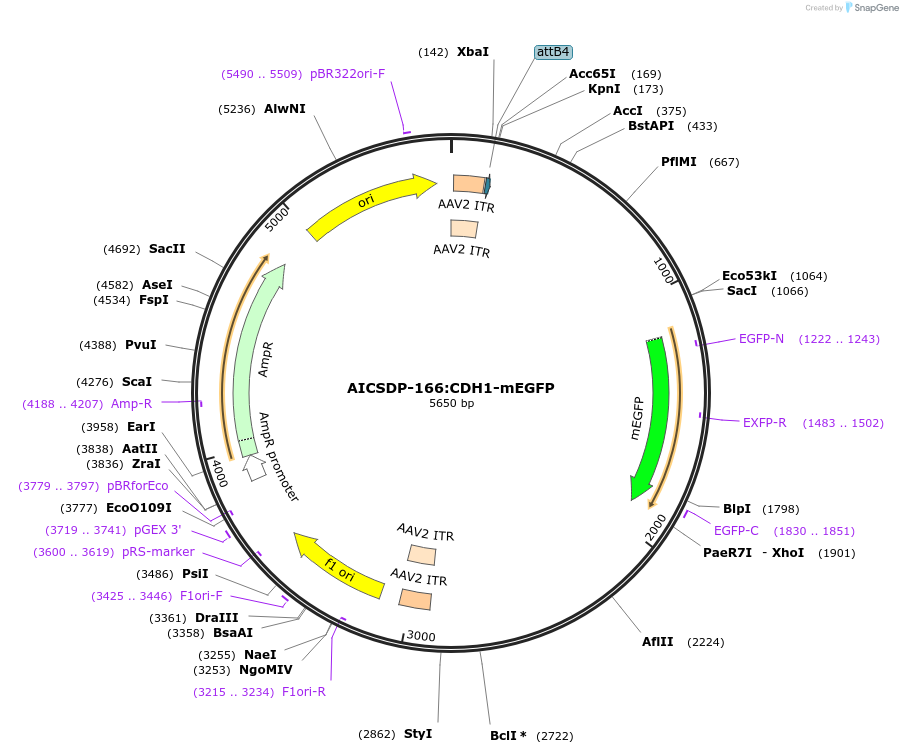
AICSDP-166:CDH1-mEGFP
Plasmid#193921PurposeHomology arms and LE-mEGFP sequence for C-terminus tagging of human cadherin 1DepositorInsertcadherin 1 Homology Arms with LE-mEGFP (CDH1 Human)
UseAAV and CRISPR; Donor templateTagslinker-mEGFPMutationhomology arms contain point mutations to disrupt …Available SinceDec. 22, 2022AvailabilityIndustry, Academic Institutions, and Nonprofits -
pGL6
Plasmid#48746PurposeExpression of luciferase driven by mPer2 promoter region (bases -1128 to +2129 with respect to transcription start site at +1)DepositorInsertmPer2 promoter/enhancer region (-1128 to +2129)
UseLuciferaseAvailable SinceDec. 19, 2013AvailabilityAcademic Institutions and Nonprofits only -
pHL1575
Plasmid#53028PurposePconNoHind:csrB, PLtetO-1:RBS(st3):csrDDepositorInsertscsrB
csrD
UseSynthetic BiologyTagsRBS (st3)ExpressionBacterialPromoterPconNoHind and pLtetO-1Available SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL1756
Plasmid#53036PurposePconNoHindM12:glgC::gfp, PconNoHind:RBS(st3):tetRDepositorInsertsGFP
tetR
UseSynthetic BiologyTagsRBS(st3) and glgC leaderExpressionBacterialPromoterpConNoHind and pConNoHindM12Available SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
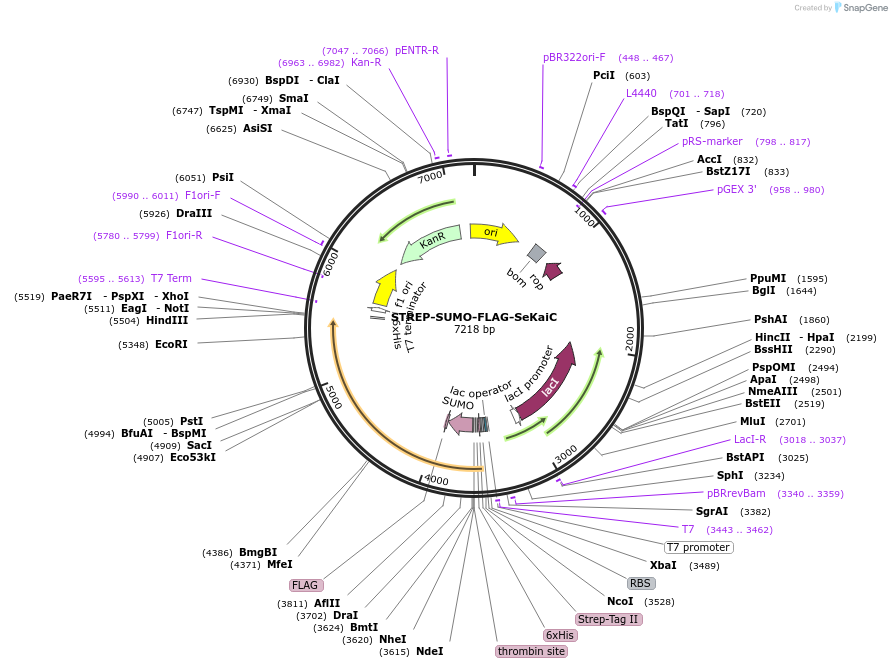
STREP-SUMO-FLAG-SeKaiC
Plasmid#218179PurposeWT SeKaiC for phosphorylation/dephosphorylation studyDepositorInsertSTREP-SUMO-FLAG-SeKaiC
TagsSTREP-TagExpressionBacterialAvailable SinceJune 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
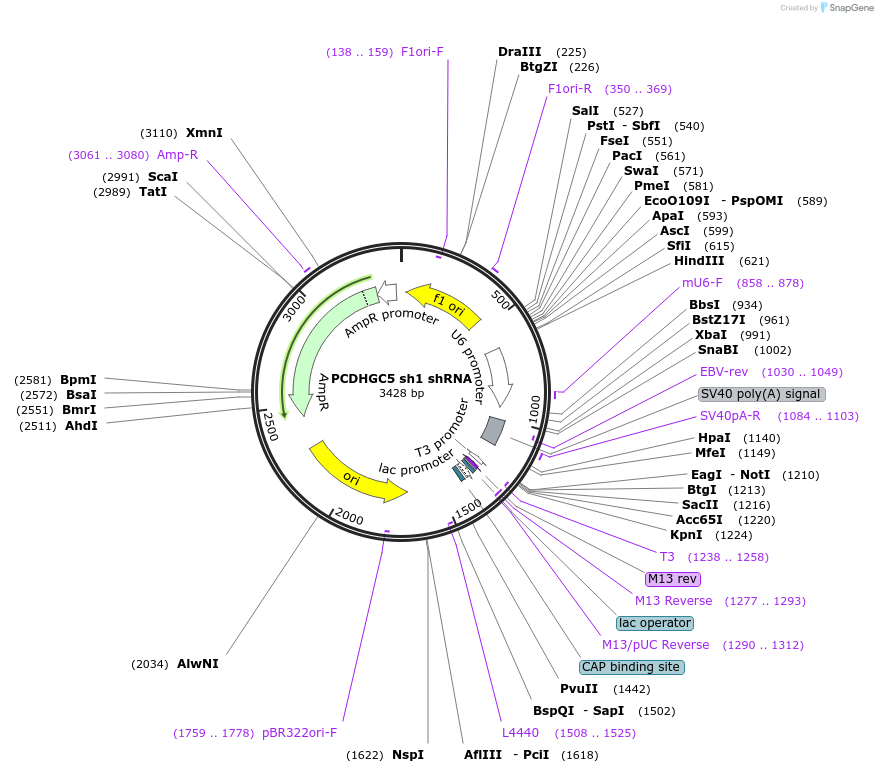
PCDHGC5 sh1 shRNA
Plasmid#122227PurposeKnock Down Protocadherin gamma C5. It targets Protocadherin gamma C5 mRNA (nucleotides 851–871 in the protein coding region).DepositorInsertPCDHGC5 shRNA (Pcdhgc5 Rat)
UseRNAiAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
pHL1757
Plasmid#53037PurposePconNoHindM2:glgC::gfp, PconNoHind:RBS(st3):tetRDepositorInsertsGFP
tetR
UseSynthetic BiologyTagsRBS(st3) and glgC leaderExpressionBacterialPromoterPconNoHindM2 and pConNoHindAvailable SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL1561
Plasmid#37571DepositorAvailable SinceAug. 14, 2012AvailabilityAcademic Institutions and Nonprofits only -
pHL1559
Plasmid#37570DepositorAvailable SinceAug. 14, 2012AvailabilityAcademic Institutions and Nonprofits only -
pHL1324
Plasmid#37638DepositorInsertsGFP
TetR
CsrA
UseSynthetic BiologyExpressionBacterialPromoterpCon and pConNoHindAvailable SinceJuly 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
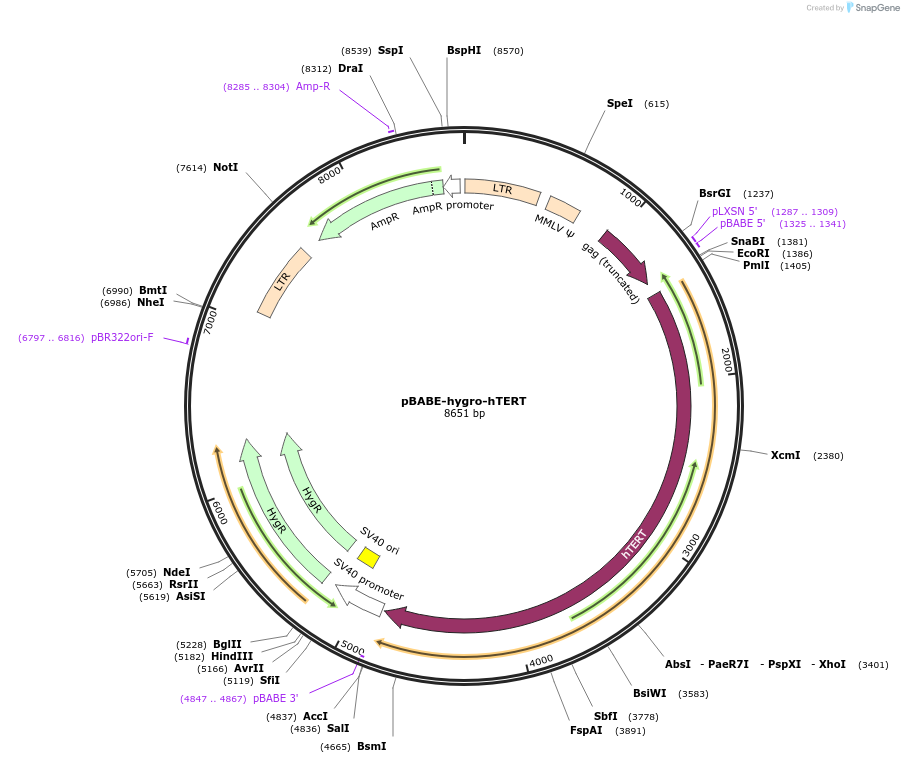
pBABE-hygro-hTERT
Plasmid#1773PurposeRetroviral expression of hTERT. Used to create immortalized cell lines.DepositorAvailable SinceJuly 18, 2006AvailabilityAcademic Institutions and Nonprofits only -
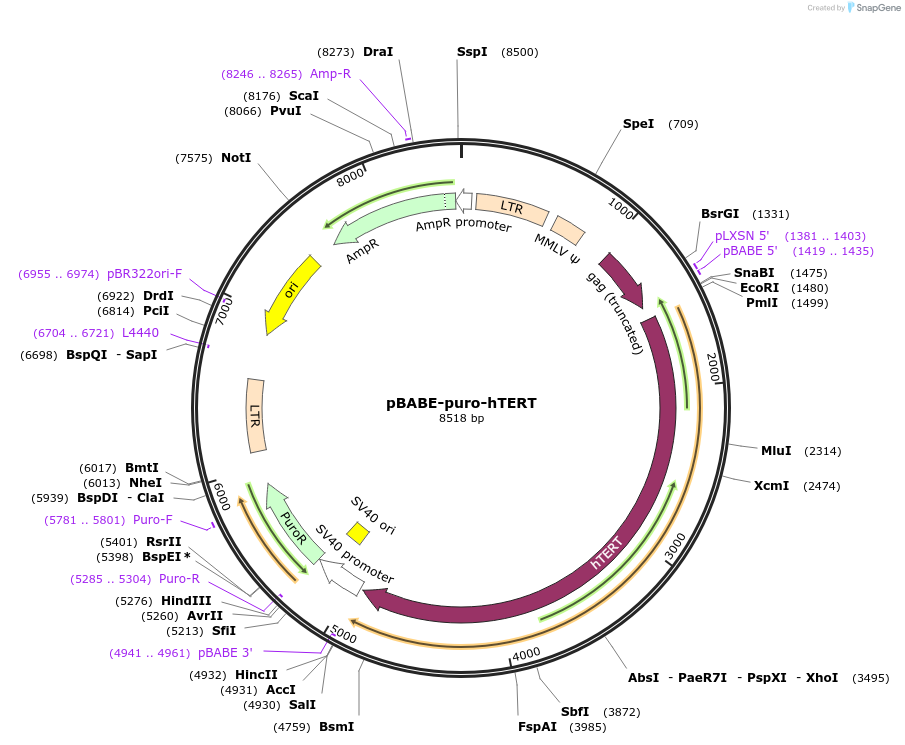
pBABE-puro-hTERT
Plasmid#1771PurposeRetroviral expression of hTERT. Used to create immortalized cell lines.DepositorAvailable SinceJune 3, 2005AvailabilityAcademic Institutions and Nonprofits only -
pGL4
Plasmid#48744PurposeExpression of luciferase driven by truncated mPer2 promoter region (bases -1128 to +1536 with respect to transcription start site at +1)DepositorInserttruncated mPer2 promoter/enhancer region (-1128 to +1536)
UseLuciferaseMutationtruncated promoter region (see pGL6)Available SinceJan. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
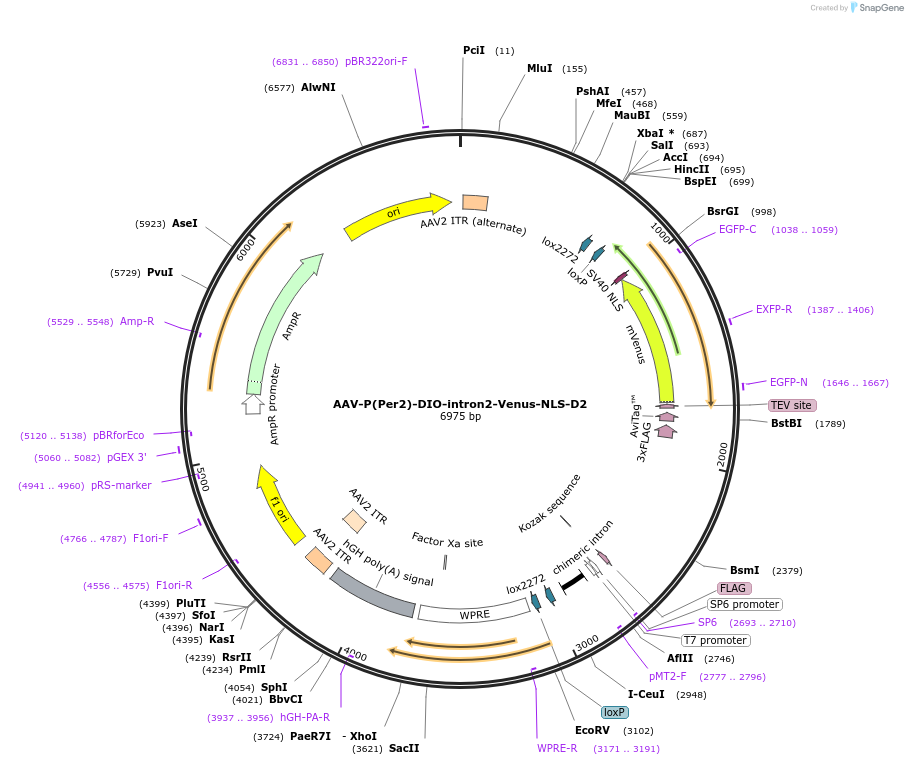
AAV-P(Per2)-DIO-intron2-Venus-NLS-D2
Plasmid#110057PurposePer2 transcription fluorescence reporterDepositorInsertPer2 promoter and Venus (Per2 Mouse)
UseAAV and Cre/LoxTagsNLS-D2ExpressionMammalianPromoterPer2Available SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
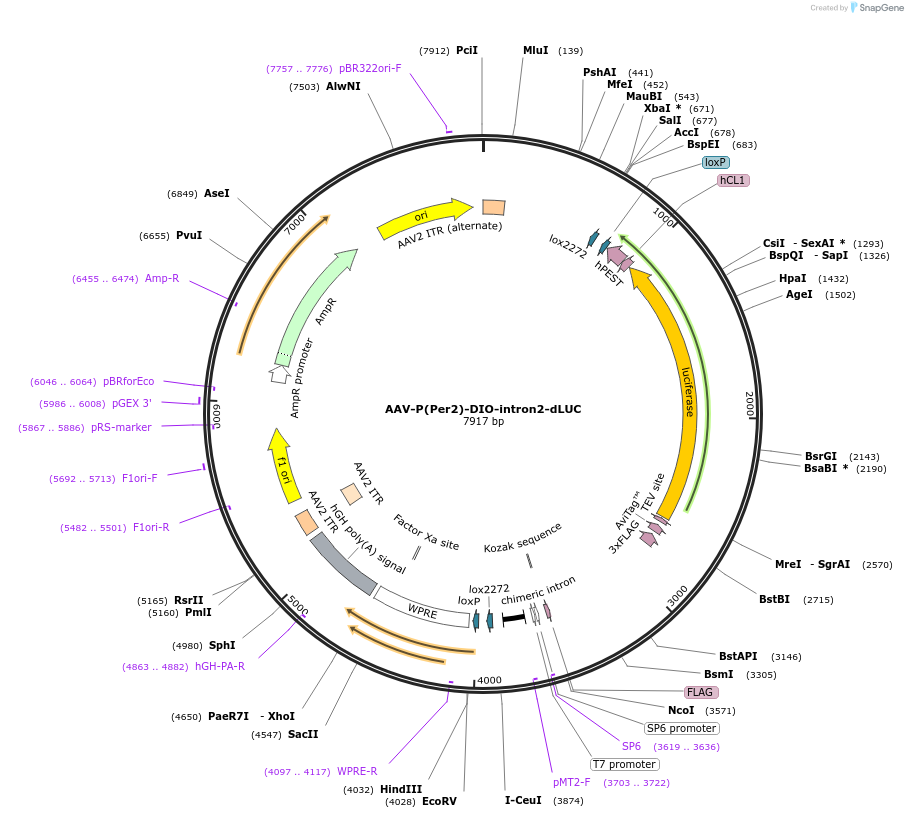
AAV-P(Per2)-DIO-intron2-dLUC
Plasmid#110059PurposePer2 transcription luciferase reporterDepositorInsertPer2 promoter and luciferase (Per2 Mouse)
UseAAV and Cre/LoxExpressionMammalianPromoterPer2Available SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
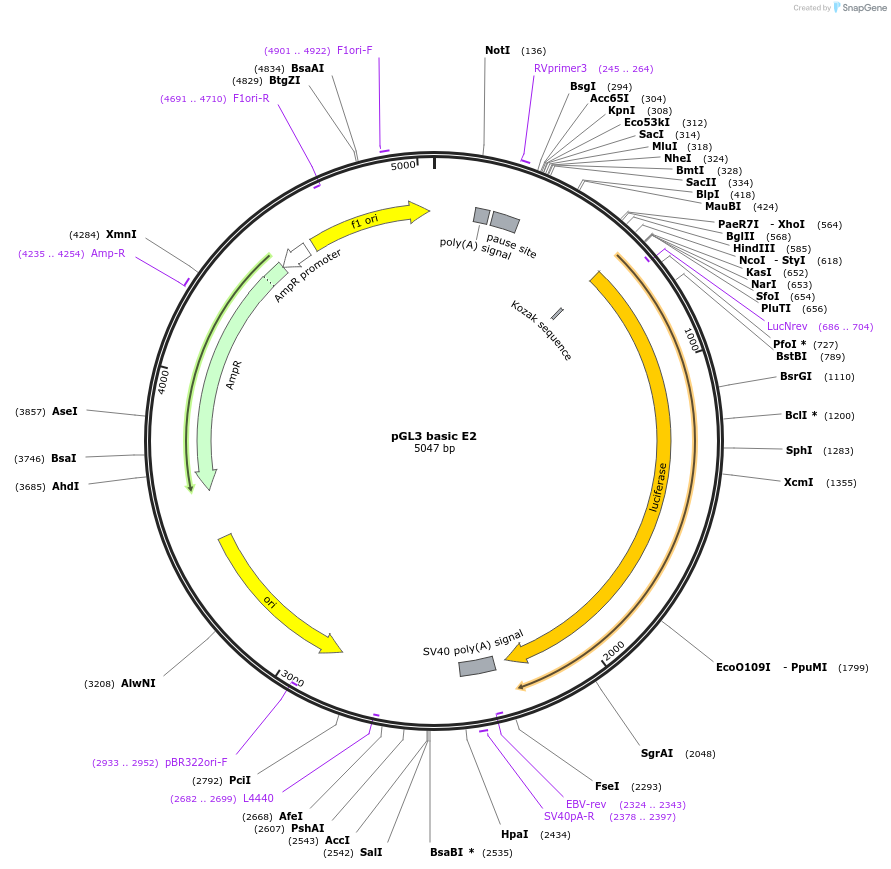
pGL3 basic E2
Plasmid#48747PurposeExpression of luciferase driven by mPer2 promoter fragment containing E-box2 (bases -112 to +98 with respect to transcription start site at +1)DepositorInsertmPer2 promoter/enhancer region containing E-box2 (-112 to +98)
UseLuciferaseAvailable SinceDec. 5, 2013AvailabilityAcademic Institutions and Nonprofits only -
pGL3
Plasmid#48743PurposeExpression of luciferase driven by truncated mPer2 promoter region (bases -1128 to +1060 with respect to transcription start site at +1)DepositorInserttruncated mPer2 promoter/enhancer region (-1128 to +1060)
UseLuciferaseMutationtruncated promoter region (see pGL6)Available SinceDec. 19, 2013AvailabilityAcademic Institutions and Nonprofits only -
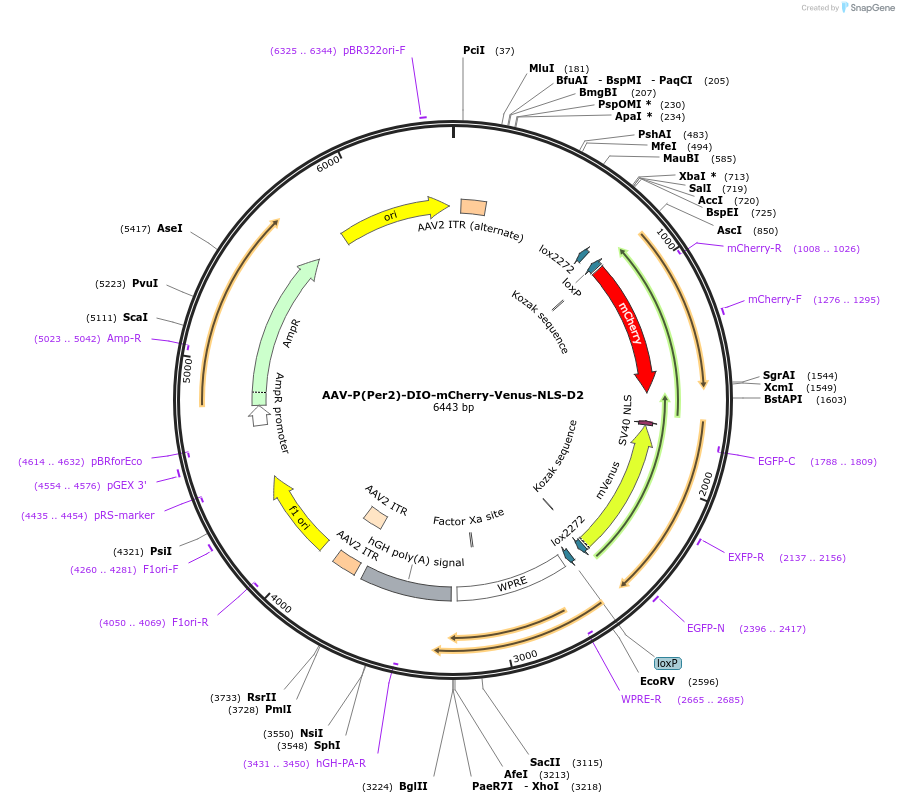
AAV-P(Per2)-DIO-mCherry-Venus-NLS-D2
Plasmid#110058PurposePer2 transcription fluorescence reporterDepositorInsertPer2 promoter and mCherry and Venus (Per2 Mouse)
UseAAV and Cre/LoxTagsNLS-D2ExpressionMammalianPromoterPer2Available SinceMay 30, 2018AvailabilityAcademic Institutions and Nonprofits only