We narrowed to 4,323 results for: Mcc;
-
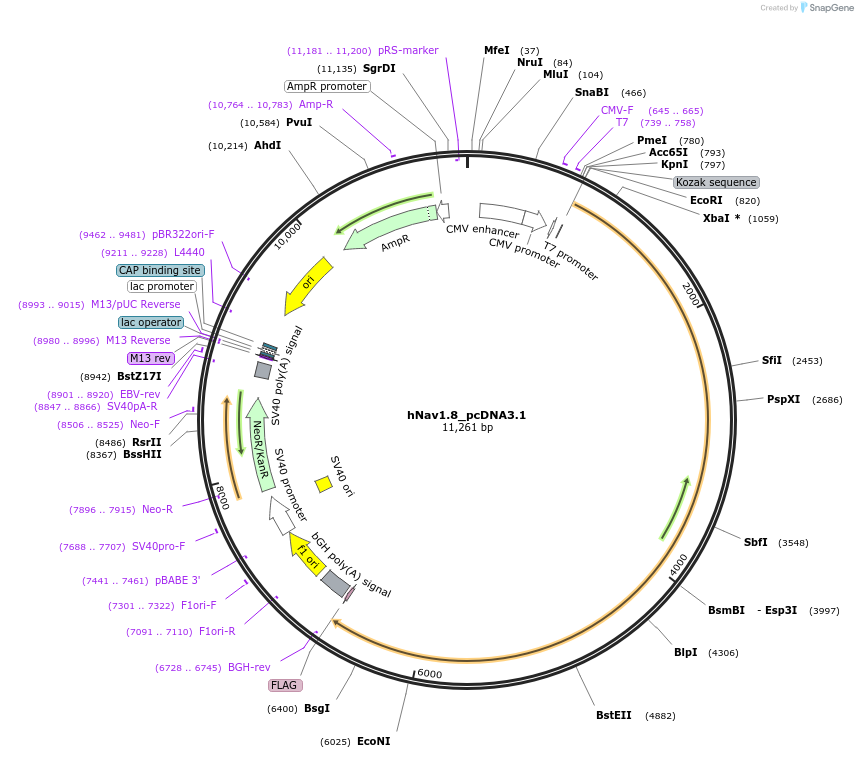
Plasmid#182847PurposeExpresses hNav1.8 in mammalian cellsDepositorInsertSCN10A (SCN10A Human)
ExpressionMammalianAvailable SinceApril 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
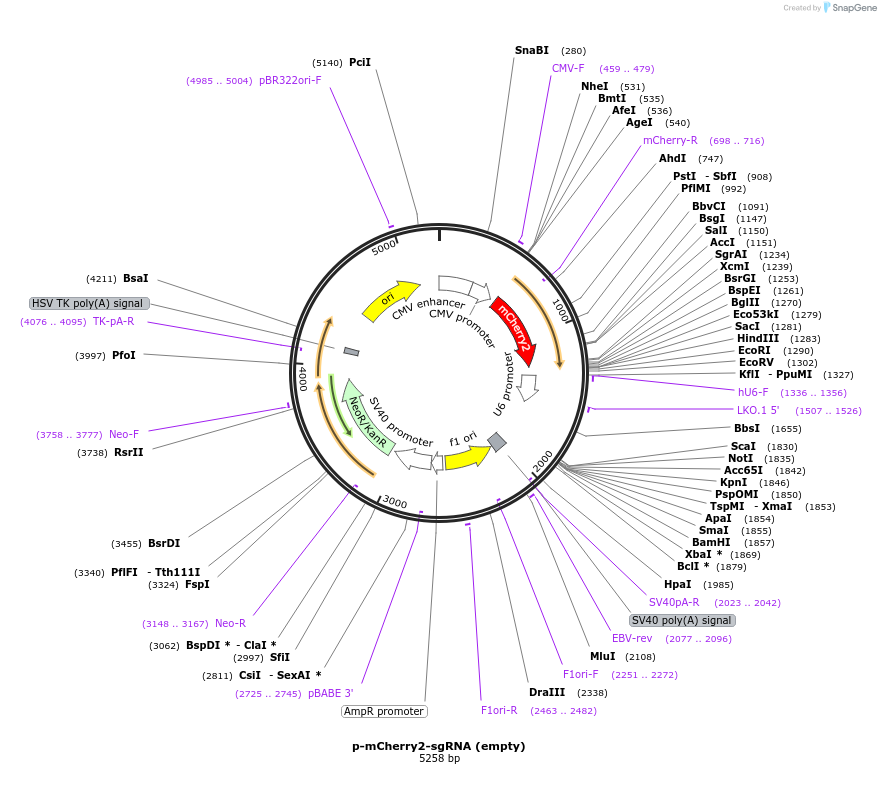
p-mCherry2-sgRNA (empty)
Plasmid#198330PurposeCustomisable sgRNA sequence with optimised sgRNA scaffold for expression under U6 promoter, with mCherry2 reporterDepositorInsertU6-sgRNA(F+E) empty
ExpressionMammalianPromoterU6Available SinceApril 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
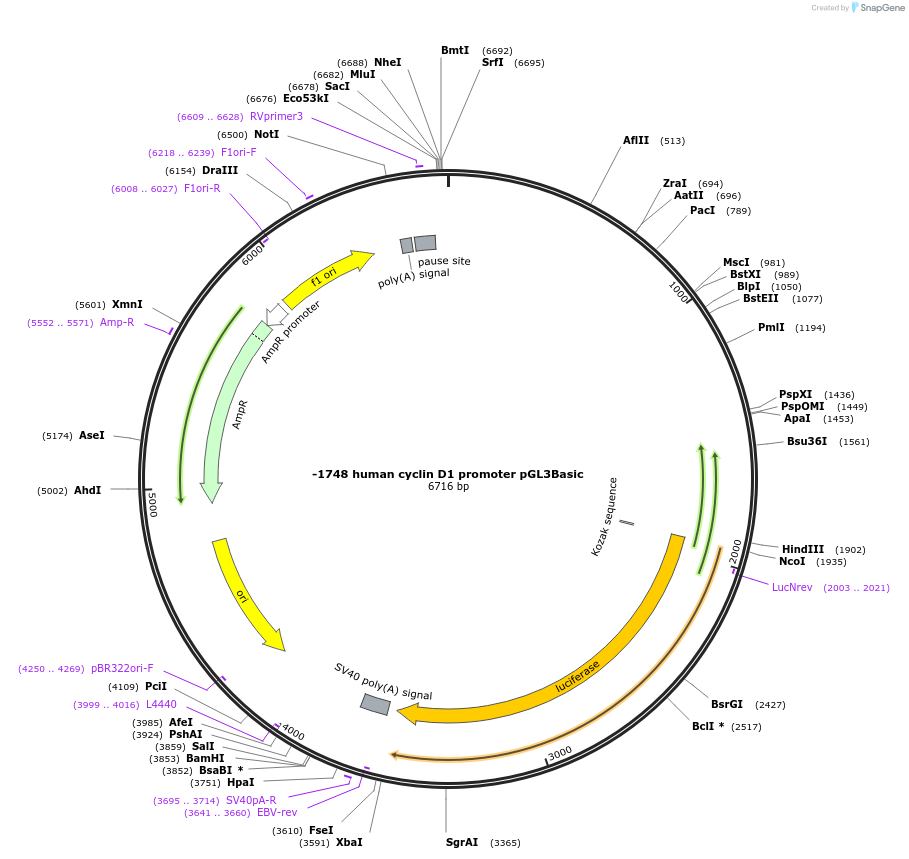
-1748 human cyclin D1 promoter pGL3Basic
Plasmid#32726DepositorAvailable SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
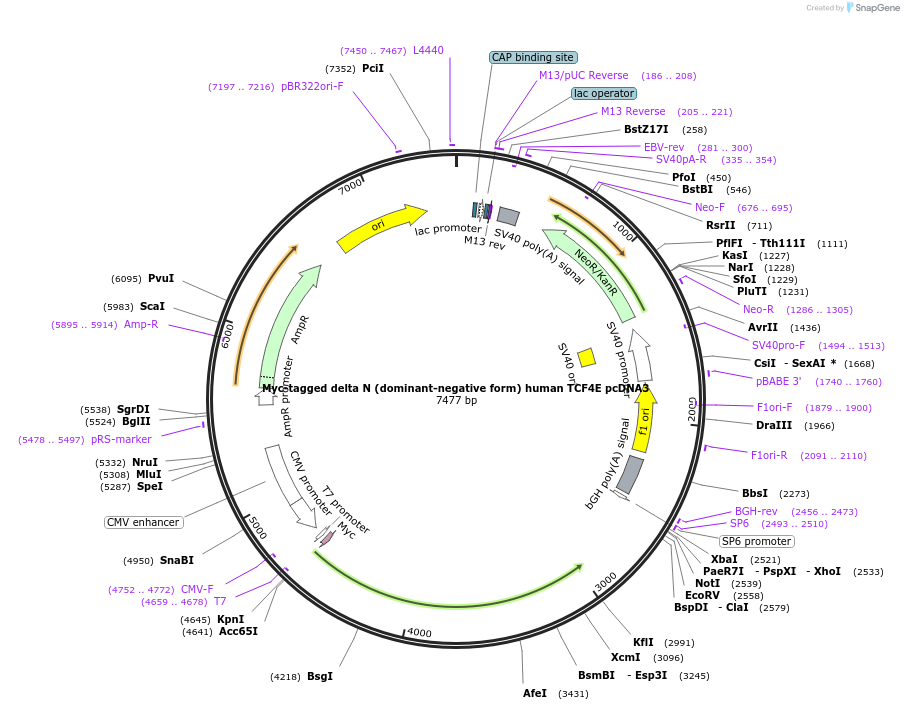
Myc-tagged delta N (dominant-negative form) human TCF4E pcDNA3
Plasmid#32739DepositorInsertTCF4E (Tcf7l2 Human)
TagsMycExpressionMammalianMutationThe N-terminal deletion form (delta N) lacks amin…Available SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
GFP SMG1 WT
Plasmid#199571PurposeExpresses WT SMG1 fused to GFPDepositorAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
-962 human cyclin D1 promoter pGL3Basic
Plasmid#32727DepositorAvailable SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
Myc-tagged human full length TCF4E pcDNA3
Plasmid#32738DepositorAvailable SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
pNL4-3 envFS eGFP Δvpu/env
Plasmid#101345PurposeDeletion from NL4-3 nt 6054-7489 encompassing all of vpu and env until before the RREDepositorInsertNL4-3
UseLentiviralMutationdelta vpu/EnvAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
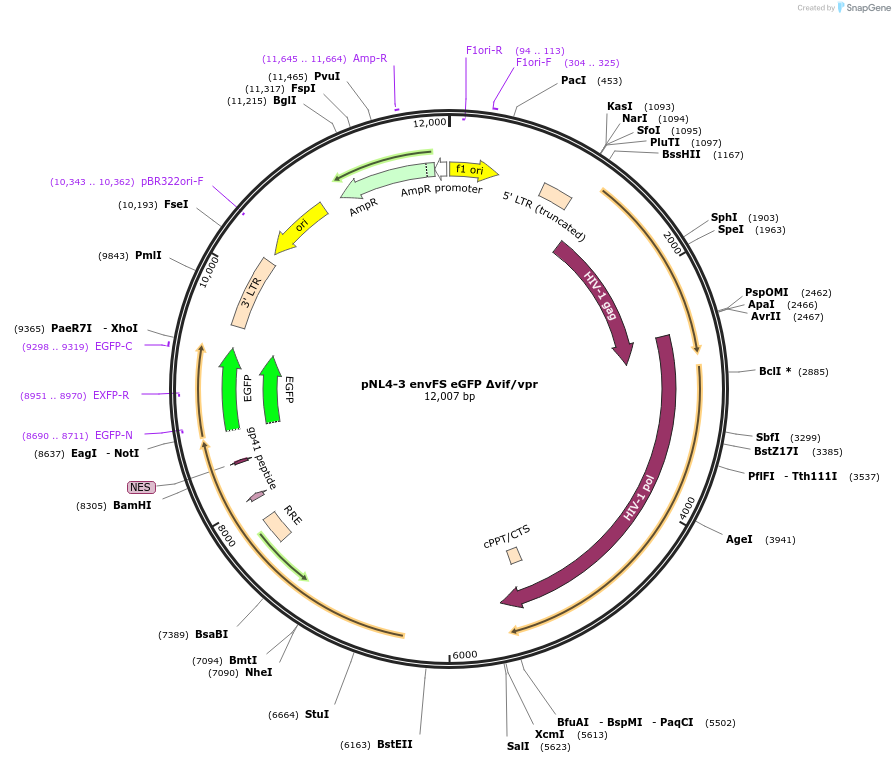
pNL4-3 envFS eGFP Δvif/vpr
Plasmid#101344PurposeDeletion from NL4-3 nt 5582-6199 encompassing Vif and Vpr coding sequenceDepositorInsertNL4-3
UseLentiviralMutationdelta vif/VprAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
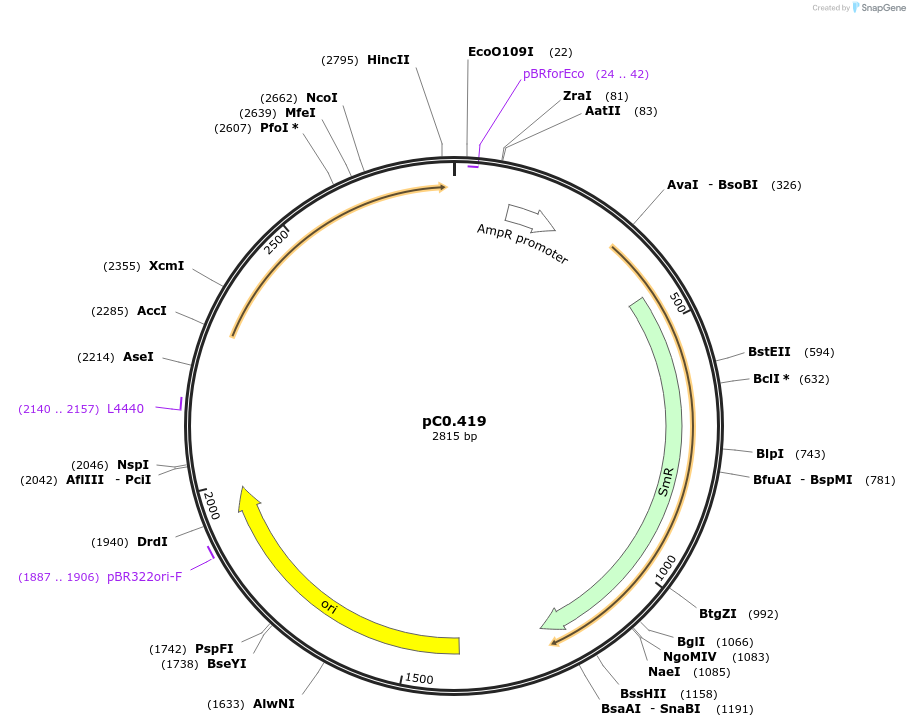
pC0.419
Plasmid#203935PurposeDown Flank. Neutral site. Down flanking sequence of type II site-specific deoxyribonuclease AquI (FEK30_10065) in Synechococcus sp. PCC 11901.DepositorInsertaquI DOWN
UseSynthetic BiologyAvailable SinceMay 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
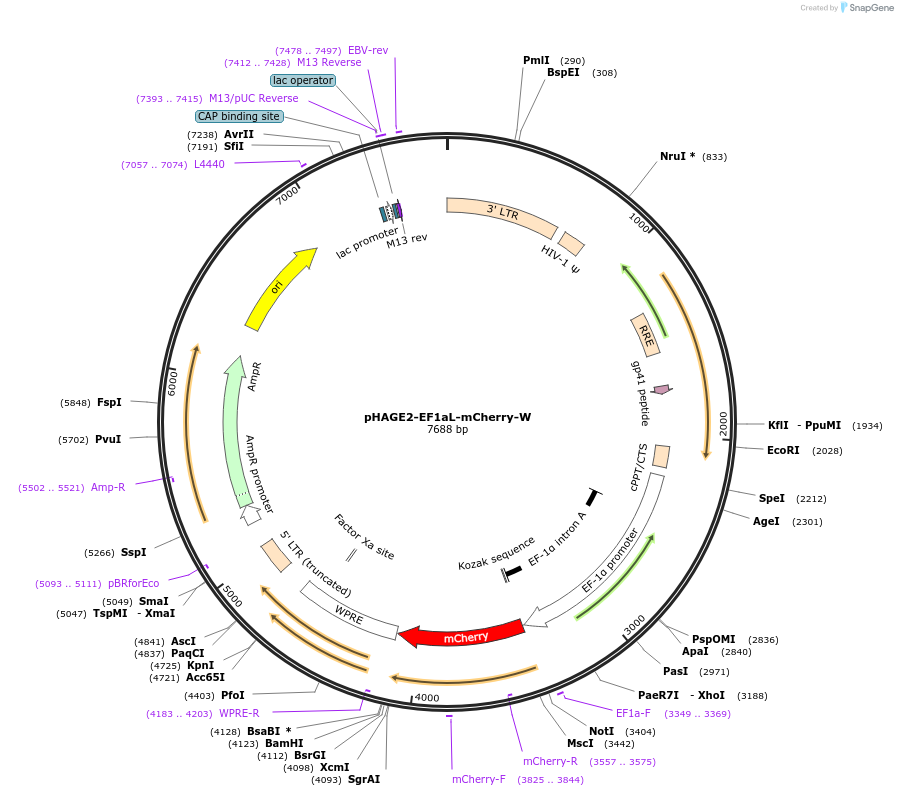
pHAGE2-EF1aL-mCherry-W
Plasmid#126703PurposeConstitutive, ubiquitous mCherry expression.DepositorInsertmCherry
UseLentiviralPromoterEf1aLAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
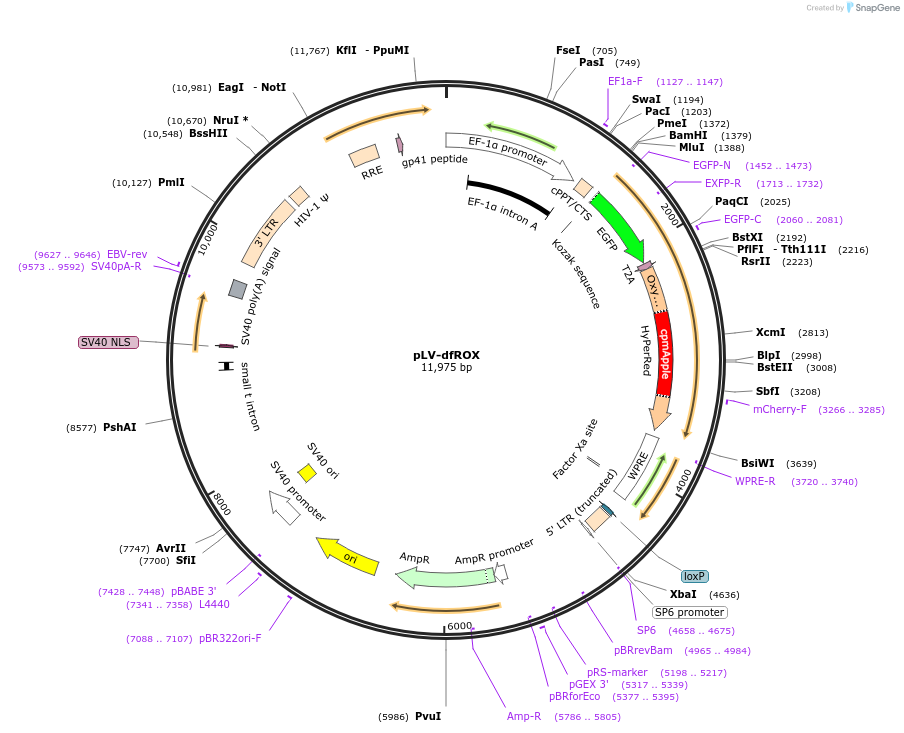
pLV-dfROX
Plasmid#137170PurposeLentiviral transfer vector for expression of roGFP2 and HyPer RedDepositorInsertroGFP2-2A-HyPer Red
UseLentiviralPromoterEF1aAvailable SinceApril 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
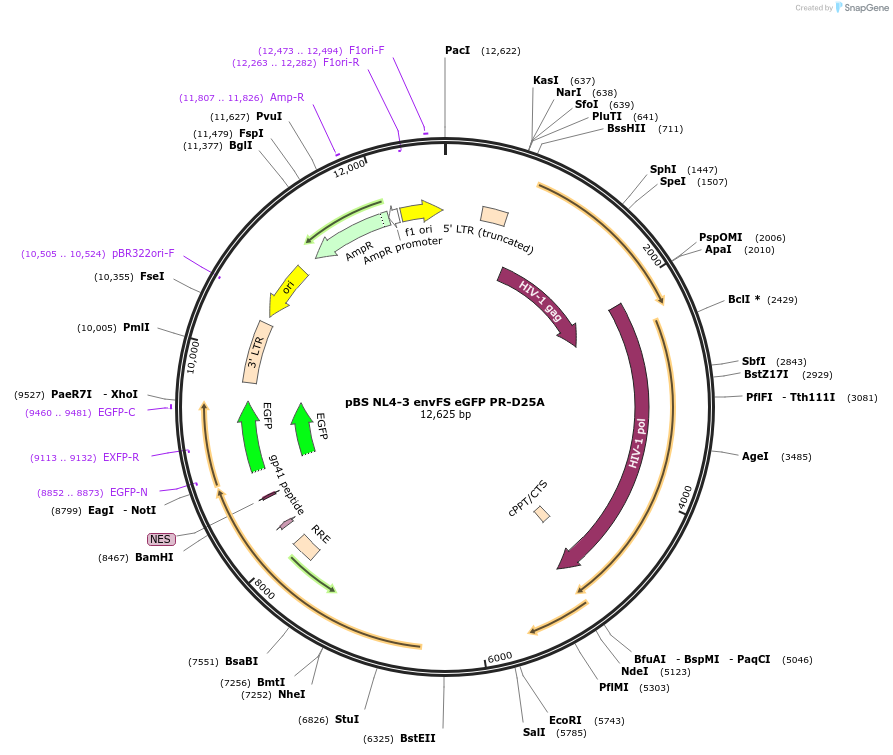
pBS NL4-3 envFS eGFP PR-D25A
Plasmid#101334PurposepBS NL4-3 envFS eGFP with mutation that disrupts Protease catalytic activityDepositorInsertNL4-3 envFS
UseLentiviralMutationProtease D25AAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
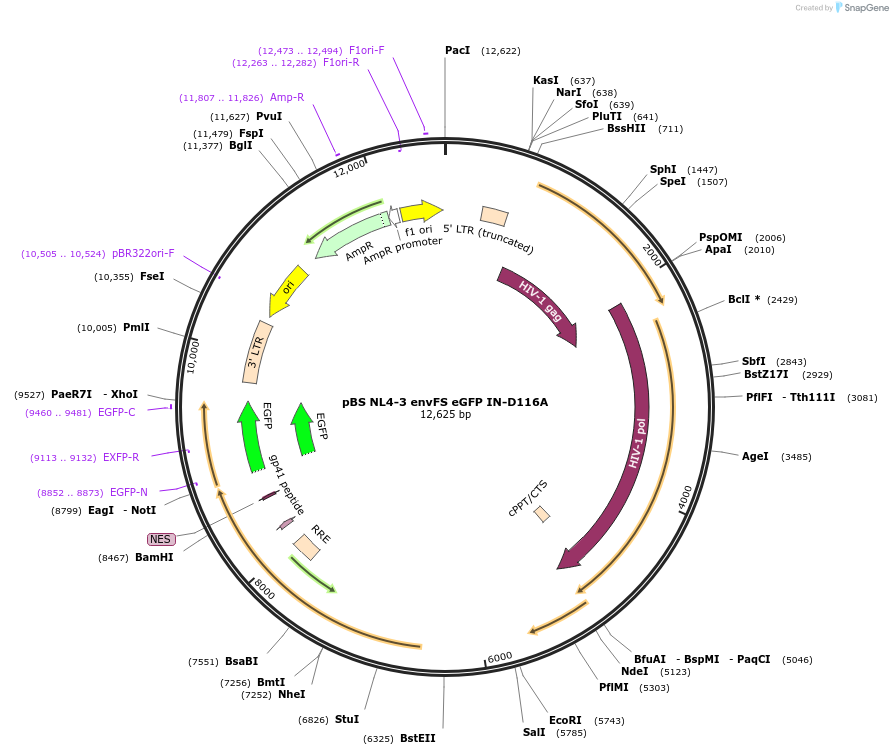
pBS NL4-3 envFS eGFP IN-D116A
Plasmid#101333PurposepBS NL4-3 envFS eGFP with mutation that disrupts IN catalytic activityDepositorInsertNL4-3 envFS
UseLentiviralMutationIntegrase D116AAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
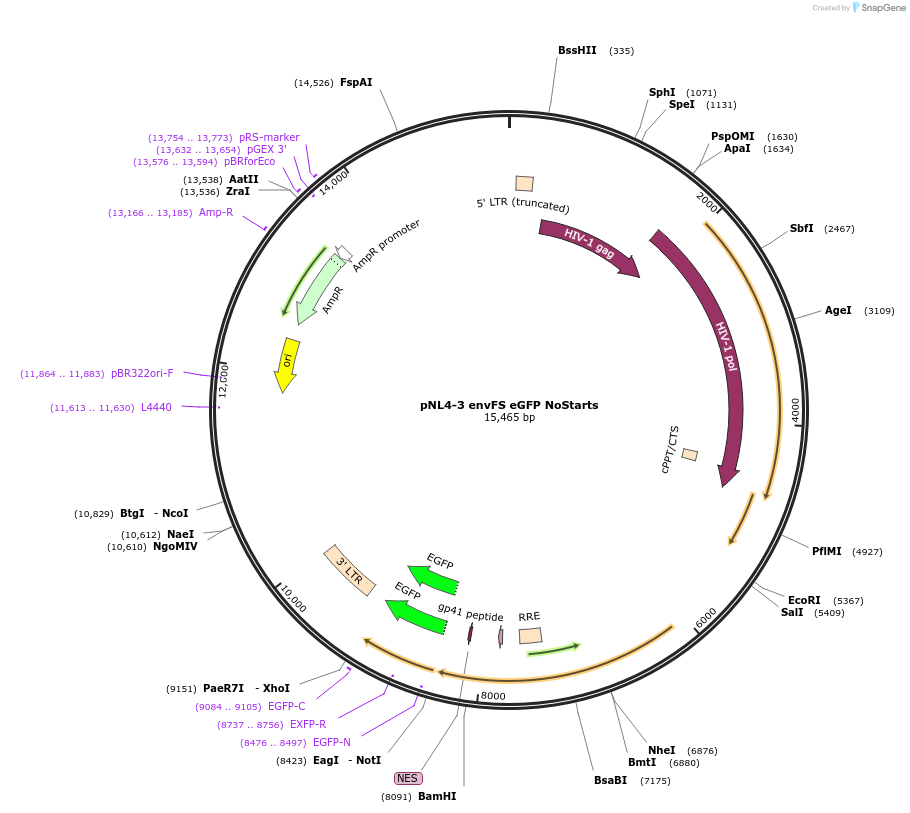
pNL4-3 envFS eGFP NoStarts
Plasmid#101342PurposeAll ATGs from the start of gag to NC mutated to ATC except the first which was mutated to ACGDepositorInsertNL4-3
UseLentiviralMutationNo methonines in GagAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
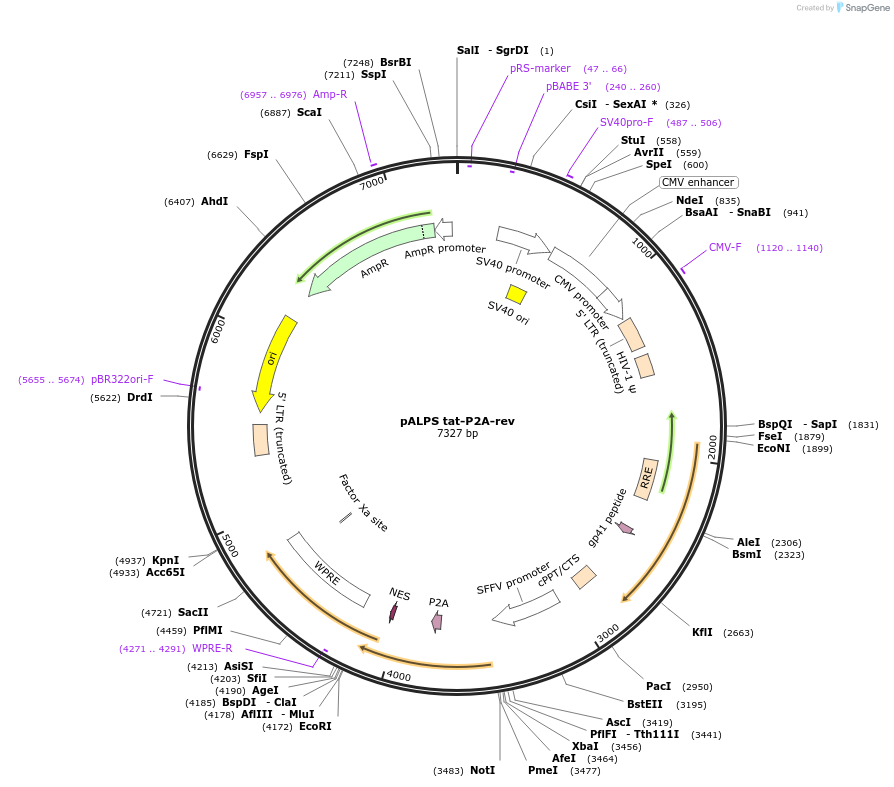
pALPS tat-P2A-rev
Plasmid#101331PurposeLentivector expressing codon optimized NL4-3 tat and rev linked by P2A peptide coding sequence (GSGATNFSLLKQAGDVEENPGP)DepositorInsertNL4-3 tat and rev
UseLentiviralAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
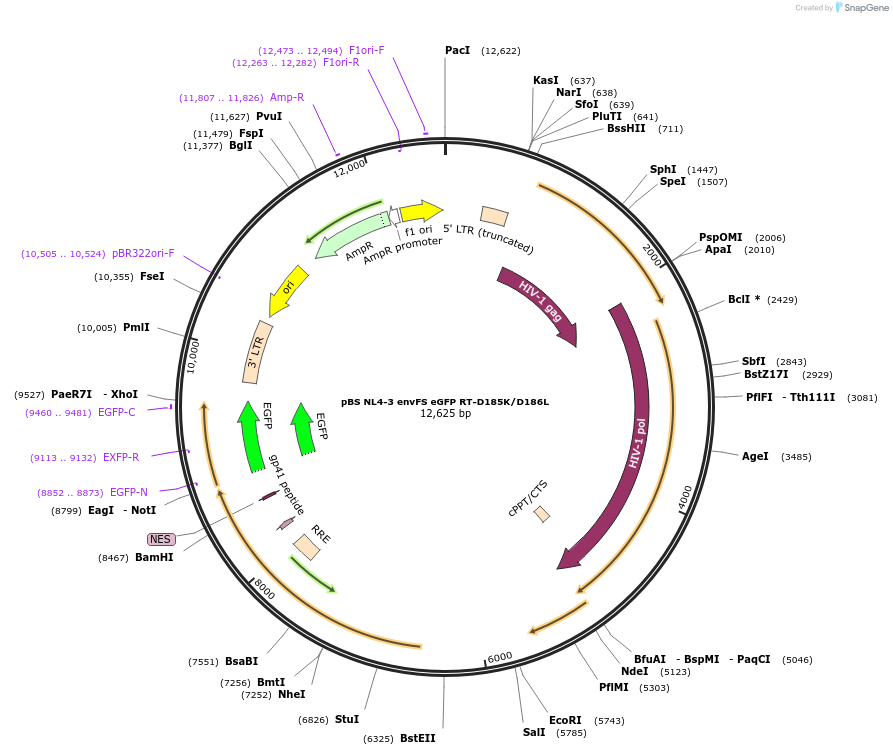
pBS NL4-3 envFS eGFP RT-D185K/D186L
Plasmid#101332PurposepBS NL4-3 envFS eGFP with mutation that disrupts RT catalytic activityDepositorInsertNL4-3 envFS
UseLentiviralMutationreverse transcriptase D185K/D186LAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
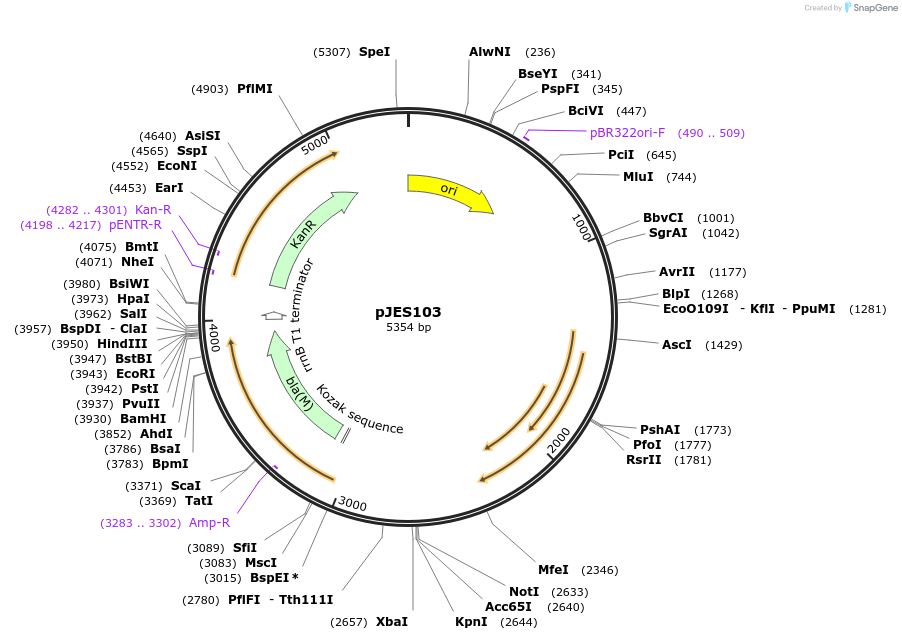
pJES103
Plasmid#135410PurposeE. coli 'blaTEM-1 with M. tuberculosis mpt63 signal sequence under the control of hsp60 promoter that can be used as a reporterDepositorInsertblaTEM-1
ExpressionBacterialMutationE. coli 'blaTEM-1 with M. tuberculosis mpt63…Promoterhsp60Available SinceFeb. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
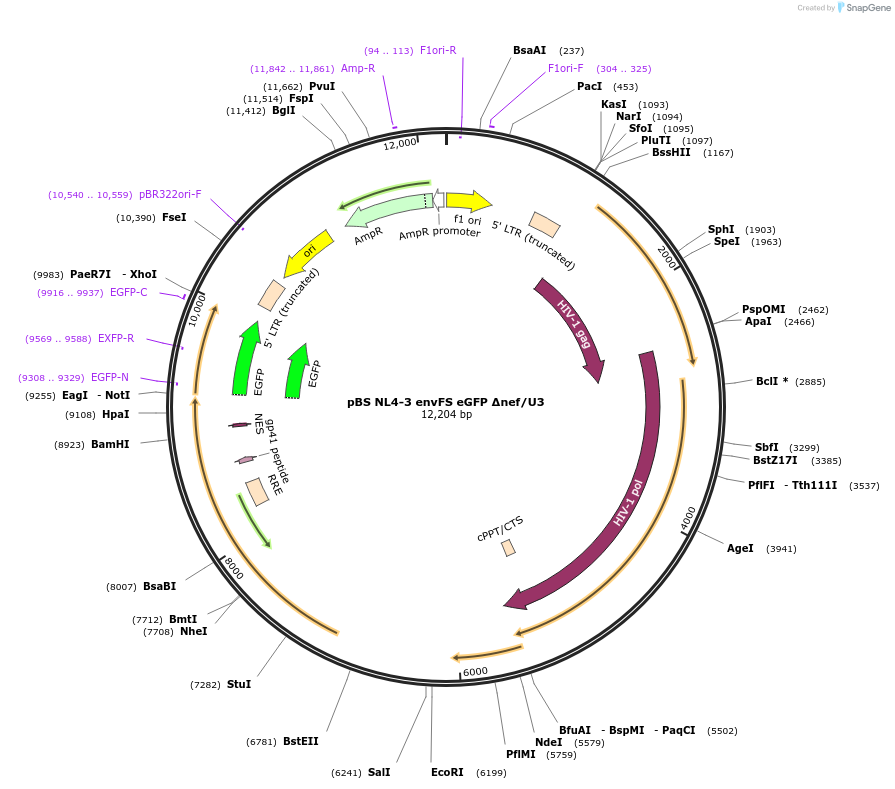
pBS NL4-3 envFS eGFP Δnef/U3
Plasmid#101346PurposeDeletion from NL4-3 nt 8911-9022 and 9088-9377. This deletes nef and U3 LTR sequencesDepositorInsertNL4-3
UseLentiviralMutationdelta nef/LTRAvailable SinceMarch 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
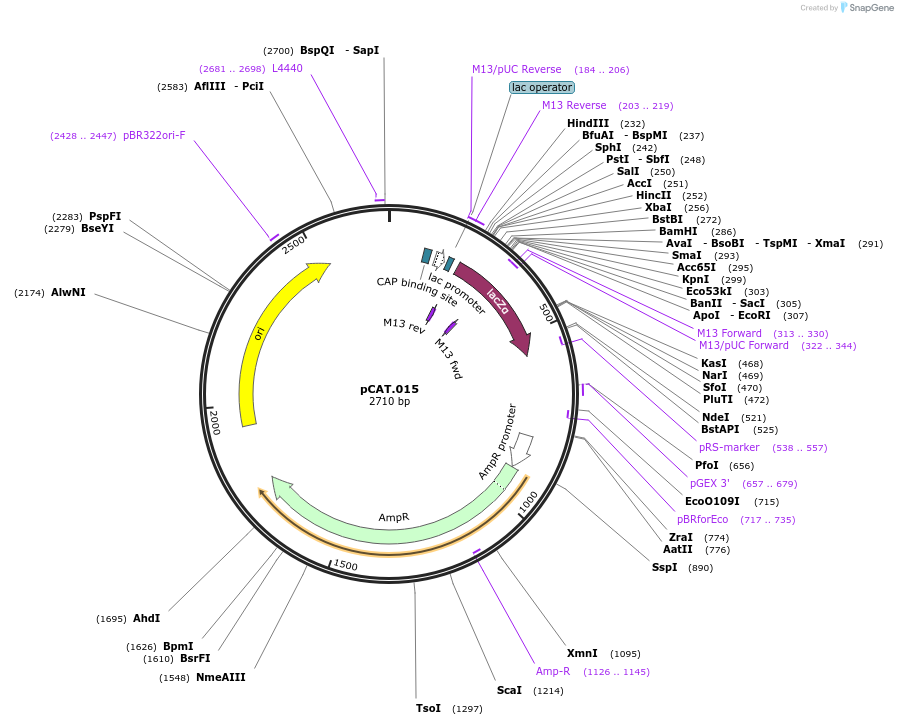
pCAT.015
Plasmid#119558PurposeLevel T acceptor vector for chromosomal integration with lacZ compatible with blue/white screening. Modified pUC19 vector.DepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only