We narrowed to 459 results for: EB1
-
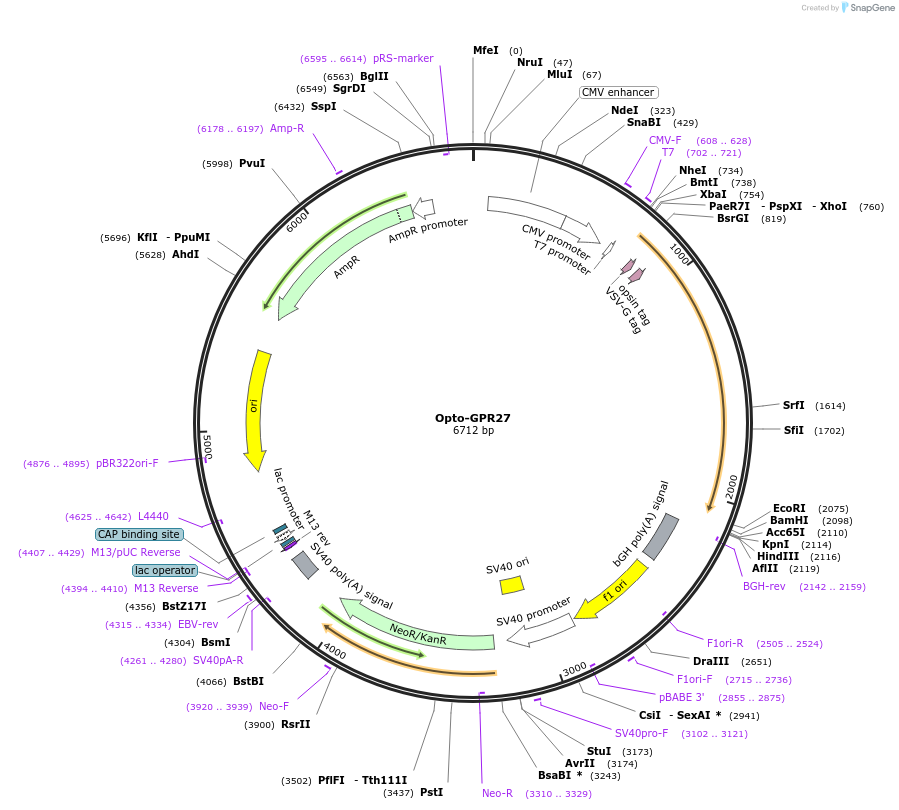
Plasmid#106013PurposeExpression of a chimeric G-protein coupled receptor (Rhodopsin-GPR27; Opto-GPR27; B3)DepositorInsertOpto-GPR27 (GPR27 Bovine, Human)
TagsRhodopsin-1D4 and VSV-GExpressionMammalianMutationChimeric receptor proteinPromoterCMVAvailable SinceJan. 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
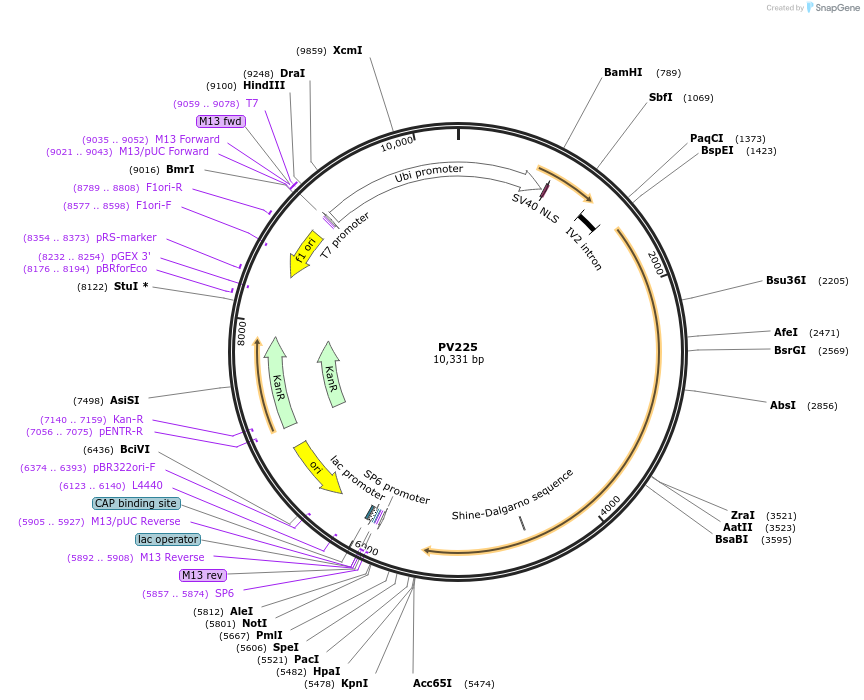
PV225
Plasmid#132334PurposedCas9 plant transcriptional activator (AT-CBF1) linked constitutive expression cassette for Zea maysDepositorInsertdCas9-AT-CBF1 (CBF1 Cas9 (Streptococcus pyogenes); AT-CBF1 (Arabidopsis thaliana))
TagsAT-CBF1ExpressionPlantMutationCodon optimize for expression and stability in Ze…Available SinceOct. 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
pChlr-8SE
Plasmid#52172PurposeEncodes module 8 with amino acids 12S-16E for recognition of nt 1GDepositorInsertPumilio homology domain amino acids 284-319 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in module 8Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SYR
Plasmid#52158PurposeEncodes module 4 with amino acids 12S-13Y-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S, 13H to 13Y, and 16Q to 16R in…Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SR
Plasmid#52162PurposeEncodes module 5 with amino acids 12S-16R for recognition of nt 4CDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16R in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SE
Plasmid#52164PurposeEncodes module 6 with amino acids 12S-16E for recognition of nt 3GDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SE
Plasmid#52152PurposeEncodes module 4 with amino acids 12S-16E for recognition of nt 5GDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 4Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-8SR
Plasmid#52174PurposeEncodes module 8 with amino acids 12S-16R for recognition of nt 1CDepositorInsertPumilio homology domain amino acids 284-319 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 8Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7CQ
Plasmid#52167PurposeEncodes module 7 with amino acids 12C-16Q for recognition of nt 2ADepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12C and 16E to 16Q in Module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7NQ
Plasmid#52169PurposeEncodes module 7 with amino acids 12N-16Q for recognition of nt 2UDepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12N and 16E to 16Q in module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7SYR
Plasmid#52170PurposeEncodes module 7 with amino acids 12S-13Y-16R for recognition of nt 2CDepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 13N to 13Y and 16E to 16R in module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SR
Plasmid#52154PurposeEncodes module 4 with amino acids 12S-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4CYQ
Plasmid#52155PurposeEncodes module 4 with amino acids 12C-13Y-16Q for recognition of nt 5ADepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C and 13H to 13Y in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SE
Plasmid#52160PurposeEncodes module 5 with amino acids 12S-16E for recognition of nt 4GDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16E in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SR
Plasmid#52166PurposeEncodes module 6 with amino acids 12S-16R for recognition of nt 3CDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
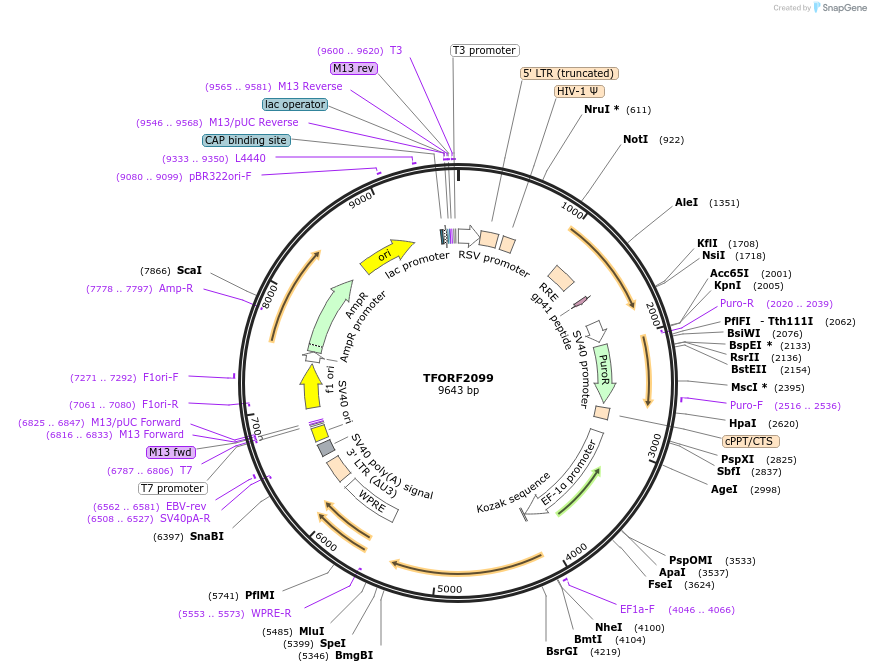
TFORF2099
Plasmid#142183PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
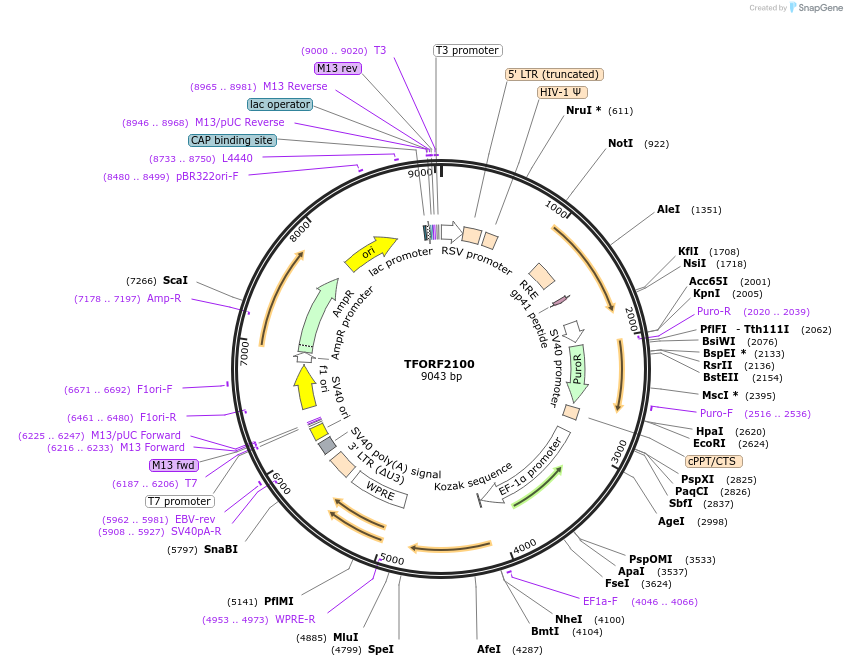
TFORF2100
Plasmid#141922PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
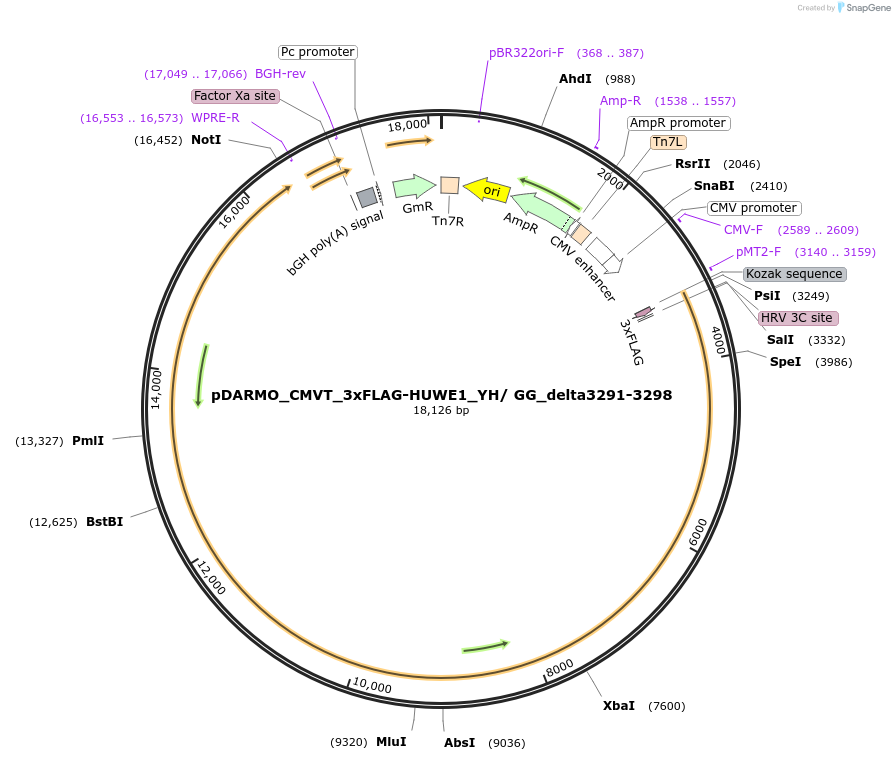
pDARMO_CMVT_3xFLAG-HUWE1_YH/ GG_delta3291-3298
Plasmid#187127Purposemammalian cell expression of FLAG-HUWE1 Y355G/H356G, delta3291-3298DepositorAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
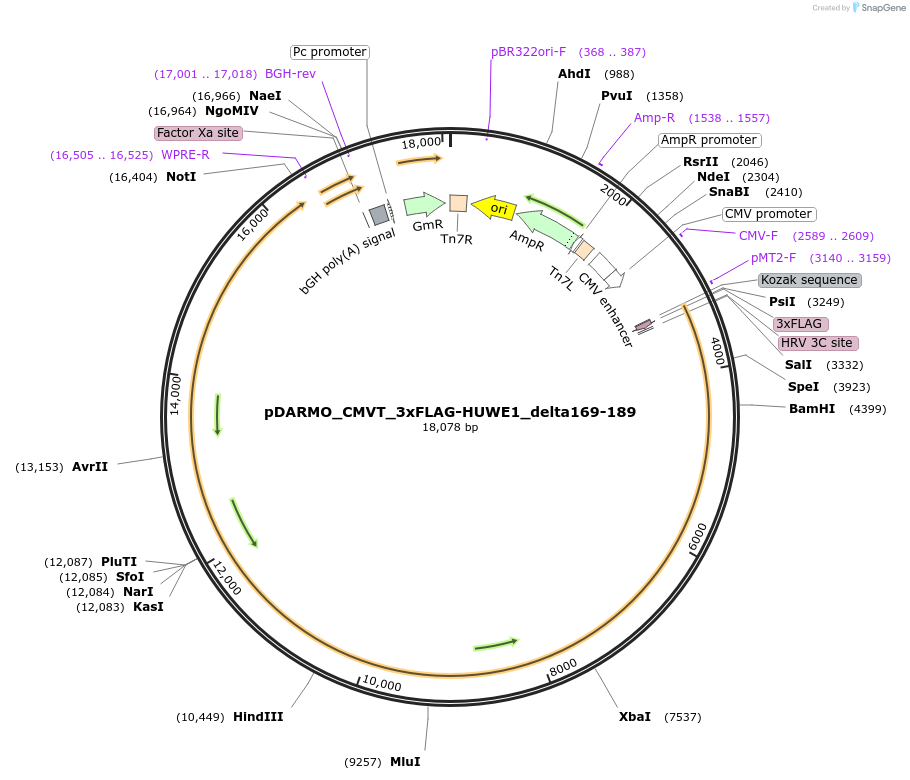
pDARMO_CMVT_3xFLAG-HUWE1_delta169-189
Plasmid#187135Purposemammalian cell expression of 3xFLAG-HUWE1 delta169-189DepositorAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
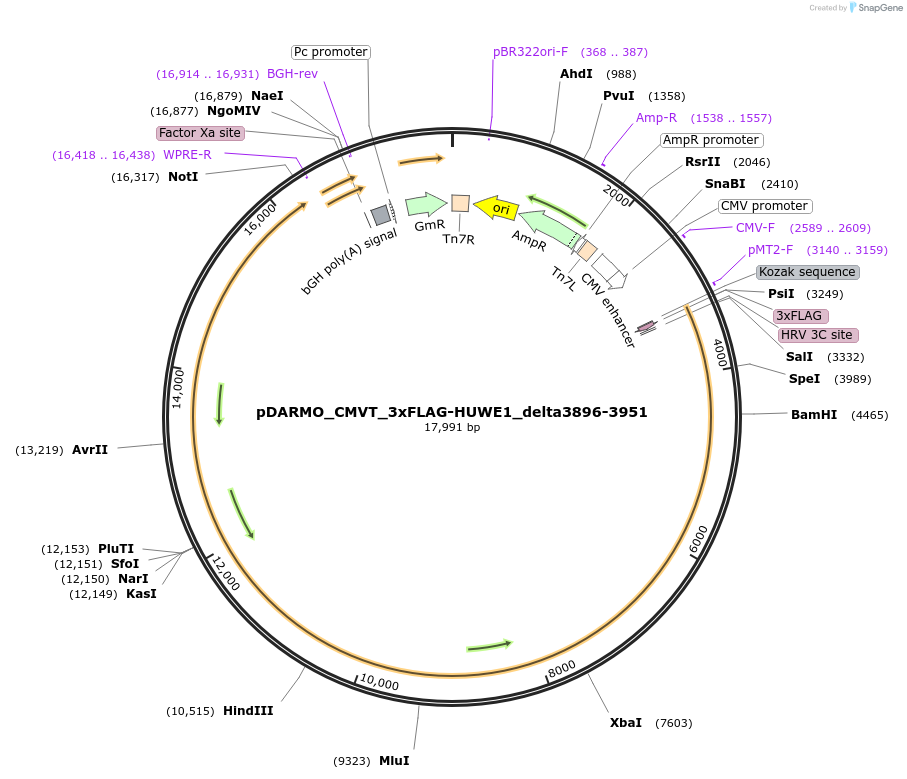
pDARMO_CMVT_3xFLAG-HUWE1_delta3896-3951
Plasmid#187133Purposemammalian cell expression of 3xFLAG-HUWE1 delta3896-3951DepositorAvailable SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only