We narrowed to 23,250 results for: ESC;
-
Pooled Library#207478PurposeHA-GD2-28z CAR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into thDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only
-
plIG-128_HA-GD2-28z_CAR_TFxTF_Library
Pooled Library#207480PurposeHA-GD2-28z CAR in combination with ~10,000 transcription factor combinations (and related proteins) and controls (GFP/RFP combinations). Library can be used to generate HDR templates for modular pooleDepositorUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-106_NY-ESO-1_TCR_SR_Library
Pooled Library#207474PurposeNY-ESO-1 TCR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T cellsDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-107_NY-ESO-1_TCR_TF_Library
Pooled Library#207475PurposeNY-ESO-1 TCR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into theDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-109_CD19-BBz_CAR_SR_Library
Pooled Library#207476PurposeCD19-BBz CAR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T cellsDepositorUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-110_CD19-BBz_CAR_TF_Library
Pooled Library#207477PurposeCD19-BBz CAR in combination with 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into theDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-124_HA-GD2-28z_CAR_SR_Library
Pooled Library#207479PurposeHA-GD2-28z CAR in combination with 129 surface receptors and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knockin (ModPoKI) into the TRAC locus of human T celDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
plIG-132_CD19-28z_CAR_TF_and_SR_Library
Pooled Library#207481PurposeCD19-28z CAR in combination with 129 surface receptors, 100 transcription factors (and related proteins) and 2 controls (GFP/RFP). Library can be used to generate HDR templates for modular pooled knocDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJan. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
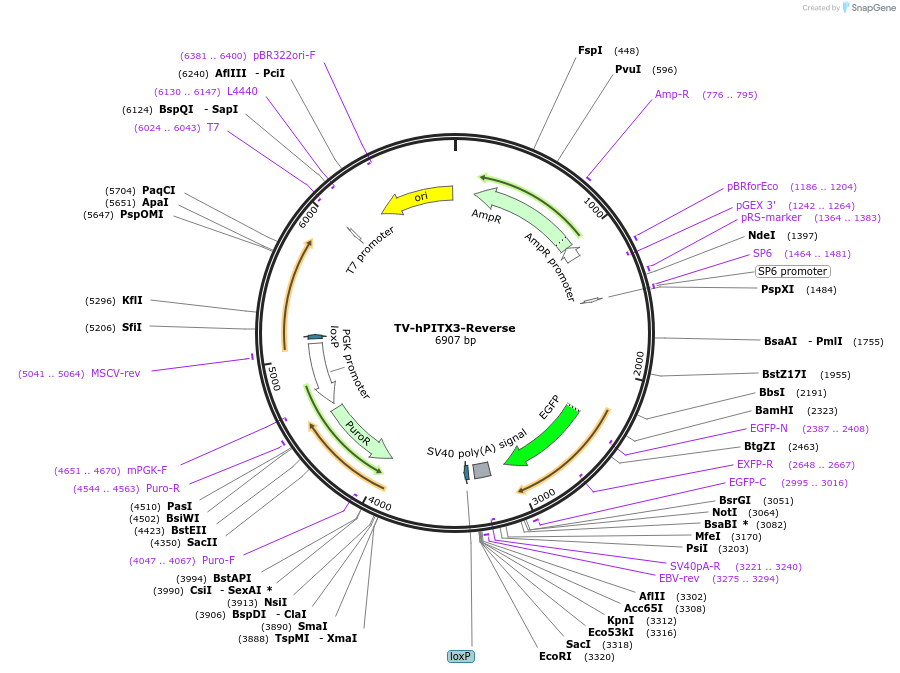
TV-hPITX3-Reverse
Plasmid#22208DepositorAvailable SinceOct. 5, 2009AvailabilityAcademic Institutions and Nonprofits only -
Jacquere (split-vector)
Pooled Library#247026PurposeHuman genome-wide CRISPR knockout library for the use of S. pyogenes Cas9. Each gene is targeted by 3 constructs.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and LentiviralAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
Jacquere (all-in-one vector)
Pooled Library#247027PurposeHuman genome-wide CRISPR knockout library for the use of S. pyogenes Cas9. Each gene is targeted by 3 constructs.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and LentiviralAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
Julianna (split-vector)
Pooled Library#247028PurposeEnriched for effective guides through prioritization of guides with strong on-target activity. Designed against current gene annotations, offering coverage of the contemporary mouse genome.DepositorExpressionMammalianSpeciesMus musculusUseCRISPR and LentiviralAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library C
Pooled Library#157973PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library B
Pooled Library#157972PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library A
Pooled Library#157971PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 1
Pooled Library#168776PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 2
Pooled Library#168777PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only