We narrowed to 798 results for: neb
-
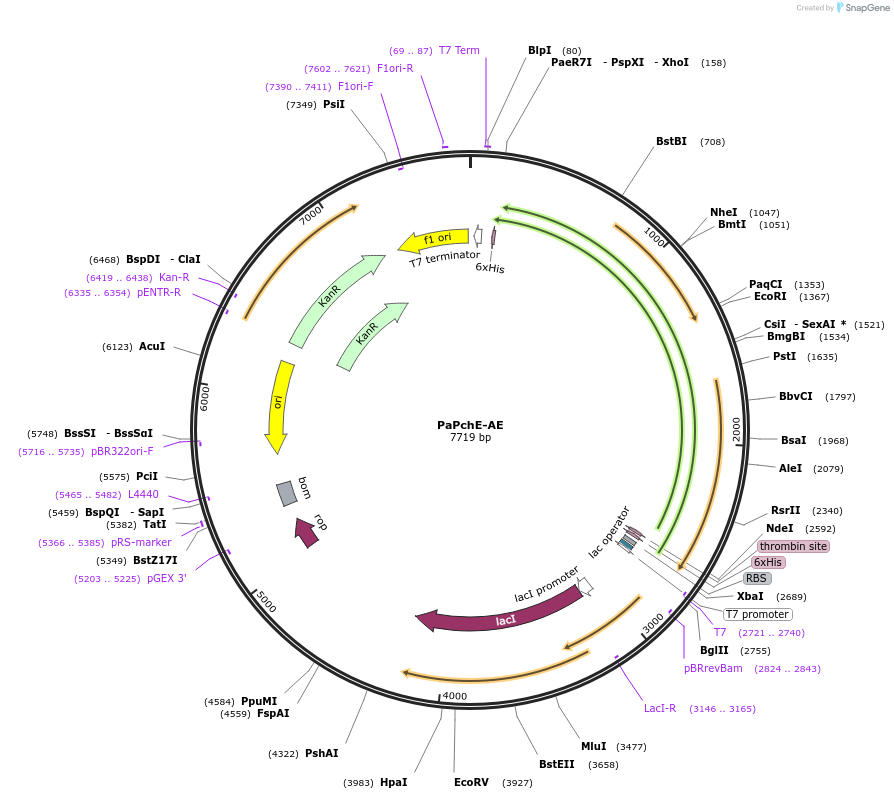
Plasmid#242103PurposeAdenylase-epimerase didomain of the nonribosomal peptide synthetase PchE from Pseudomonas aeruginosa for the production of pyochelinDepositorInsertPaPchE-AE
Tags6xHisExpressionBacterialAvailable SinceSept. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
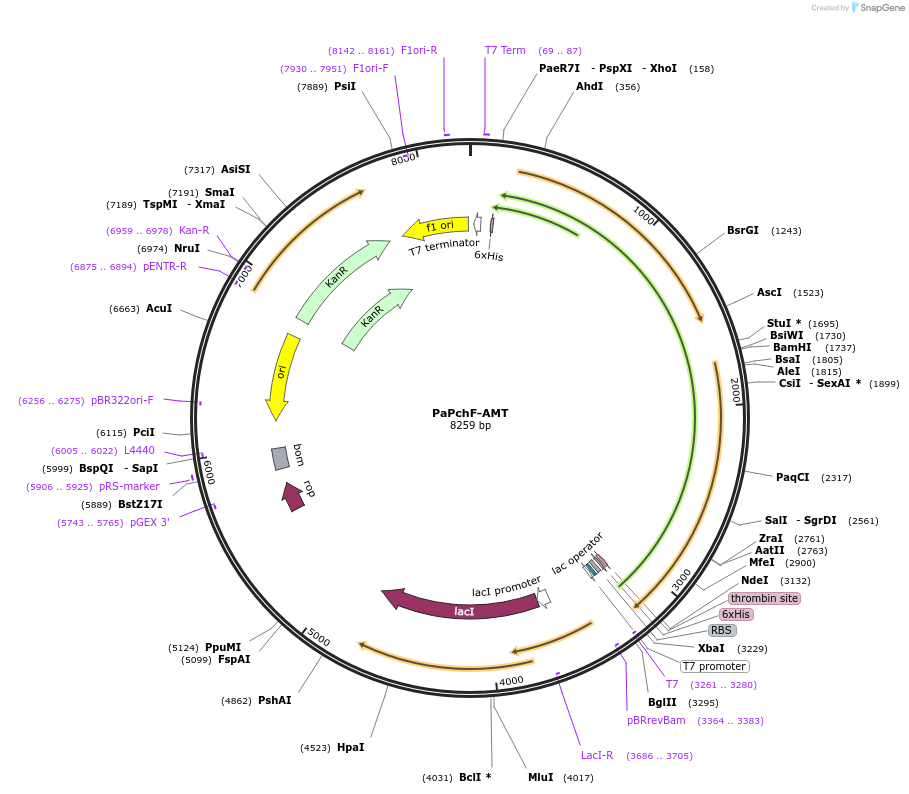
PaPchF-AMT
Plasmid#242104PurposeAdenylase-methyltransferase didomain of the nonribosomal peptide synthetase Pchf from Pseudomonas aeruginosa for the production of pyochelinDepositorInsertPaPchF-AMT
Tags6xHisExpressionBacterialAvailable SinceSept. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
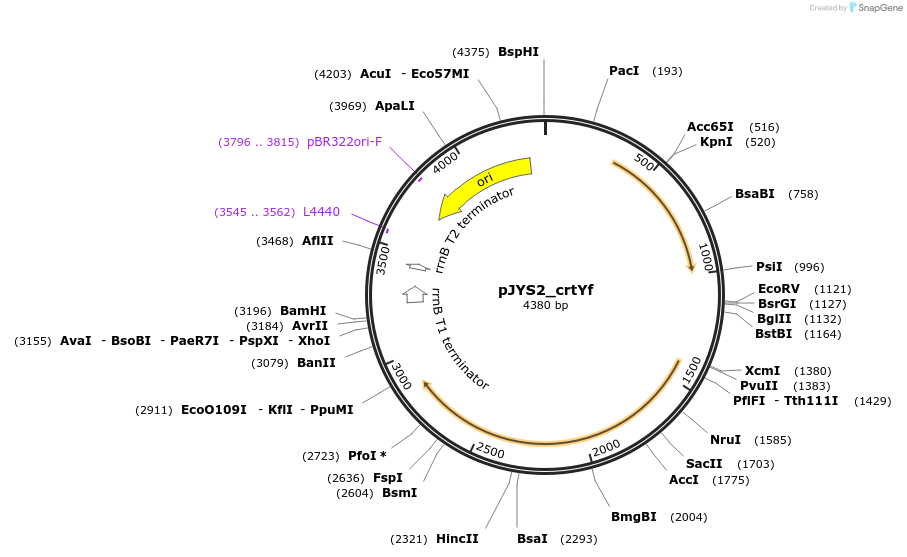
pJYS2_crtYf
Plasmid#85544PurposeConstitutive transcription of crRNA of crtYf, Spectinomycin resistanceDepositorInsertcrRNA of crtYf
UseCRISPR; Shuttle vector corynebacterium glutamicum…ExpressionBacterialAvailable SinceMay 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
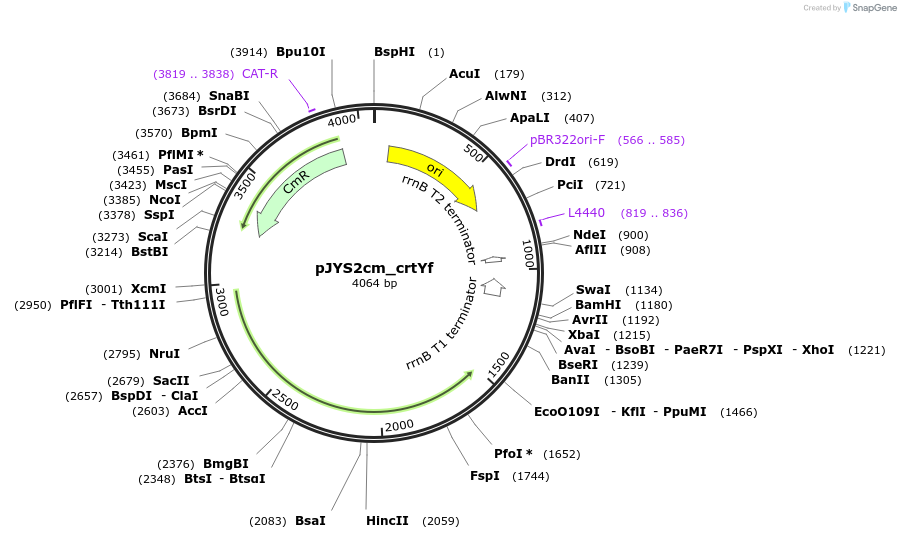
pJYS2cm_crtYf
Plasmid#85543PurposeConstitutive transcription of crRNA of crtYf, Chloramphenicol resistanceDepositorInsertcrRNA of crtYf
UseCRISPR; Shuttle vector corynebacterium glutamicum…ExpressionBacterialAvailable SinceMay 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
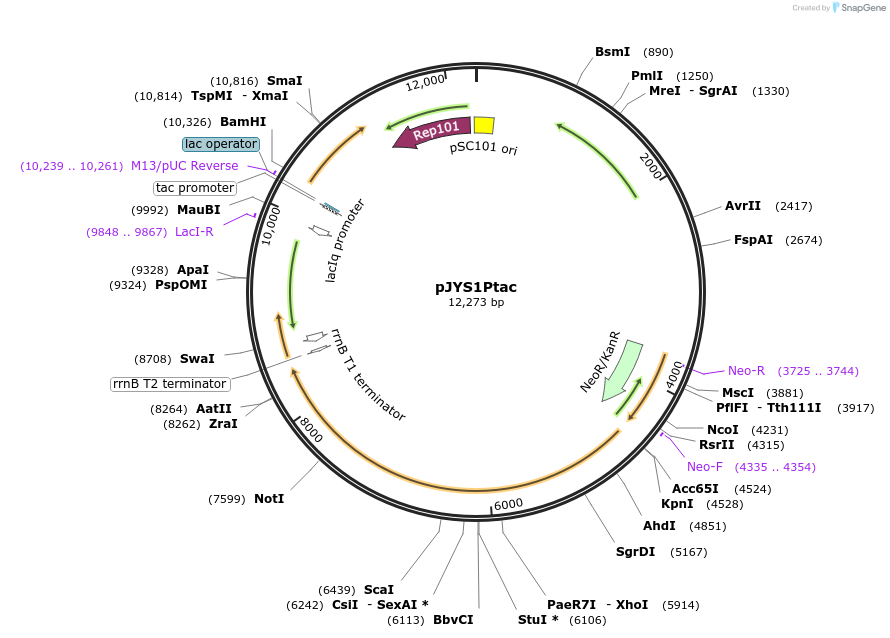
pJYS1Ptac
Plasmid#85545PurposeConstitutive trancription of FnCpf1 and recT in C.glutamitum, tac promoterDepositorInsertsCpf1
recT
UseCRISPR; Shuttle vector corynebacterium glutamicum…ExpressionBacterialPromotertac and unknownAvailable SinceMay 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
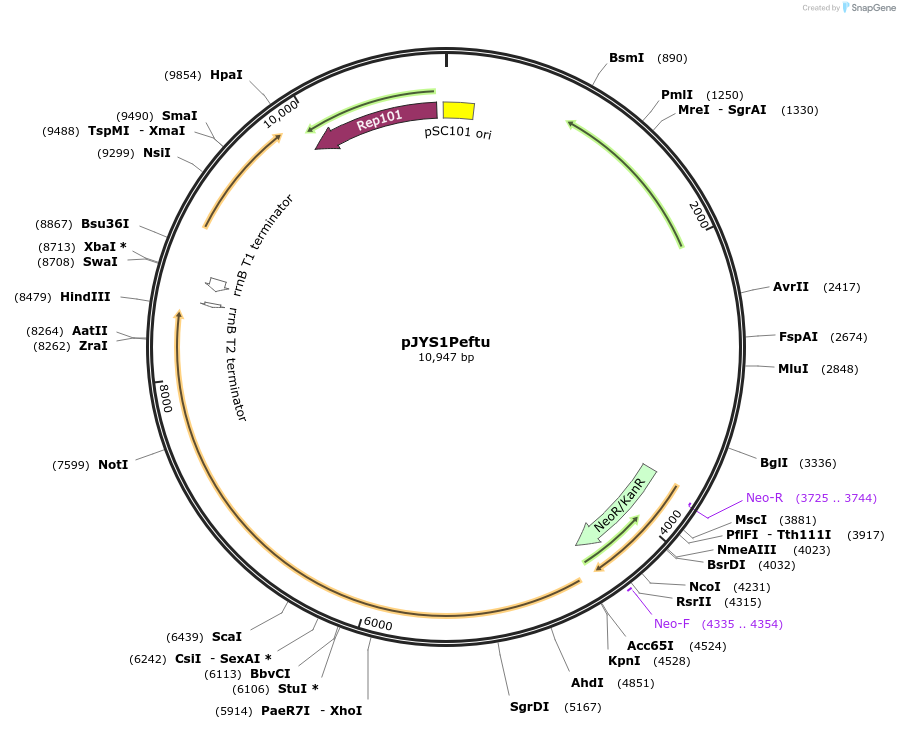
pJYS1Peftu
Plasmid#85546PurposeConstitutive expression of FnCpf1 and recT in C.glutamitum, etfu promoterDepositorInsertsCpf1
recT
UseCRISPR; Shuttle vector corynebacterium glutamicum…ExpressionBacterialPromoteretfu and unknownAvailable SinceMay 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
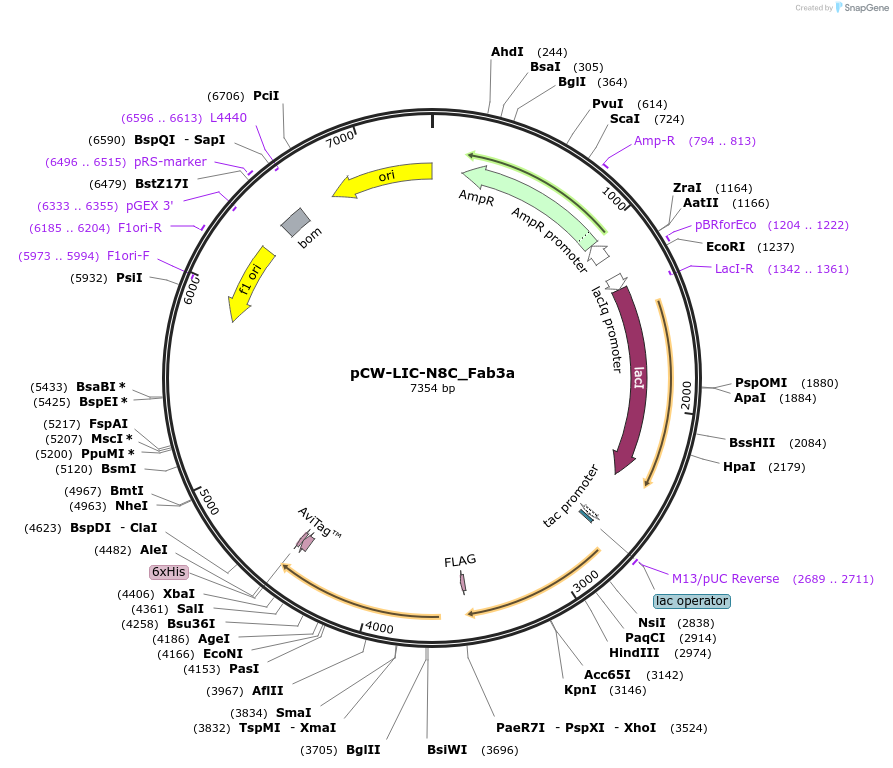
pCW-LIC-N8C_Fab3a
Plasmid#203689PurposeExpresses N8C_Fab3aDepositorInsertN8C_Fab3a
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
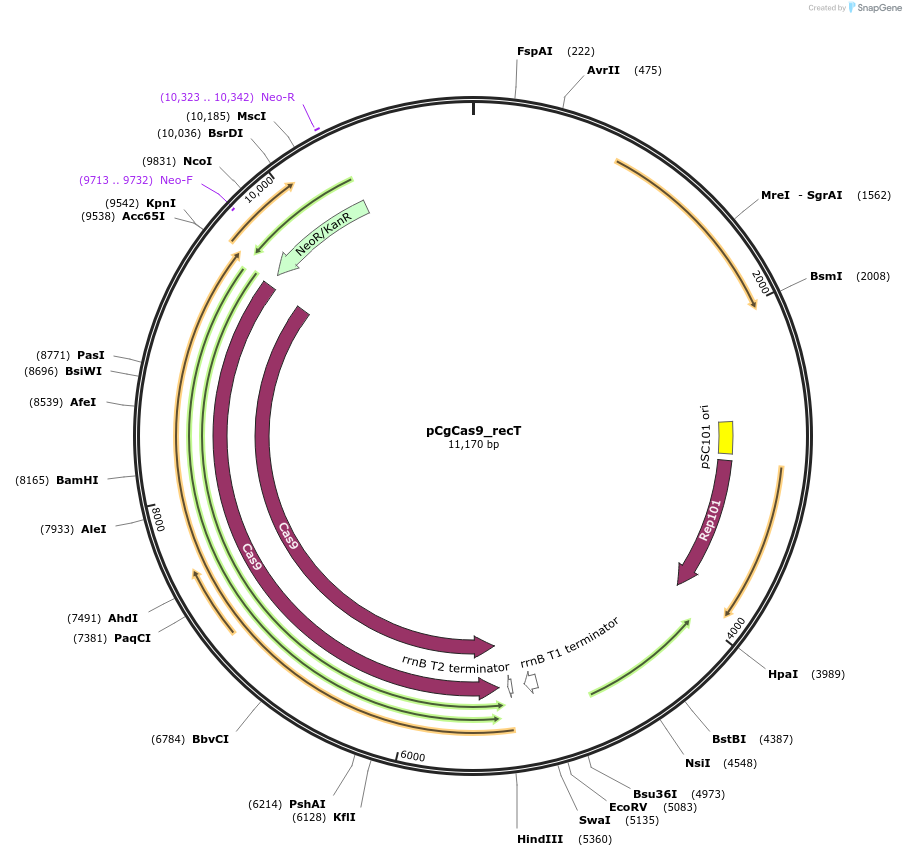
pCgCas9_recT
Plasmid#153337PurposeDouble-plasmid-based CRISPR-Cas9 system in Corynebacterium glutamicum; pBL1ts oriVC. glutamicum; pSC101 oriVE. coli PlacM-SpCas9 Streptomyces coelicolor codon-optimized, Peftu-RecT; KanrDepositorInsertCas9
UseCRISPRExpressionBacterialAvailable SinceJuly 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
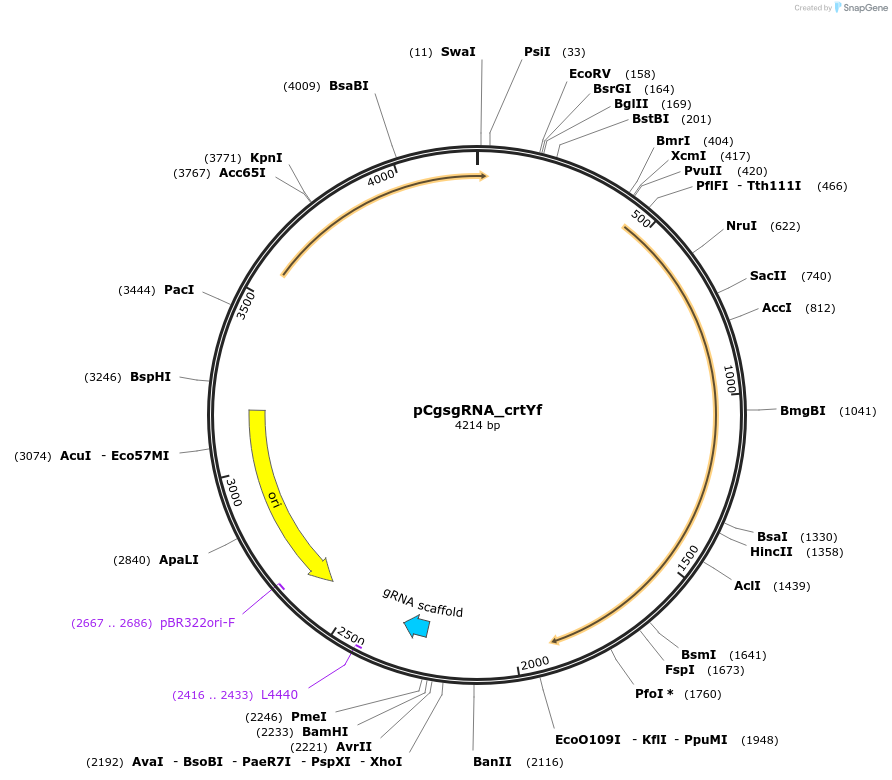
pCgsgRNA_crtYf
Plasmid#153338PurposeDouble-plasmid-based CRISPR-Cas9 system in Corynebacterium glutamicum; rep oriVC. glutamicum; pMB1 oriVE. Coli; Pj23119-crRNA targeting crtYf; SpcrDepositorInsertcrRNA targeting crtYf
UseCRISPRExpressionBacterialAvailable SinceJuly 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
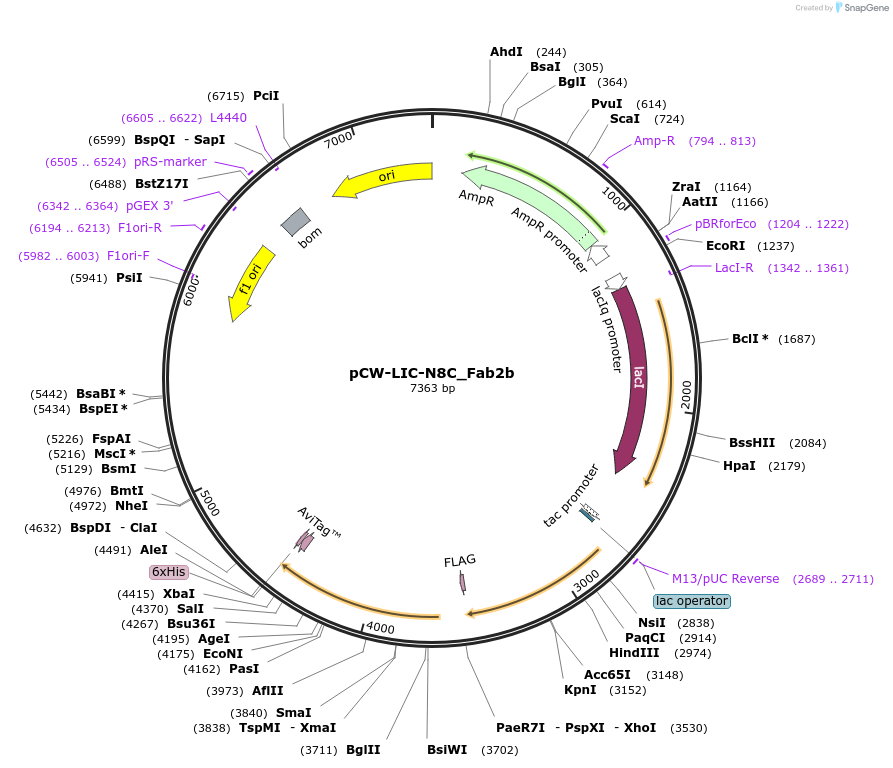
pCW-LIC-N8C_Fab2b
Plasmid#203692PurposeExpresses N8C_Fab2bDepositorInsertN8C_Fab2b
ExpressionBacterialAvailable SinceSept. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
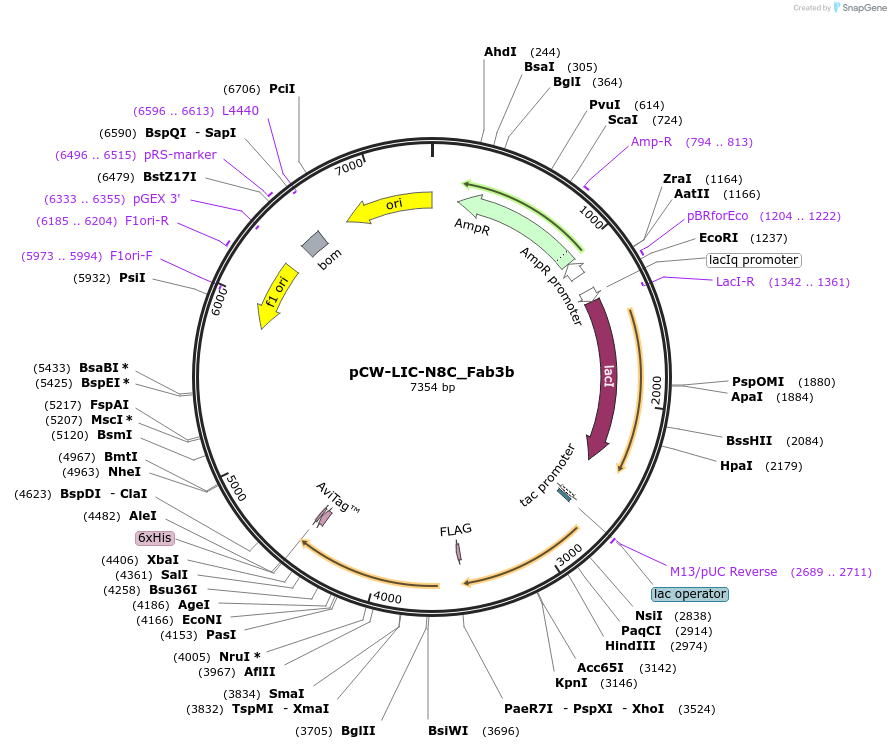
pCW-LIC-N8C_Fab3b
Plasmid#203685PurposeExpresses N8C_Fab3bDepositorInsertN8C_Fab3b
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
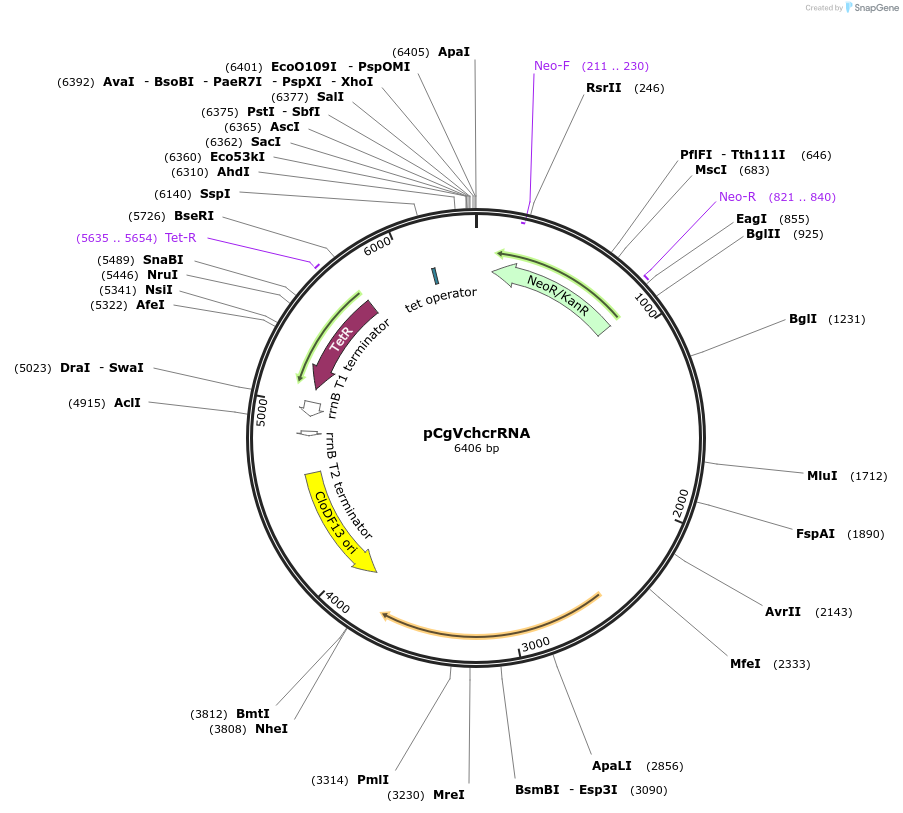
pCgVchcrRNA
Plasmid#203815PurposeExpression of CRISPR RNA, shuttle vector for Corynebacterium glutamicum / E. coliDepositorInsertCRISPR(BsaI)
UseCRISPRExpressionBacterialAvailable SinceAug. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
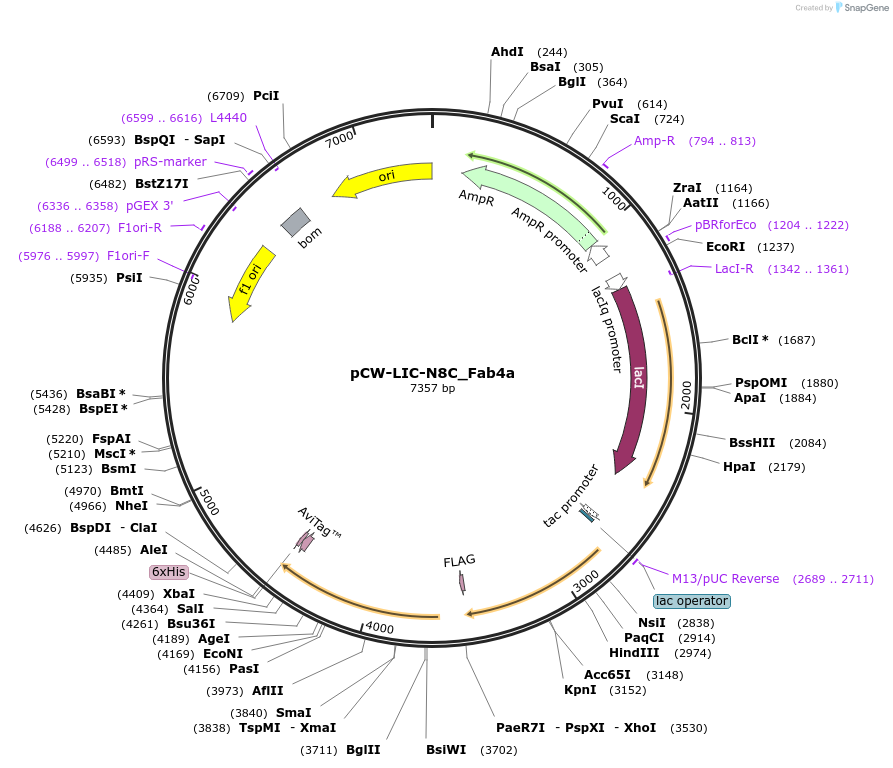
pCW-LIC-N8C_Fab4a
Plasmid#203690PurposeExpresses N8C_Fab4aDepositorInsertN8C_Fab4a
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
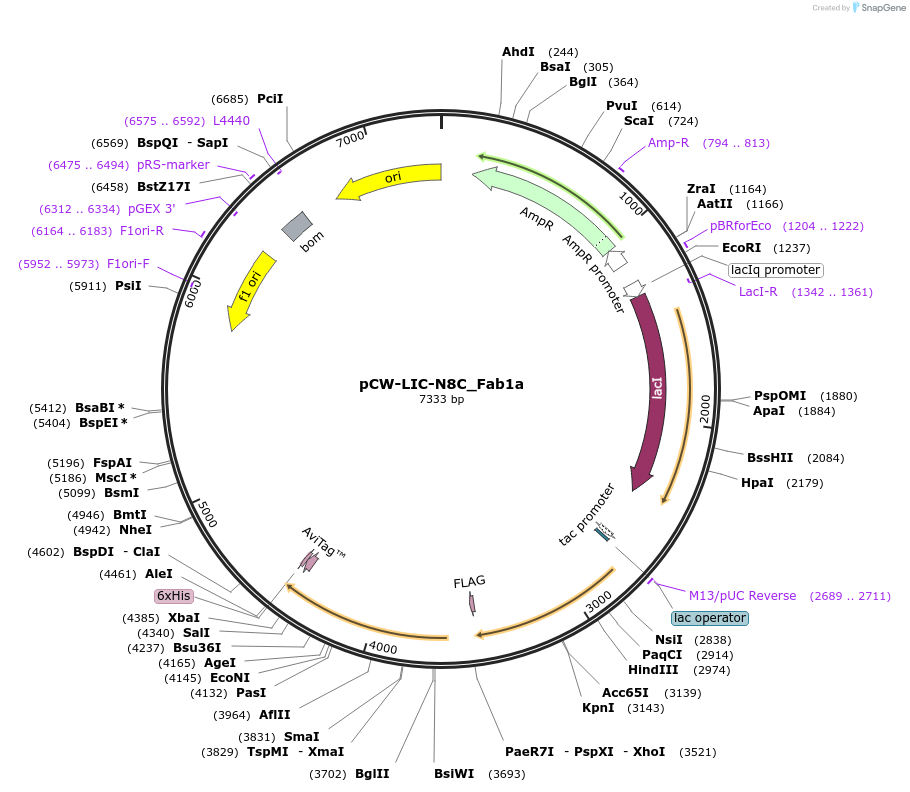
pCW-LIC-N8C_Fab1a
Plasmid#203687PurposeExpresses N8C_Fab1aDepositorInsertN8C_Fab1a
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
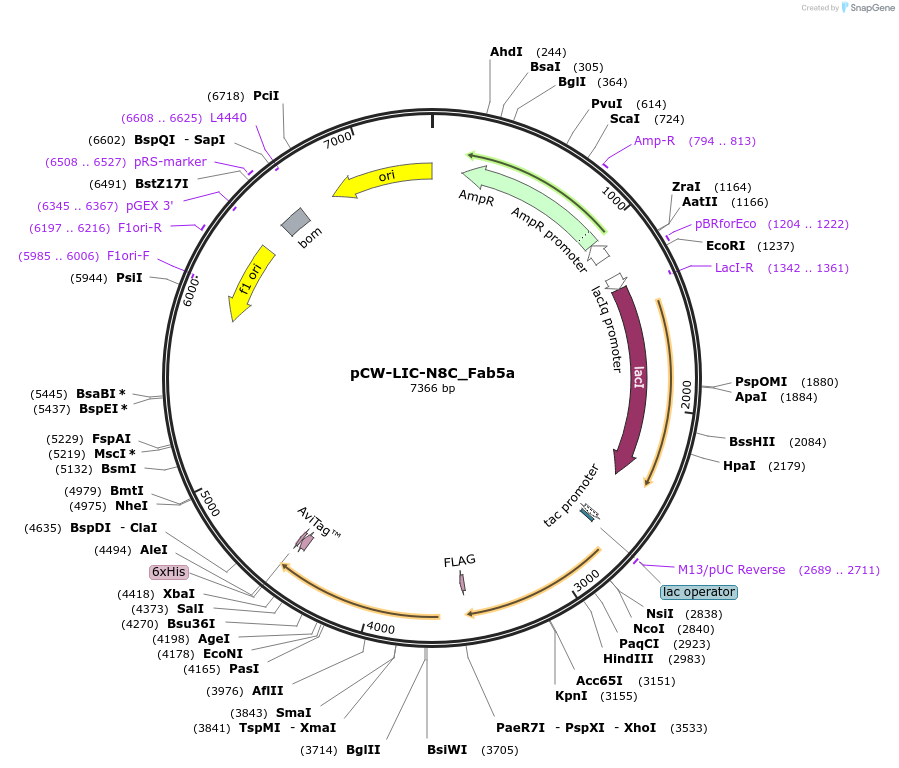
pCW-LIC-N8C_Fab5a
Plasmid#203693PurposeExpresses N8C_Fab5aDepositorInsertN8C_Fab5a
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
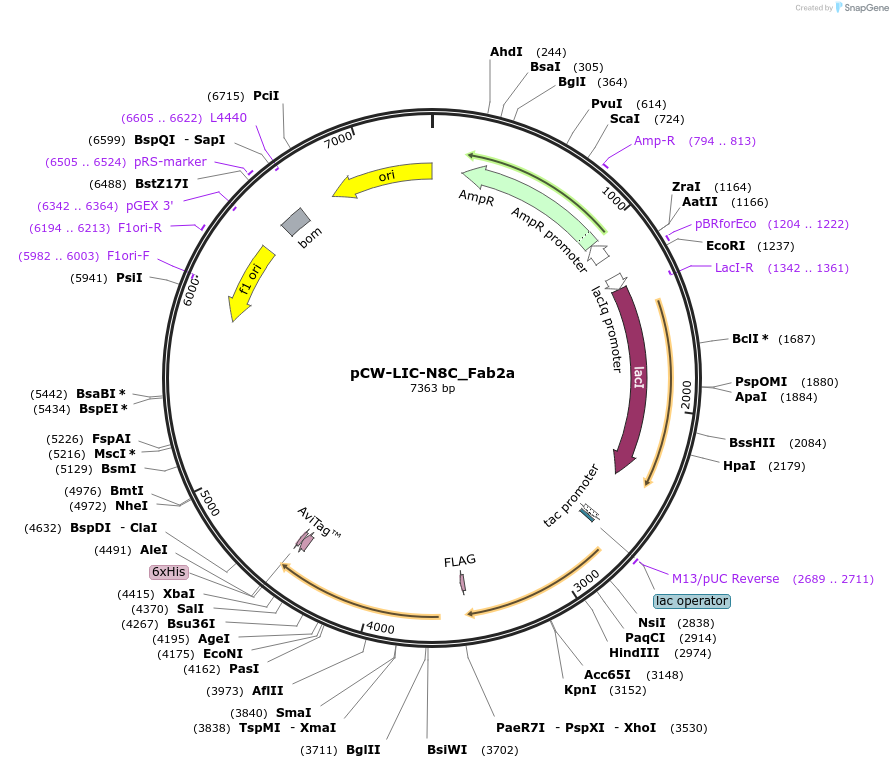
pCW-LIC-N8C_Fab2a
Plasmid#203688PurposeExpresses N8C_Fab2aDepositorInsertN8C_Fab2a
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
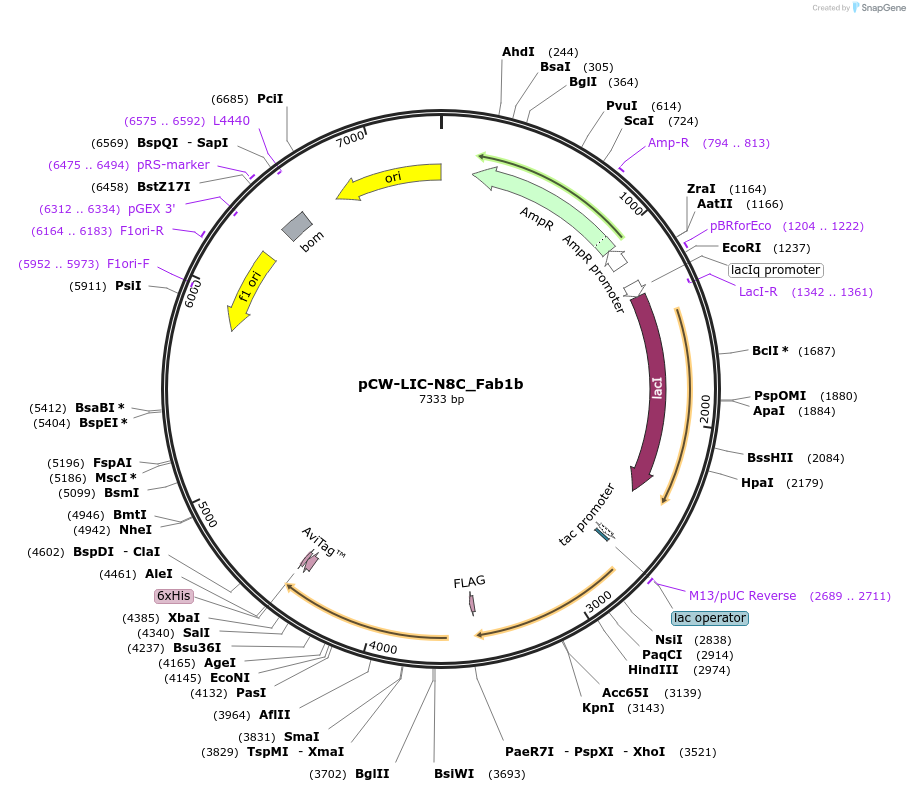
pCW-LIC-N8C_Fab1b
Plasmid#203691PurposeExpresses N8C_Fab1bDepositorInsertN8C_Fab1b
ExpressionBacterialAvailable SinceAug. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
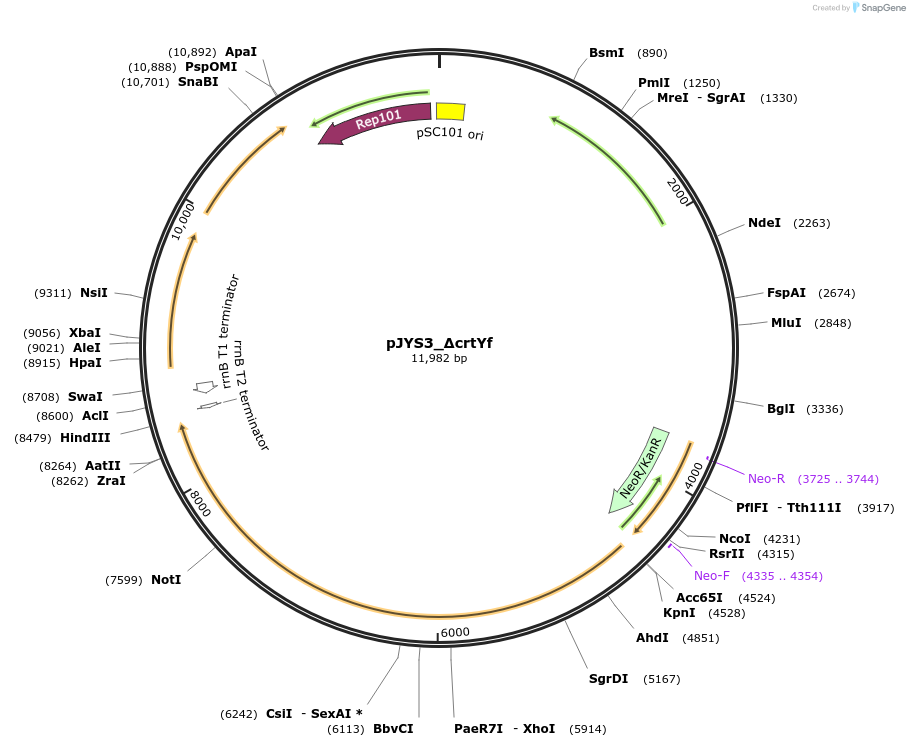
pJYS3_ΔcrtYf
Plasmid#85542PurposeConstitutive transcription of FnCpf1 and crRNA of crtYfDepositorInsertsCpf1
crRNA of crtYf
UseCRISPR; Shuttle vector corynebacterium glutamicum…ExpressionBacterialAvailable SinceMay 8, 2017AvailabilityAcademic Institutions and Nonprofits only -
pEGFP-WRAP53beta
Plasmid#64676Purposeexpresses Wild type WRAP53betaDepositorAvailable SinceMay 22, 2015AvailabilityAcademic Institutions and Nonprofits only -
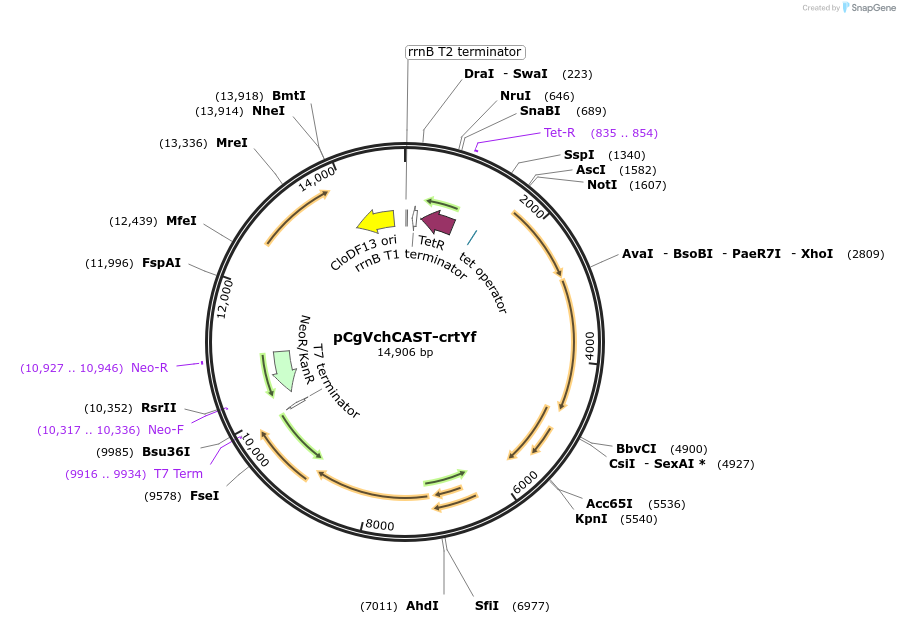
pCgVchCAST-crtYf
Plasmid#203816PurposeExpression of CRISPR-associated transposases, shuttle vector for Corynebacterium glutamicum / E. coliDepositorInsertVchTniQ, VchCas5/8, VchCas7, VchCas6, VchTnsABC, CRISPR(crtYf)
UseCRISPRExpressionBacterialAvailable SinceAug. 7, 2023AvailabilityAcademic Institutions and Nonprofits only