We narrowed to 75,518 results for: LAS
-
Plasmid#45292DepositorInsertTLOC1 (SEC62 Human)
UseLentiviralTagsV5ExpressionMammalianMutationdeleted 27 c-terminal amino acidsAvailable SinceMay 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
pLEX_970_puro_DEST_V5_TLOC1(220aa)deltaN149
Plasmid#45294DepositorAvailable SinceMay 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
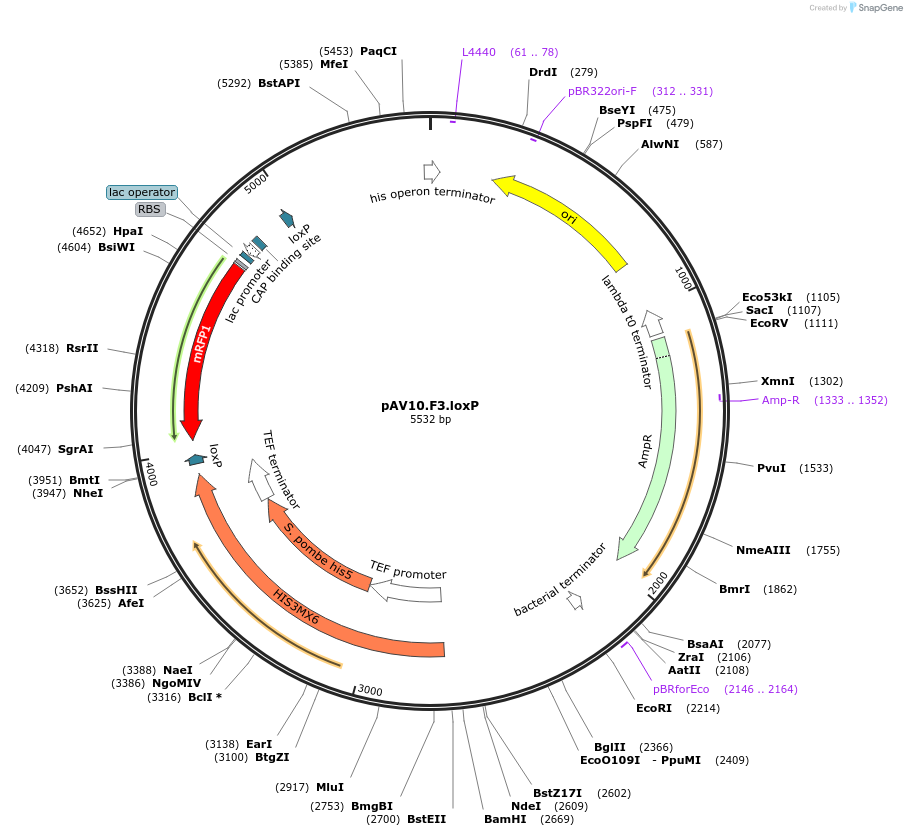
pAV10.F3.loxP
Plasmid#63200PurposepCACS backbone (Amp), SpHIS5, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into chrVI: 97873-98803 (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
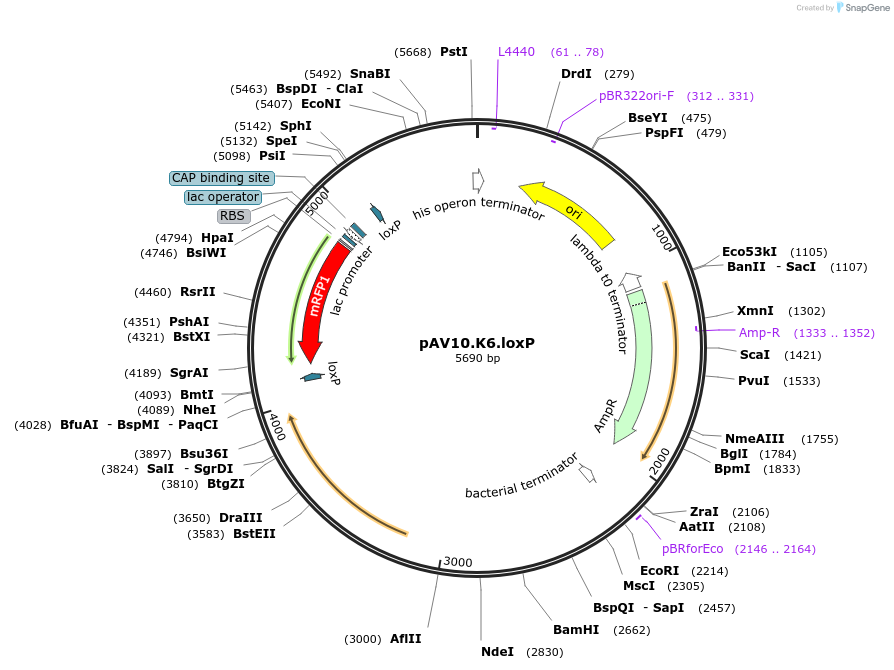
pAV10.K6.loxP
Plasmid#63203PurposepCACS backbone (Amp), KlURA3, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into YKL162C-A (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
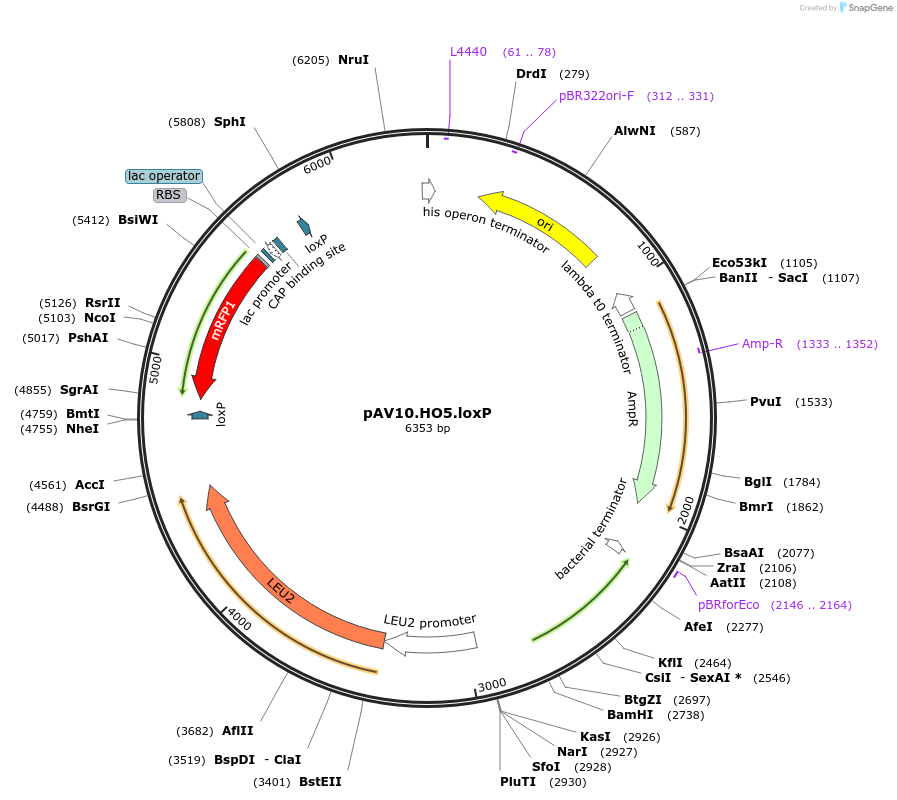
pAV10.HO5.loxP
Plasmid#63206PurposepCACS backbone (Amp), LEU2, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into HO locus (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 1
Pooled Library#168776PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 2
Pooled Library#168777PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCVSN2c-MCP-oScarlet-KASH-6xMS2 Barcode Library
Pooled Library#227672PurposeRVdG genome plasmids that carry an error-correctable barcode compatible with 3’ capture single cell or single nuclei RNA sequencing readout. For the production of CVS N2c strain G-deleted rabies virusDepositorExpressionMammalianSpeciesSyntheticAvailable SinceFeb. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
EGFP miSFIT Lineage Barcoding Library
Pooled Library#229136PurposeThe EGFP_miSFIT_Lineage_BC_lib library is a transcribed lineage barcoding vector library.DepositorExpressionMammalianUseLentiviralAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
ALL-PAM sensor Library
Pooled Library#179219Purpose10 human and mouse sgRNAs each where for each sgRNA the tethered target site has 1 of 64 possible 3 bp PAM sequences.DepositorExpressionMammalianUseLentiviralAvailable SinceJune 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
DH10beta F' DOT sbcC (donor strain)
Bacterial Strain#12082DepositorBacterial ResistanceTetracyclineAvailable SinceMarch 1, 2007AvailabilityAcademic Institutions and Nonprofits only -
FD1
Bacterial Strain#125258PurposeDerivative of MG1655 used for the screening of clones able to survive the bad seed toxicity of dCas9, while still efficiently silencing the expression of mcherryDepositorBacterial ResistanceNoneAvailable SinceAug. 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
N4177 strain
Bacterial Strain#13333DepositorBacterial ResistanceCoumermycinAvailable SinceOct. 27, 2006AvailabilityAcademic Institutions and Nonprofits only -
Human Base Editing Sensor Library
Pooled Library#179218PurposesgRNAs that target frequently occurring cancer associated mutations identified in the MSK IMPACT dataset that can be modeled with cytosine base editing.DepositorExpressionMammalianUseLentiviralAvailable SinceJune 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
Mouse Base Editing Sensor Library
Pooled Library#179217PurposesgRNAs that target frequently occurring cancer associated mutations identified in the MSK IMPACT dataset that can be modeled with cytosine base editing.DepositorExpressionMammalianUseLentiviralAvailable SinceJune 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
Strain CSH100
Bacterial Strain#21875DepositorBacterial ResistanceNoneAvailable SinceNov. 3, 2009AvailabilityAcademic Institutions and Nonprofits only