We narrowed to 24,746 results for: CHI
-
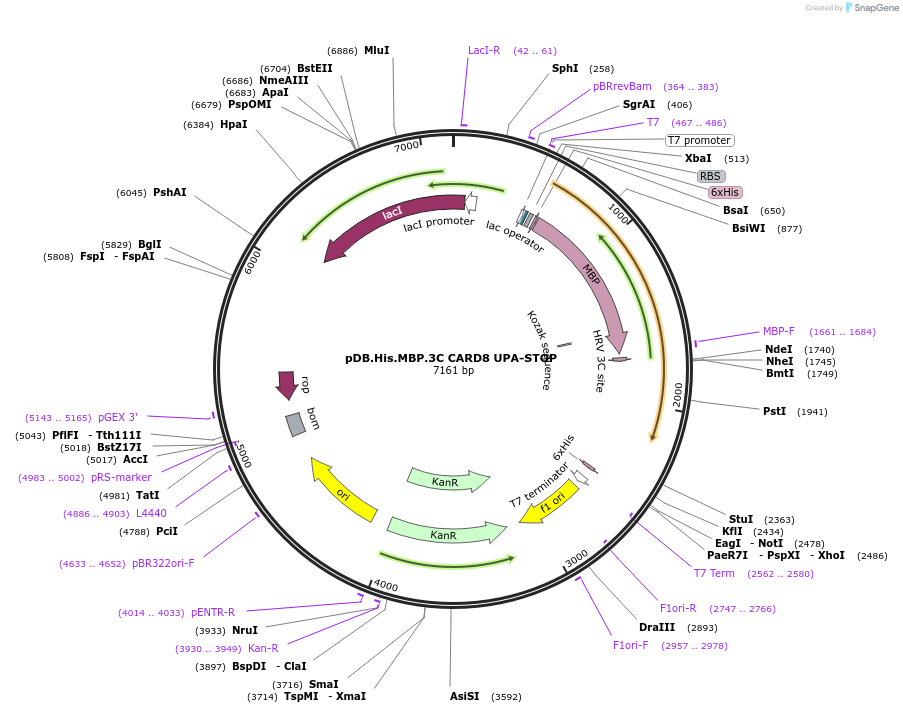
Plasmid#164031PurposeBacterial expression vector encoding a HRV 3C-cleavable His-MBP-tagged CARD8 UPADepositorAvailable SinceFeb. 19, 2021AvailabilityAcademic Institutions and Nonprofits only
-
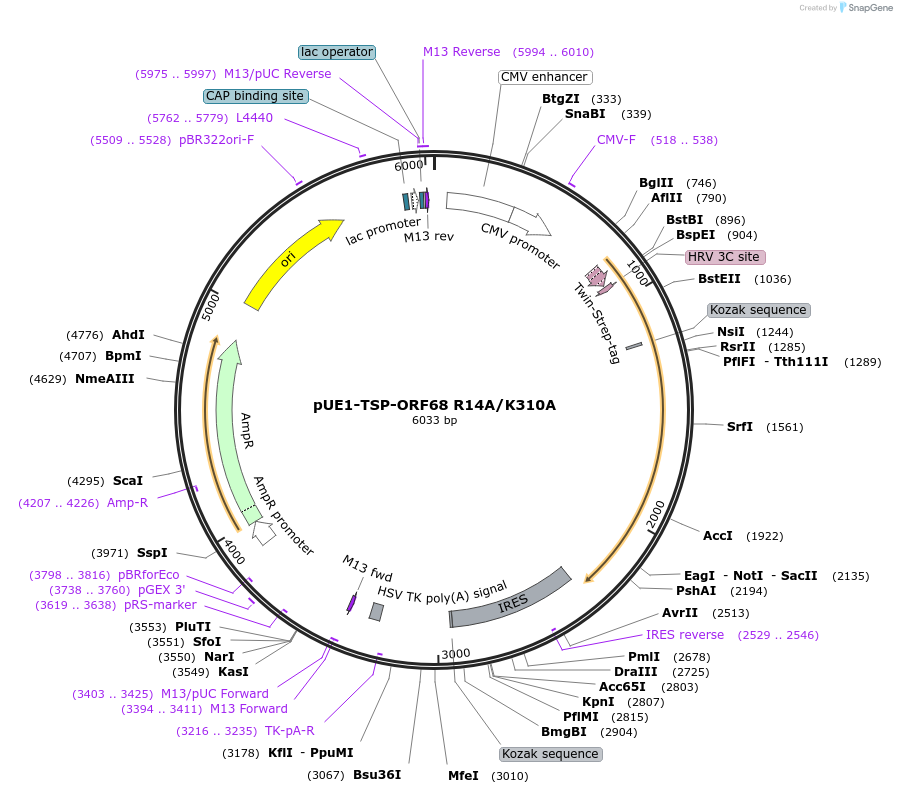
pUE1-TSP-ORF68 R14A/K310A
Plasmid#162655PurposeExpresses ORF68 R14A/K310A in mammalian cells with an N-terminal TwinStrep tag and HRV 3C protease cleavage siteDepositorInsertORF68
TagsN-terminal 2xStrep-Tag II and HRV 3C siteExpressionMammalianMutationAmino acids R14 and K310 mutated to alaninePromoterCMVAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
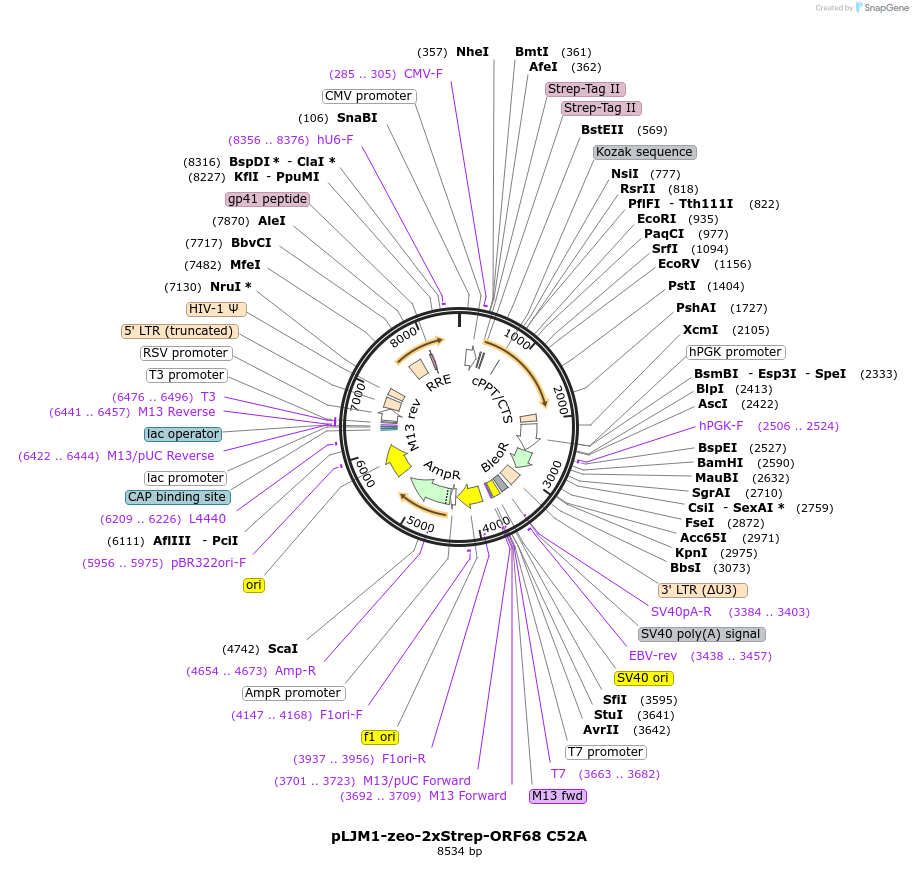
pLJM1-zeo-2xStrep-ORF68 C52A
Plasmid#162660PurposeLentiviral vector for stable expression of N-terminal 2xStrep-tagged ORF68 C52A in mammalian cellsDepositorInsertORF68
UseLentiviralTagsN-terminal 2xStrep-Tag IIExpressionMammalianMutationAmino acid C52 mutated to alaninePromoterCMVAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
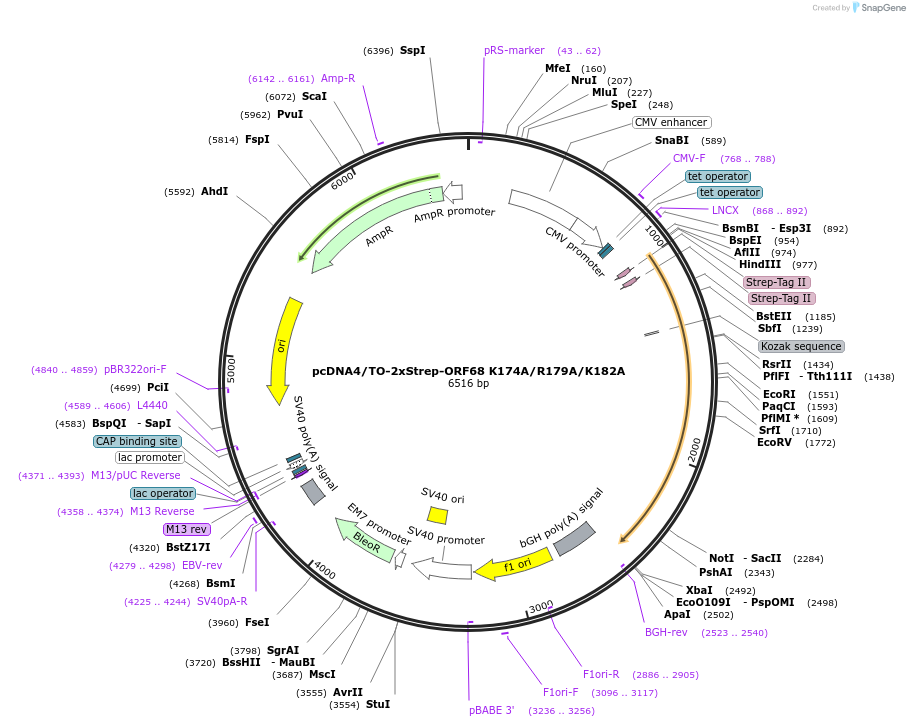
pcDNA4/TO-2xStrep-ORF68 K174A/R179A/K182A
Plasmid#162639PurposeExpresses N-terminal 2x-Strep-tagged ORF68 K174A/R179A/K182A in mammalian cellsDepositorInsertORF68
TagsN-terminal 2xStrep-Tag IIExpressionMammalianMutationAmino acids K174, R179, and K182 mutated to alani…PromoterCMVAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
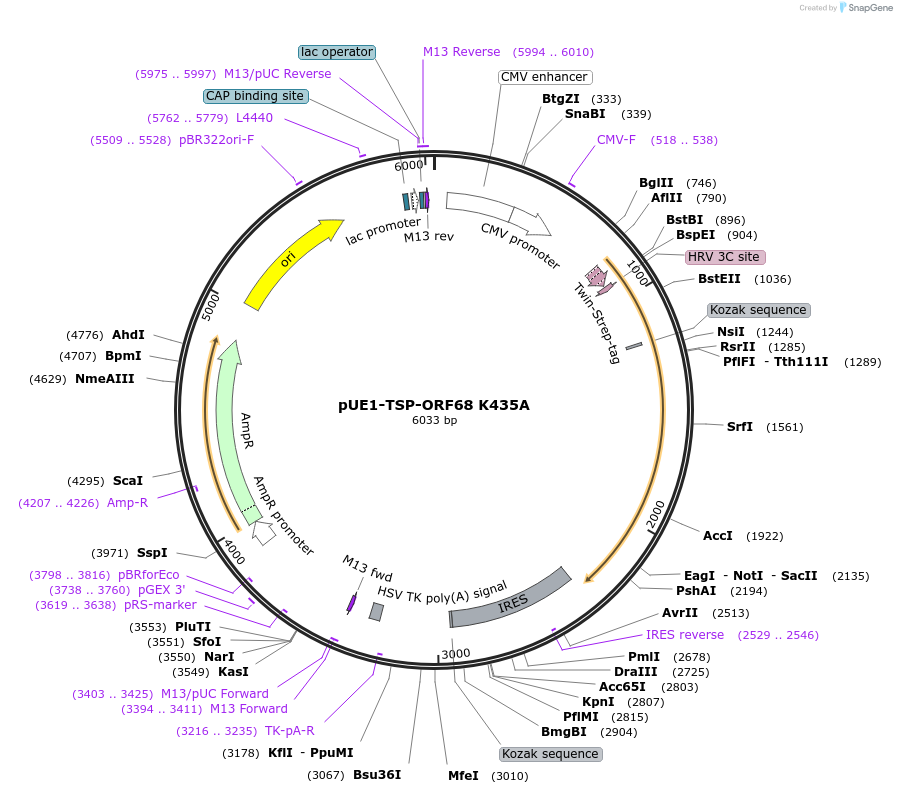
pUE1-TSP-ORF68 K435A
Plasmid#162651PurposeExpresses ORF68 K435A in mammalian cells with an N-terminal TwinStrep tag and HRV 3C protease cleavage siteDepositorInsertORF68
TagsN-terminal 2xStrep-Tag II and HRV 3C siteExpressionMammalianMutationAmino acid K435 mutated to alaninePromoterCMVAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
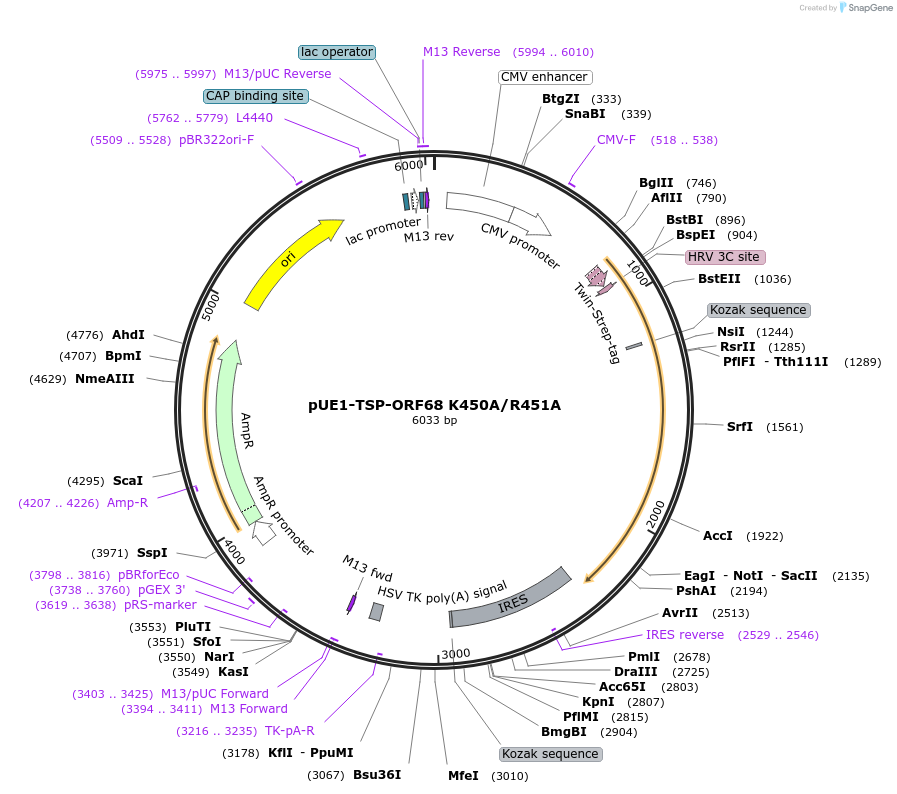
pUE1-TSP-ORF68 K450A/R451A
Plasmid#162653PurposeExpresses ORF68 K450A/R451A in mammalian cells with an N-terminal TwinStrep tag and HRV 3C protease cleavage siteDepositorInsertORF68
TagsN-terminal 2xStrep-Tag II and HRV 3C siteExpressionMammalianMutationAmino acids K450 and R451 mutated to alaninePromoterCMVAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
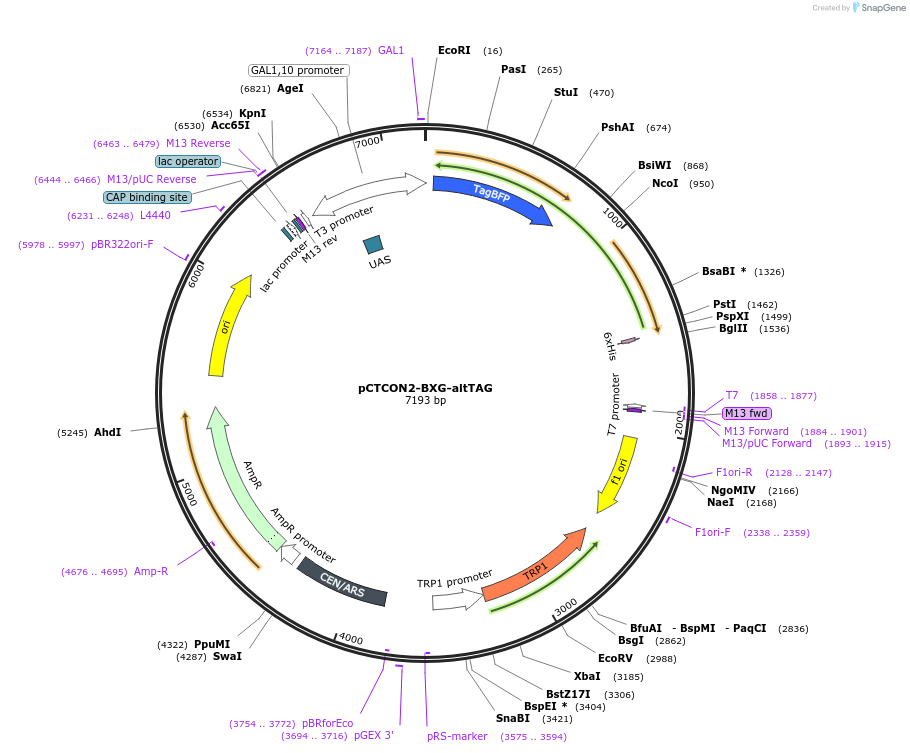
pCTCON2-BXG-altTAG
Plasmid#158132PurposeDual fluorescent protein reporter (BFP-GFP) for ncAA incorporation (TAG construct)DepositorInsertBFP-GFP fragment
ExpressionYeastMutationCloned in a TAG codon at the first serine residue…PromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
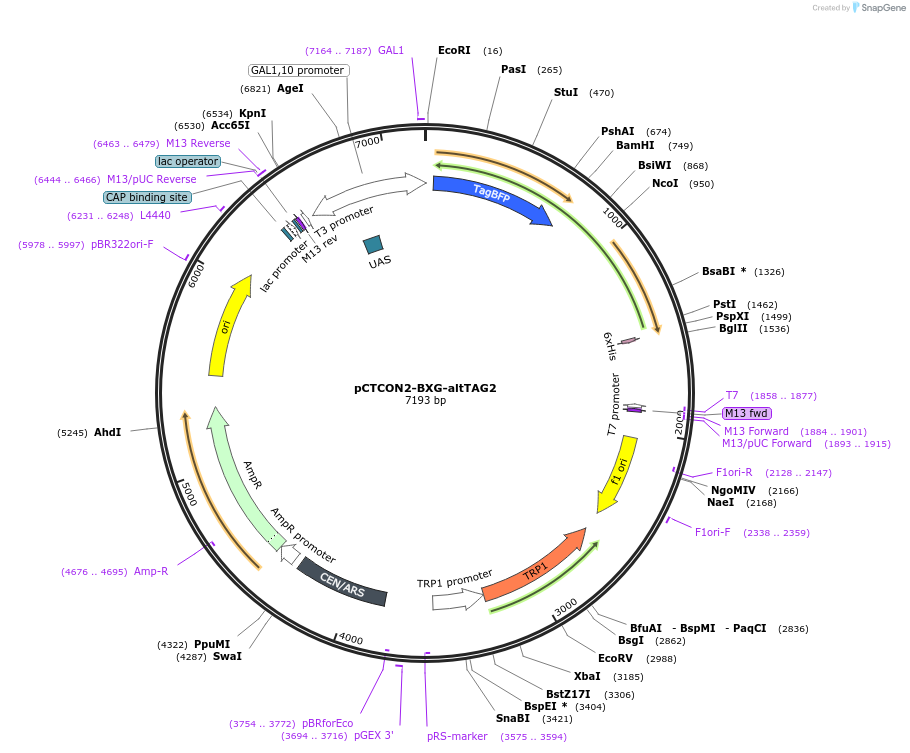
pCTCON2-BXG-altTAG2
Plasmid#158134PurposeDual fluorescent protein reporter (BFP-GFP) for ncAA incorporation (TAG construct)DepositorInsertBFP-GFP fragment
ExpressionYeastMutationCloned in a TAG codon at the second serine residu…PromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
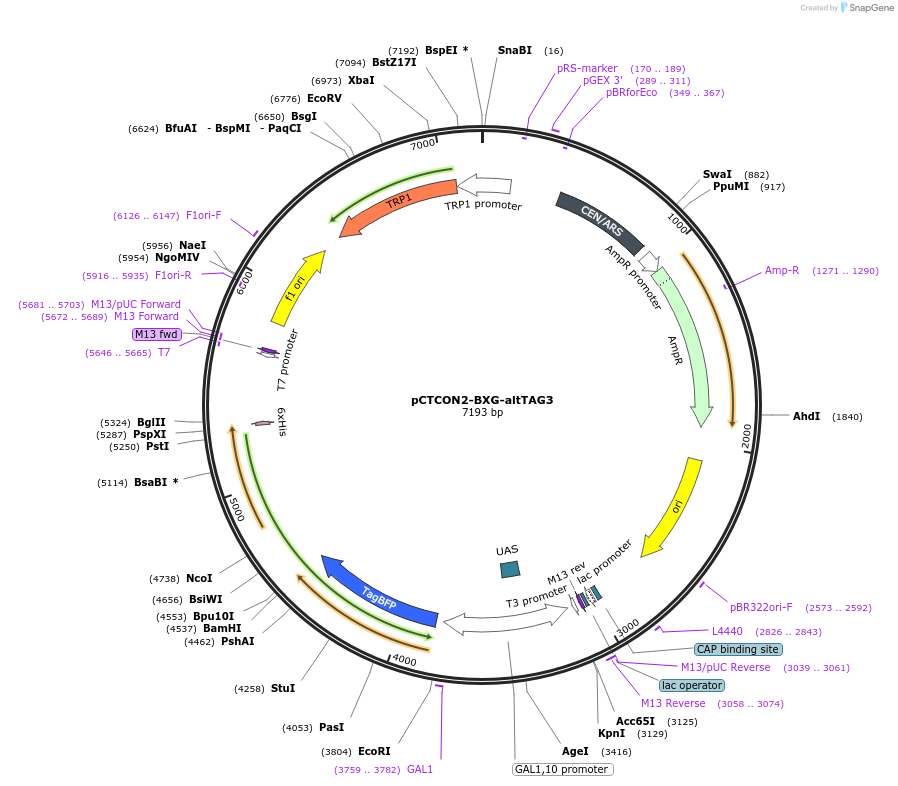
pCTCON2-BXG-altTAG3
Plasmid#158136PurposeDual fluorescent protein reporter (BFP-GFP) for ncAA incorporation (TAG construct)DepositorInsertBFP-GFP fragment
ExpressionYeastMutationCloned in a TAG codon at the third alanine residu…PromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
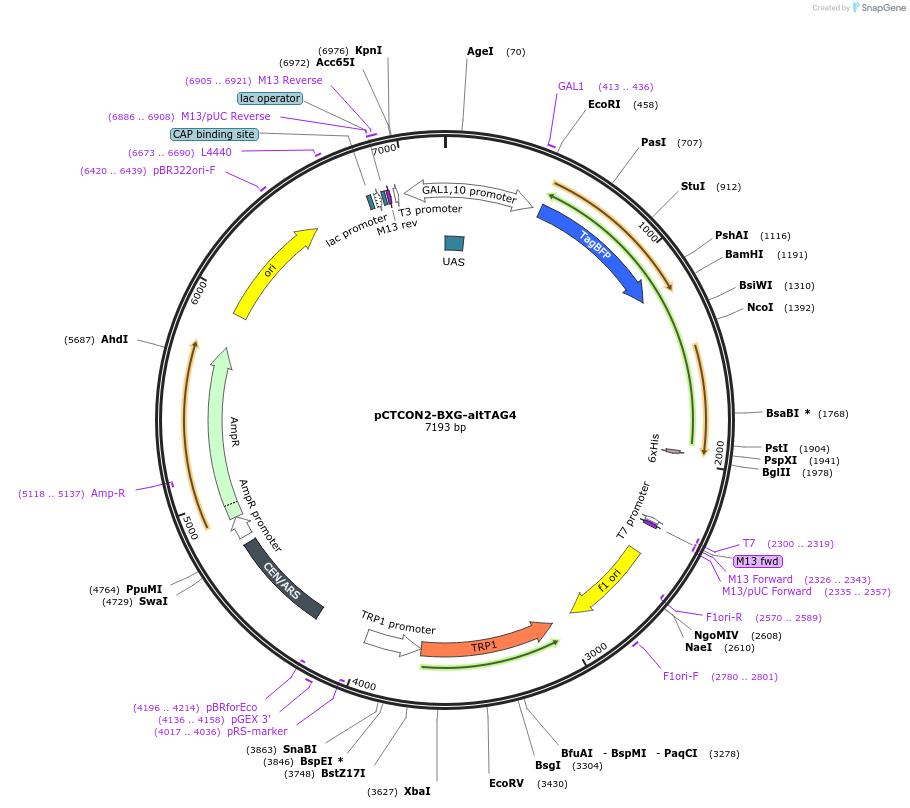
pCTCON2-BXG-altTAG4
Plasmid#158137PurposeDual fluorescent protein reporter (BFP-GFP) for ncAA incorporation (TAG construct)DepositorInsertBFP-GFP fragment
ExpressionYeastMutationCloned in a TAG codon at the third serine residue…PromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
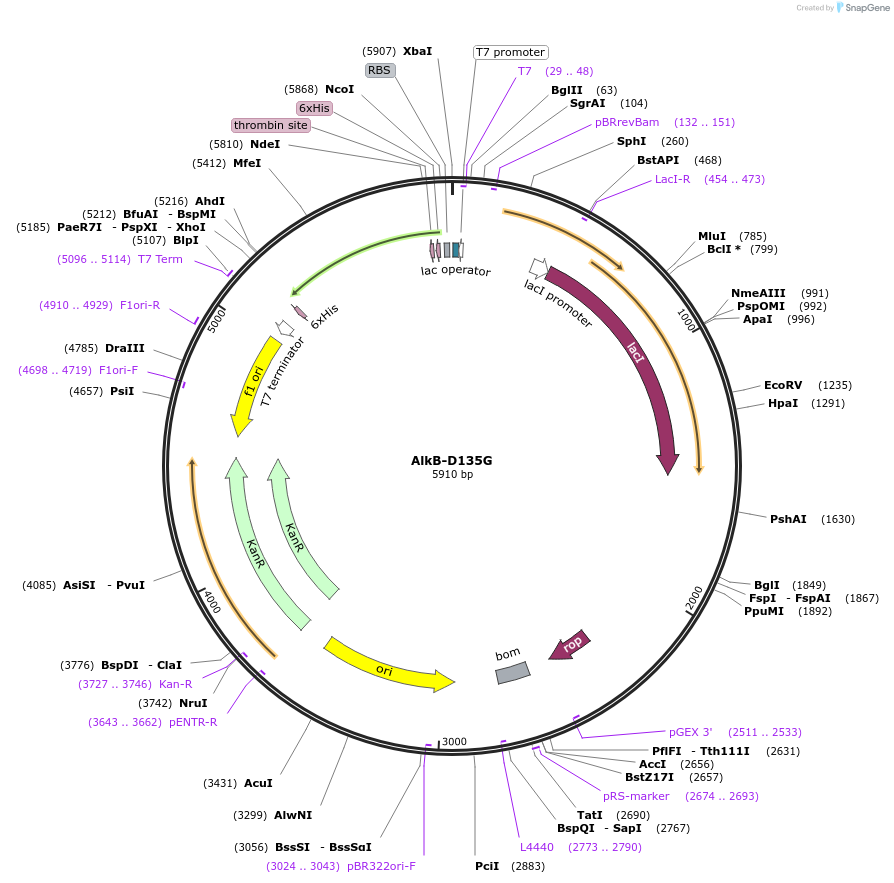
AlkB-D135G
Plasmid#160434PurposeA plasmid for expression in E. coli to produce His-tag fusion protein AlkB with Asp at position 135 mutated to GlyDepositorInsertE. coli AlkB
TagsHis-tagExpressionBacterialMutationChanged Aspartic acid 135 to Glycine; deleted the…PromoterT7Available SinceDec. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
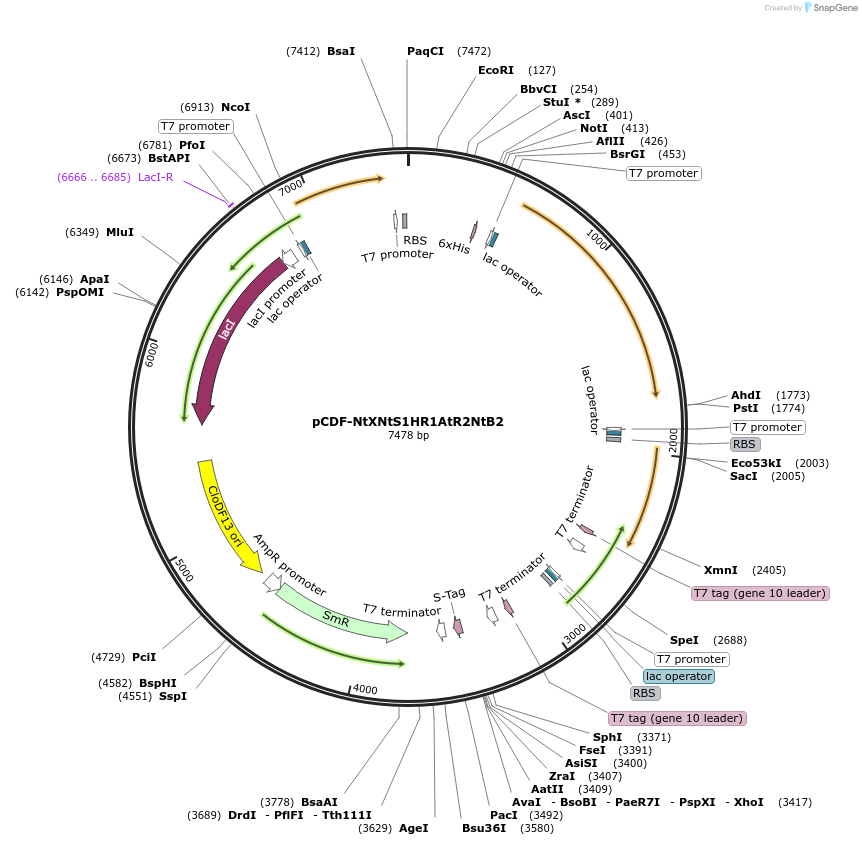
pCDF-NtXNtS1HR1AtR2NtB2
Plasmid#160890PurposeExpression of N. tabacum Rubisco small subunit (rbcS-SH1), Rubisco chaperone protiens rbcX, raf1, bsd2 and A. thaliana Rubisco chaperone proteins raf2 in E. coli.DepositorInsertPT7-Nt-rbcS-S1H
Tags6xHisExpressionBacterialPromoterrbcS-SH1: T7, chaperones: T13Available SinceDec. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
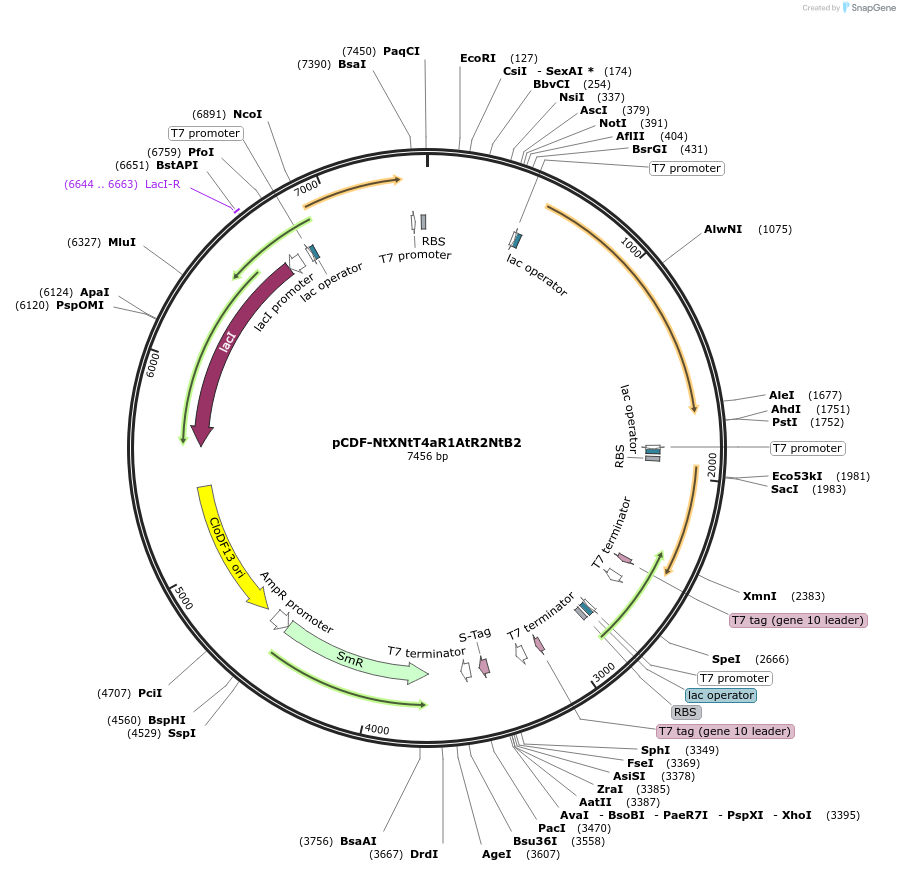
pCDF-NtXNtT4aR1AtR2NtB2
Plasmid#160887PurposeExpression of N. tabacum Rubisco small subunit (rbcS-T4a), Rubisco chaperone protiens rbcX, raf1, bsd2 and A. thaliana Rubisco chaperone proteins raf2 in E. coli.DepositorInsertPT7-Nt-rbcS-T4a
ExpressionBacterialPromoterrbcS-T4a: T7, chaperones: T10Available SinceDec. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
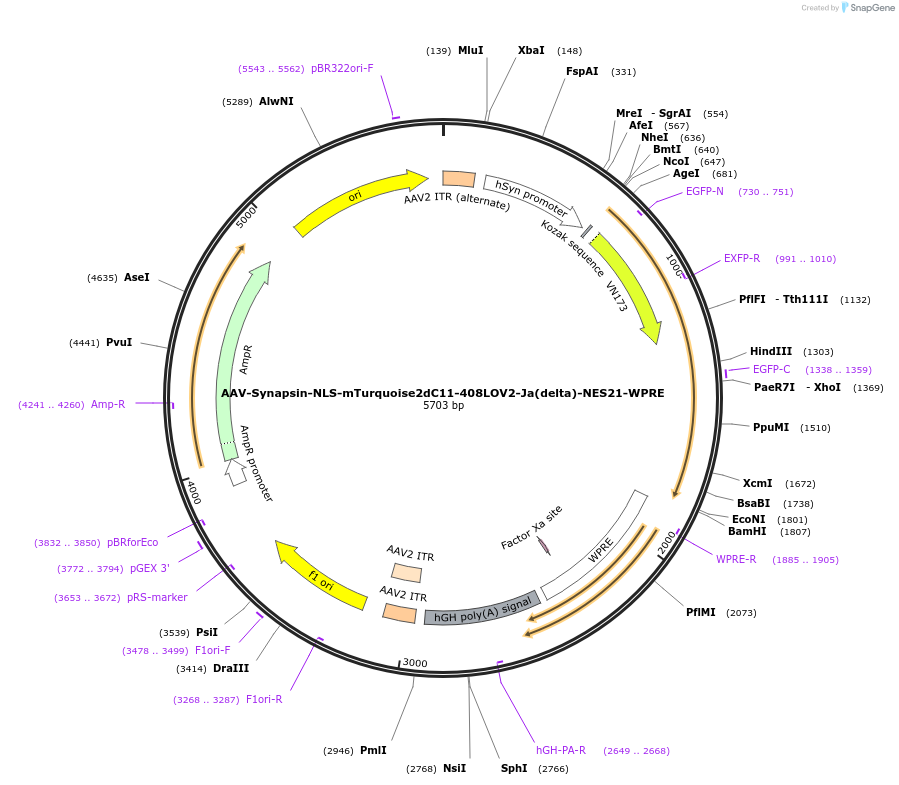
AAV-Synapsin-NLS-mTurquoise2dC11-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159961PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC11-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
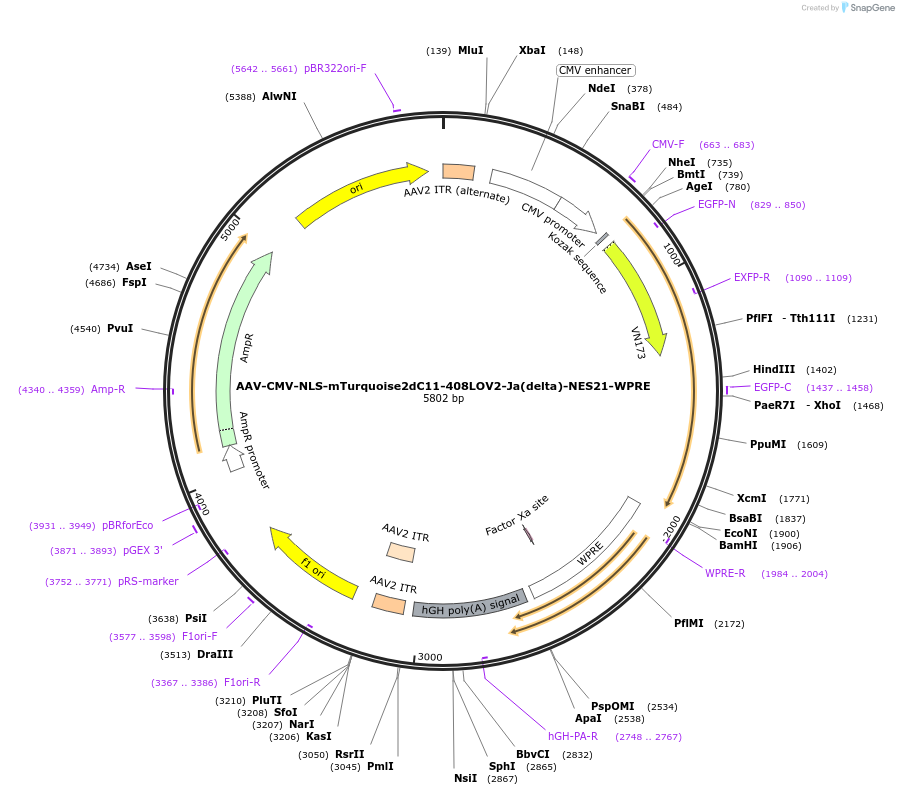
AAV-CMV-NLS-mTurquoise2dC11-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159965PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC11-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterCMV-IEAvailable SinceOct. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
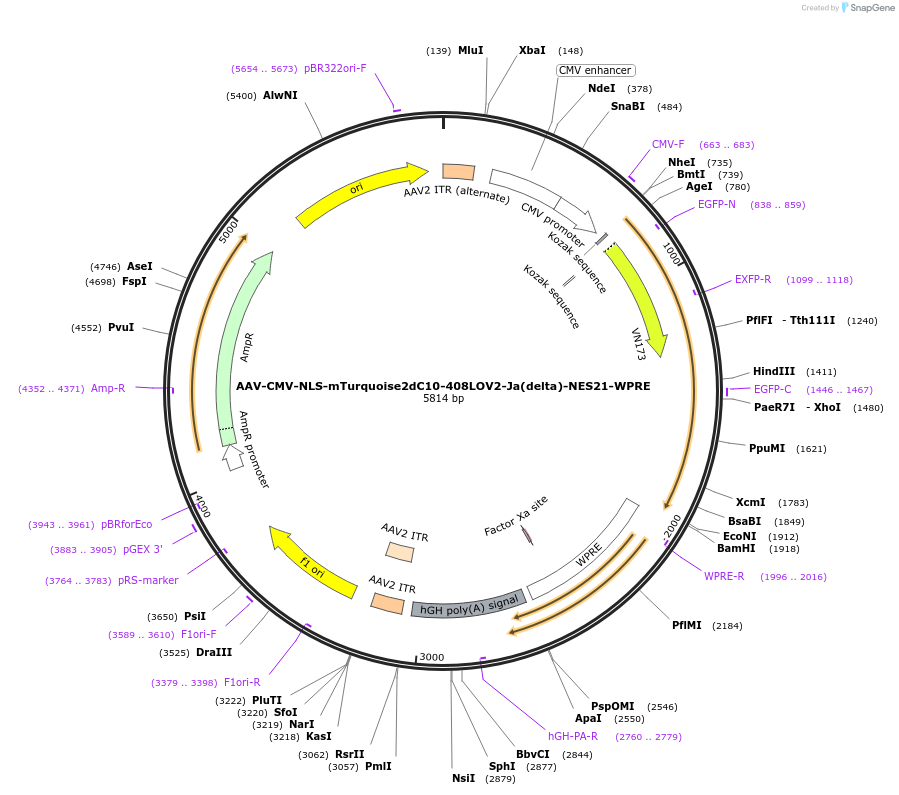
AAV-CMV-NLS-mTurquoise2dC10-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159964PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC10-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterCMV-IEAvailable SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
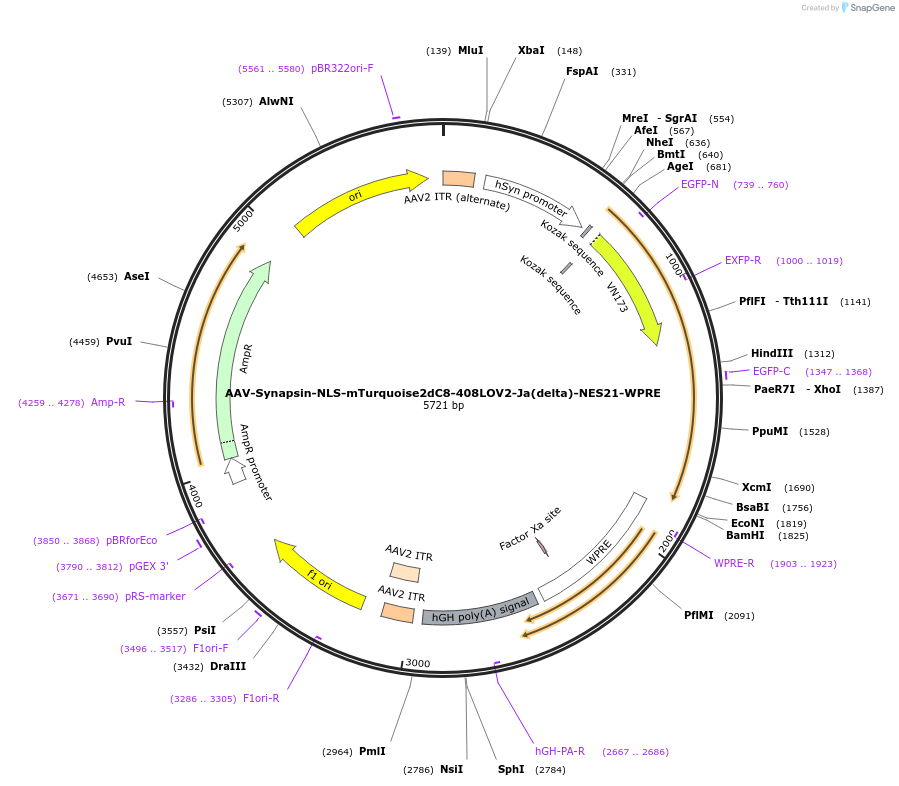
AAV-Synapsin-NLS-mTurquoise2dC8-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159958PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC8-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
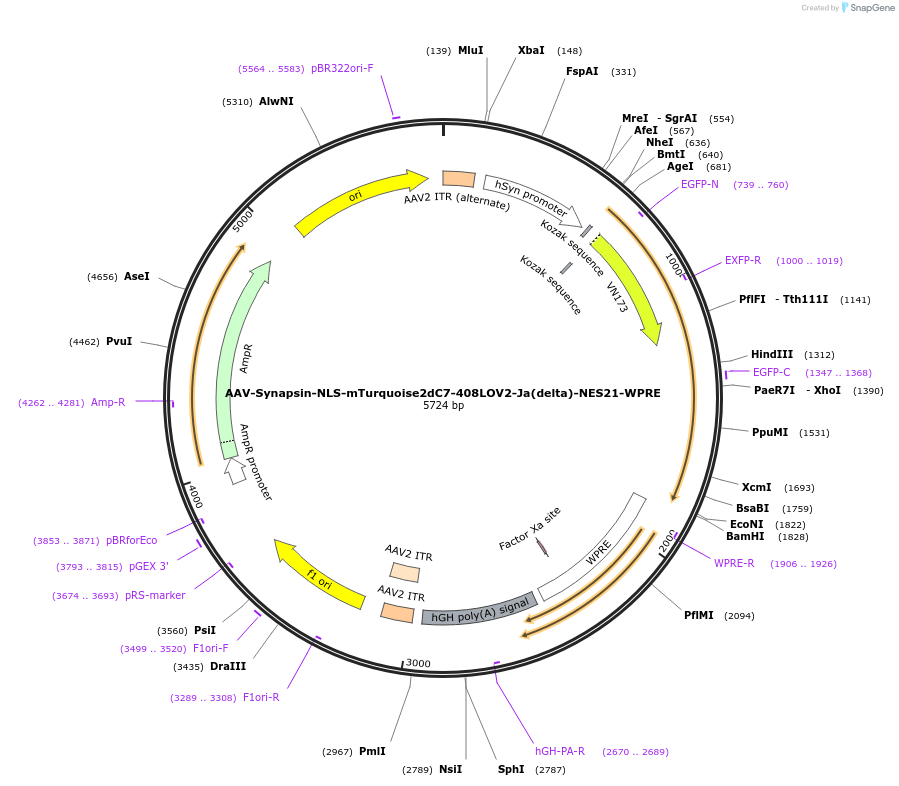
AAV-Synapsin-NLS-mTurquoise2dC7-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159957PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC7-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
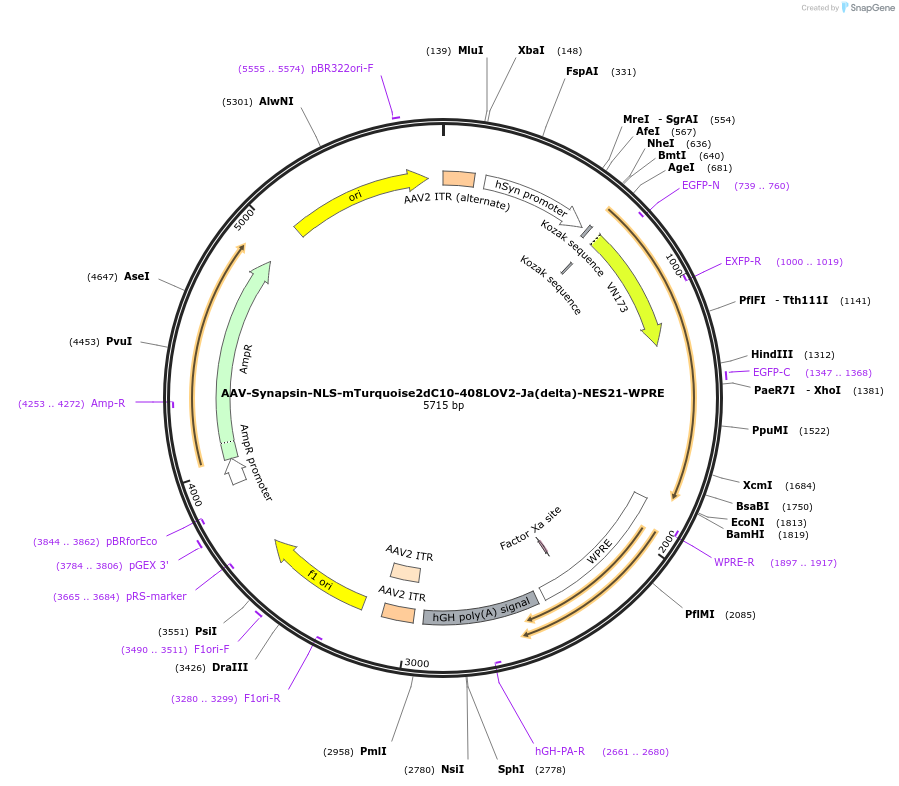
AAV-Synapsin-NLS-mTurquoise2dC10-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159960PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC10-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
AAV-Synapsin-NLS-mTurquoise2dC9-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159959PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC9-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
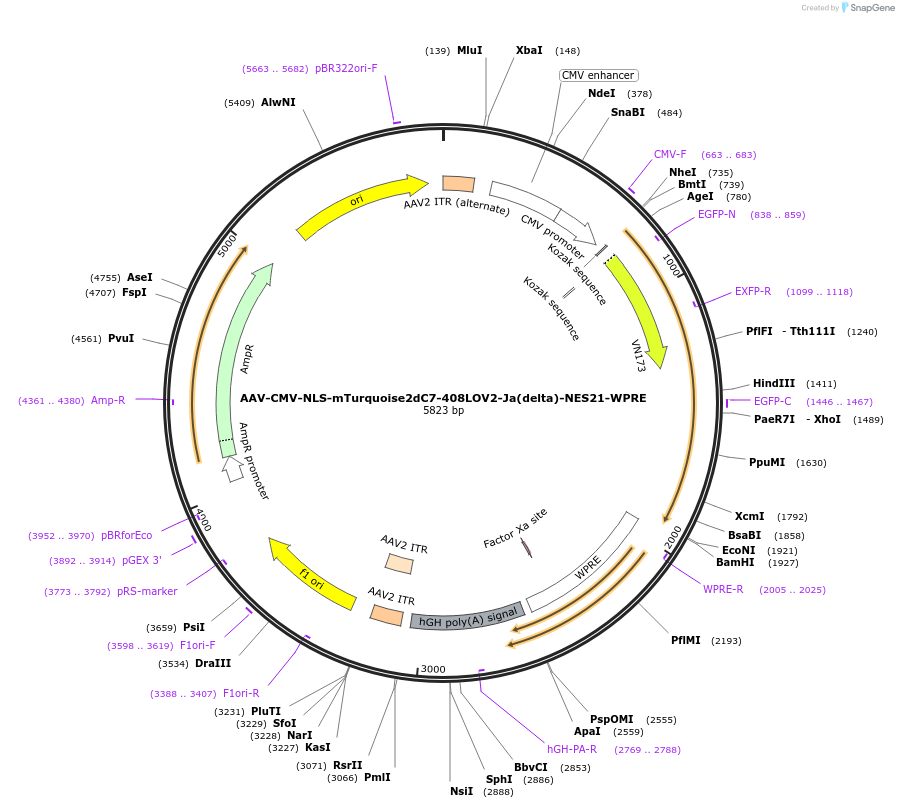
AAV-CMV-NLS-mTurquoise2dC7-408LOV2-Ja(delta)-NES21-WPRE
Plasmid#159963PurposeoptoNES construct with modified mTurquoise2-LOV2 RETDepositorInsertNLS-mTurquoise2dC7-408LOV2-Ja(delta)-NES21
UseAAVExpressionMammalianMutationsyntheticPromoterCMV-IEAvailable SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
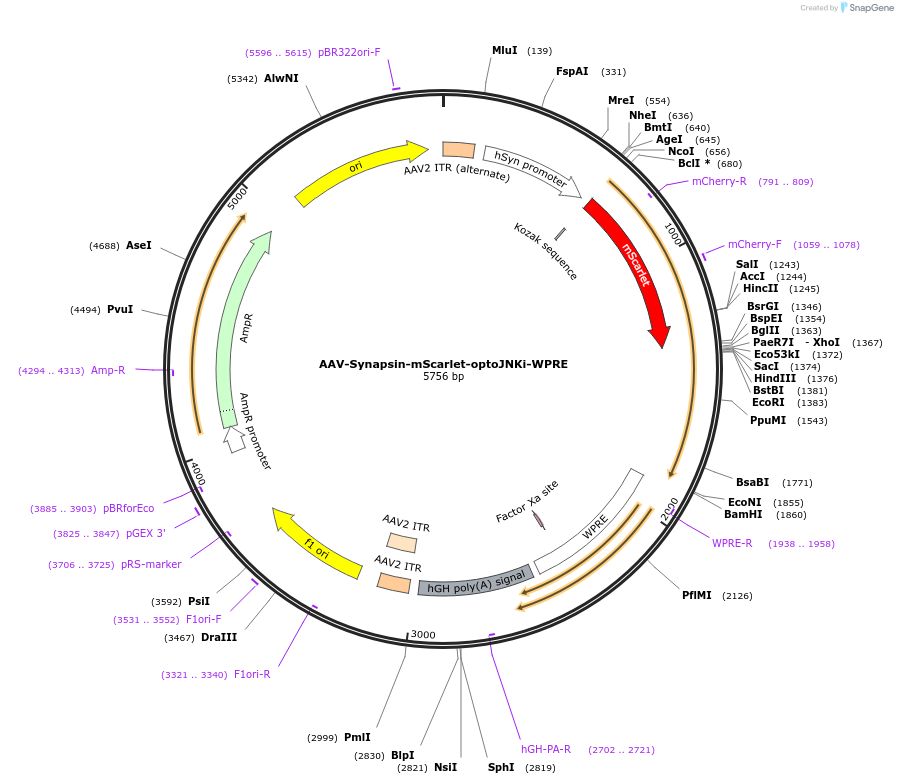
AAV-Synapsin-mScarlet-optoJNKi-WPRE
Plasmid#159968Purposeoptogenetic JNK inhibitor with increased RET from LOV2 to reduce actuation sensitivity, detectable without activationDepositorInsertmScarlet-optoJNKi
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
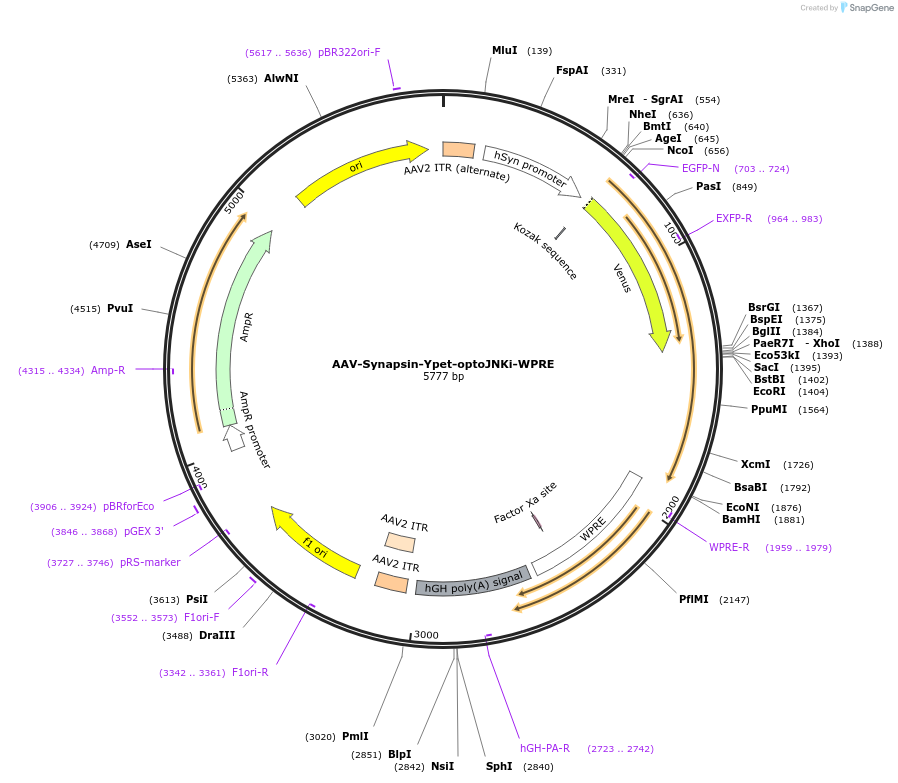
AAV-Synapsin-Ypet-optoJNKi-WPRE
Plasmid#159967Purposeoptogenetic JNK inhibitor with increased RET from LOV2 to reduce actuation sensitivity, detectable without activationDepositorInsertYpet-optoJNKi
UseAAVExpressionMammalianMutationsyntheticPromoterhSyn1Available SinceOct. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
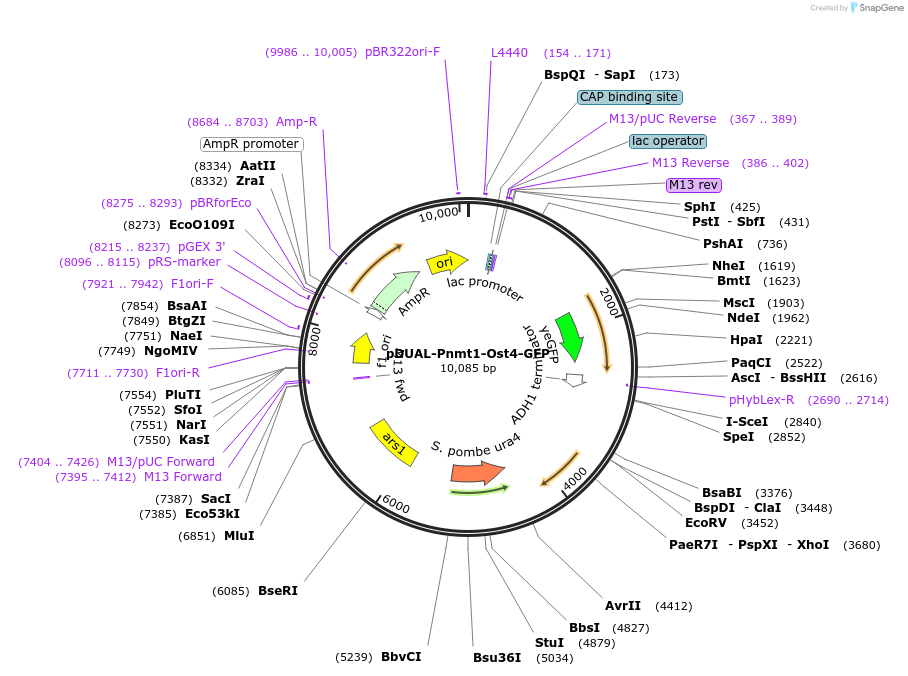
pDUAL-Pnmt1-Ost4-GFP
Plasmid#154368PurposeYeast expression of Ost4DepositorInsertOst4 (ost4 Fission Yeast)
ExpressionYeastAvailable SinceOct. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
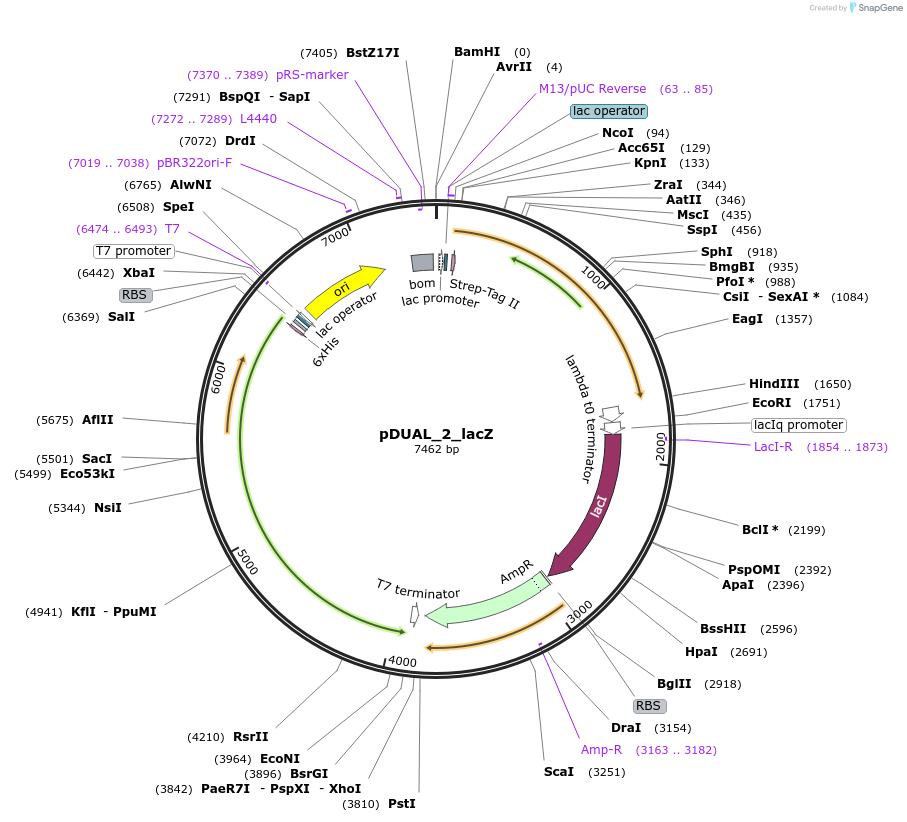
pDUAL_2_lacZ
Plasmid#155173Purposesee pDUAL_lacZ: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceOct. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
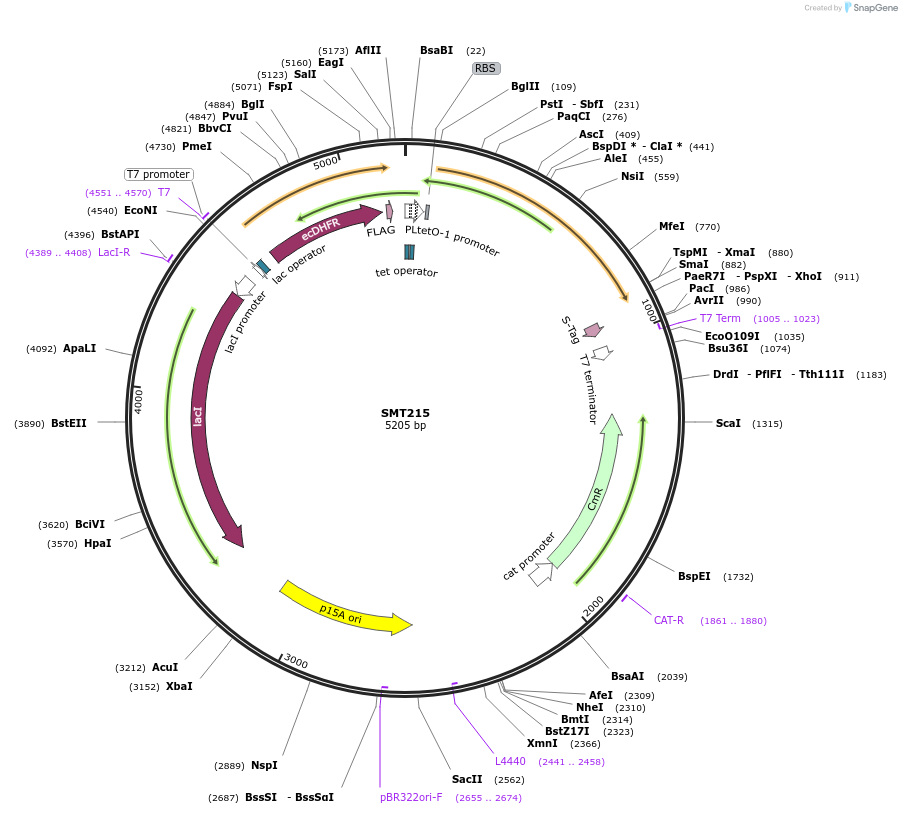
SMT215
Plasmid#134818PurposeSMT205 with DHFR-FLAG-tag, western blotDepositorInsertsDihydrofolate reductase
Thymidylate synthase
TagsFLAGExpressionBacterialMutationSilent mutations to eliminate internal restrictio…PromoterT7 and TETAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
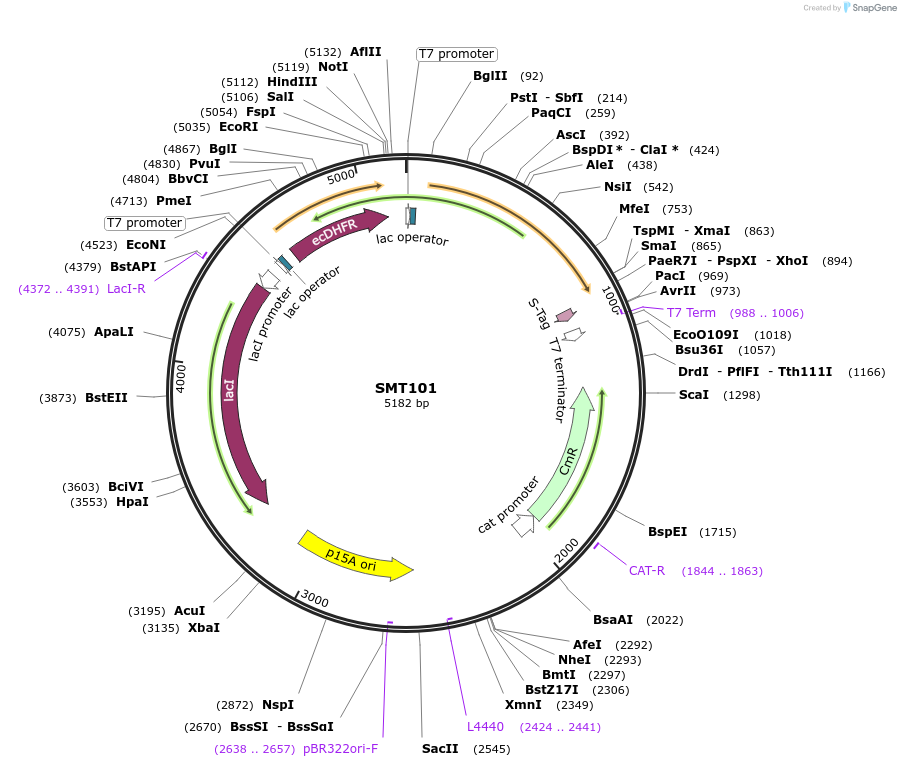
SMT101
Plasmid#134815PurposeDual expression of DHFR and TYMS, in vivo assaysDepositorInsertsDihydrofolate reductase
Thymidylate synthase
ExpressionBacterialMutationSilent mutations to eliminate internal restrictio…PromoterT7Available SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
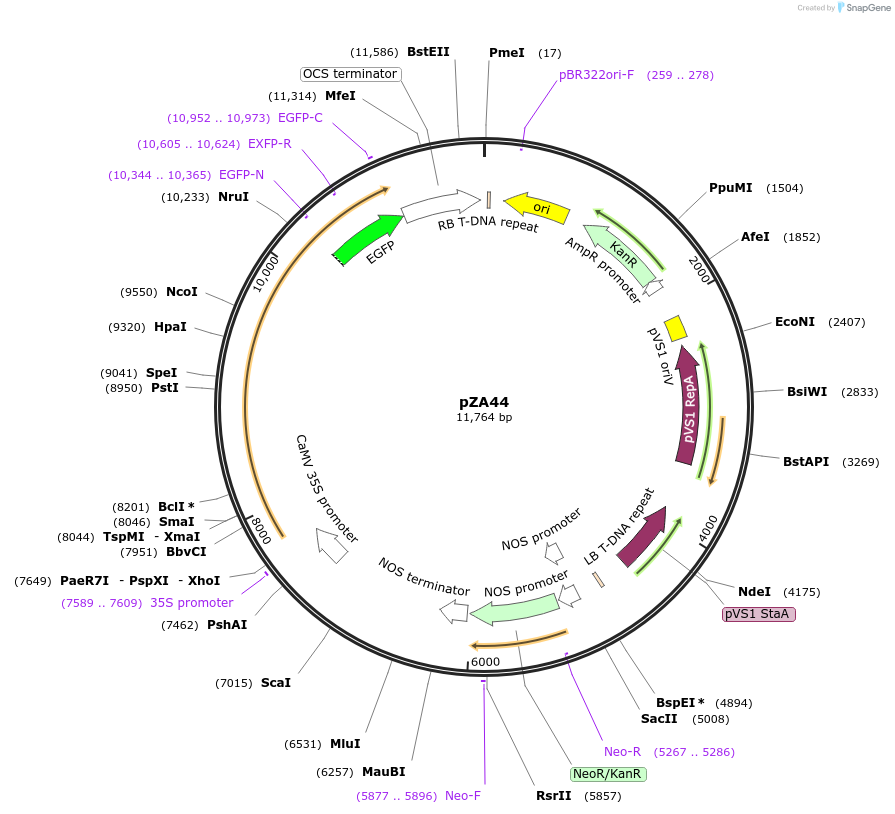
pZA44
Plasmid#158522PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V/L17E-eGFP. Km plant selection marker.DepositorAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
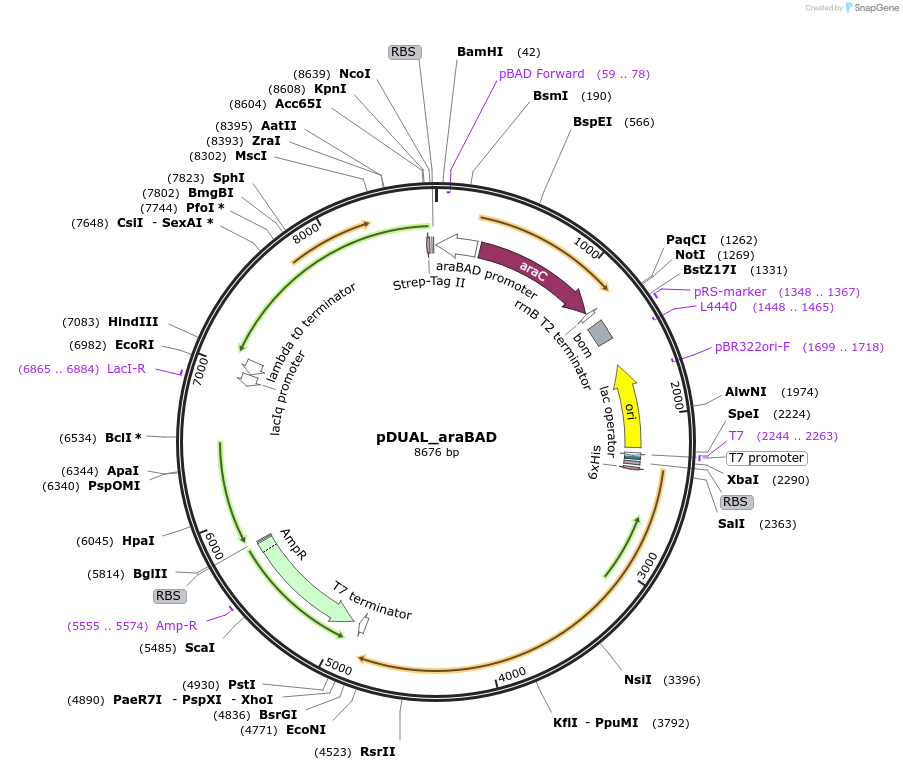
pDUAL_araBAD
Plasmid#155172PurposeBaSP and CuCBP co-expression: araBAD promoter controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
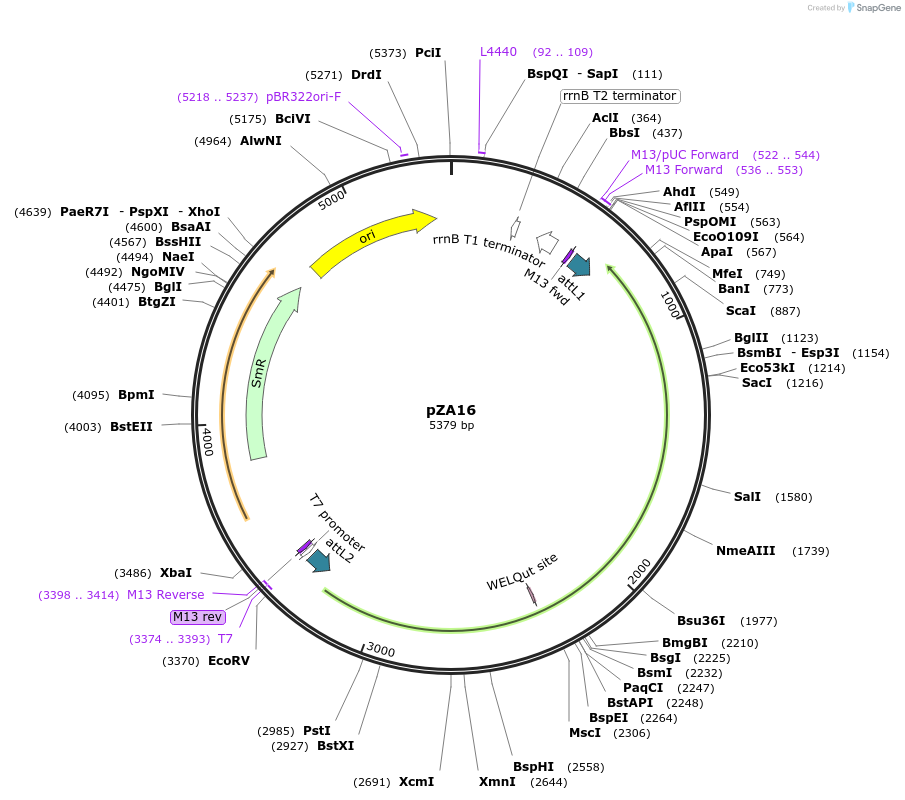
pZA16
Plasmid#158494PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V/L17E without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, D481V, no STOP codonAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
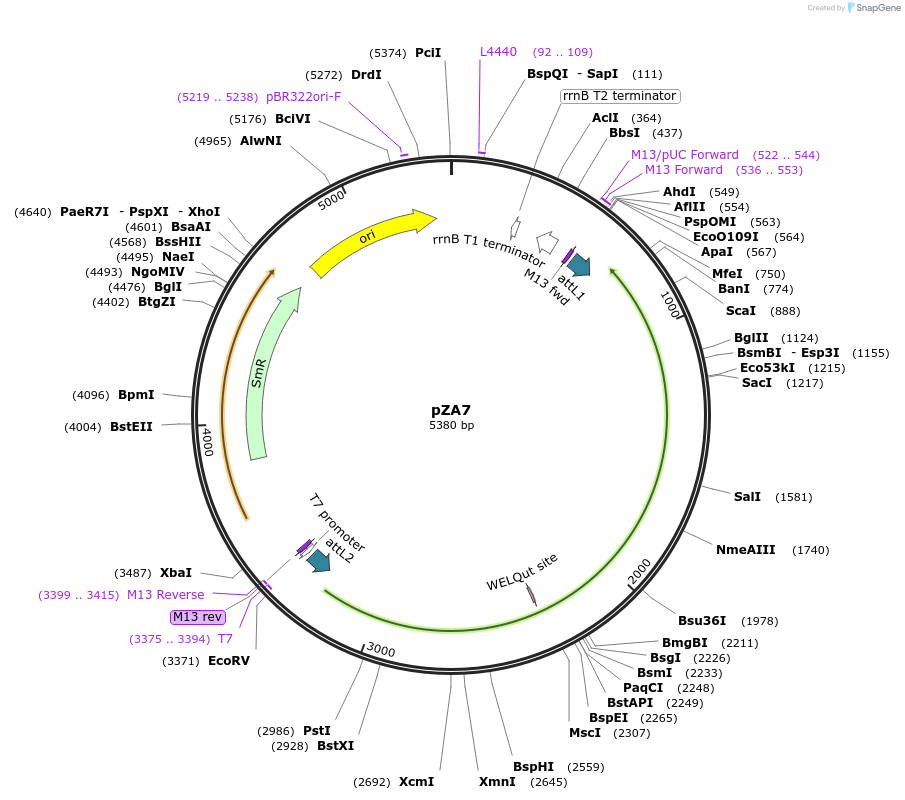
pZA7
Plasmid#158485PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
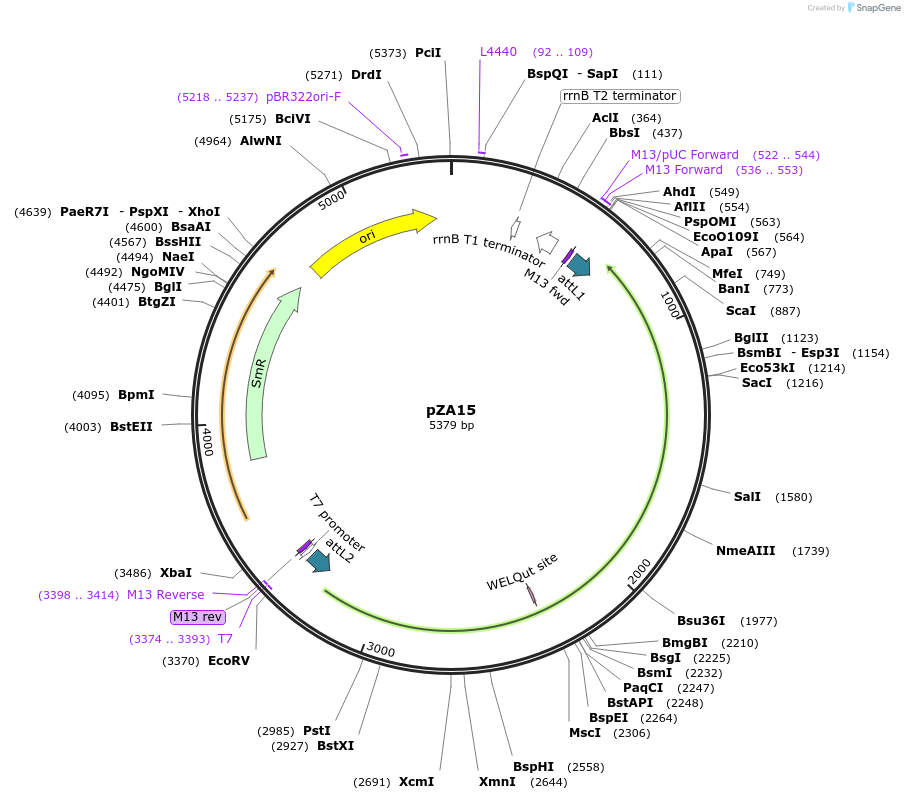
pZA15
Plasmid#158493PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, no STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
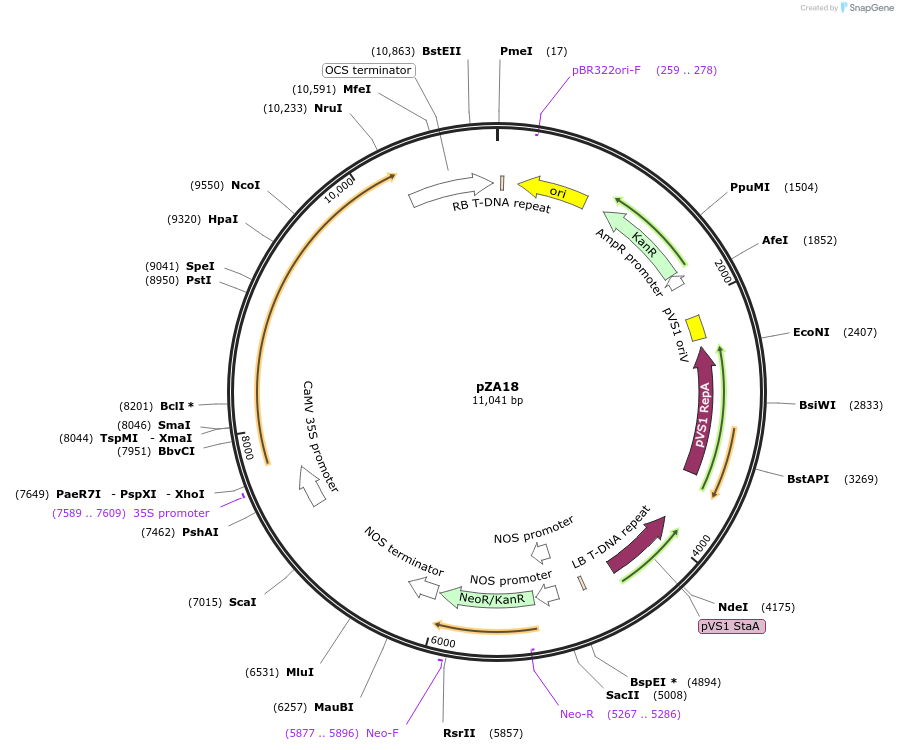
pZA18
Plasmid#158496PurposeIn planta gene expression of Golden gate compatibleA. thaliana ZAR1 D489V. Km plant selection marker.DepositorAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
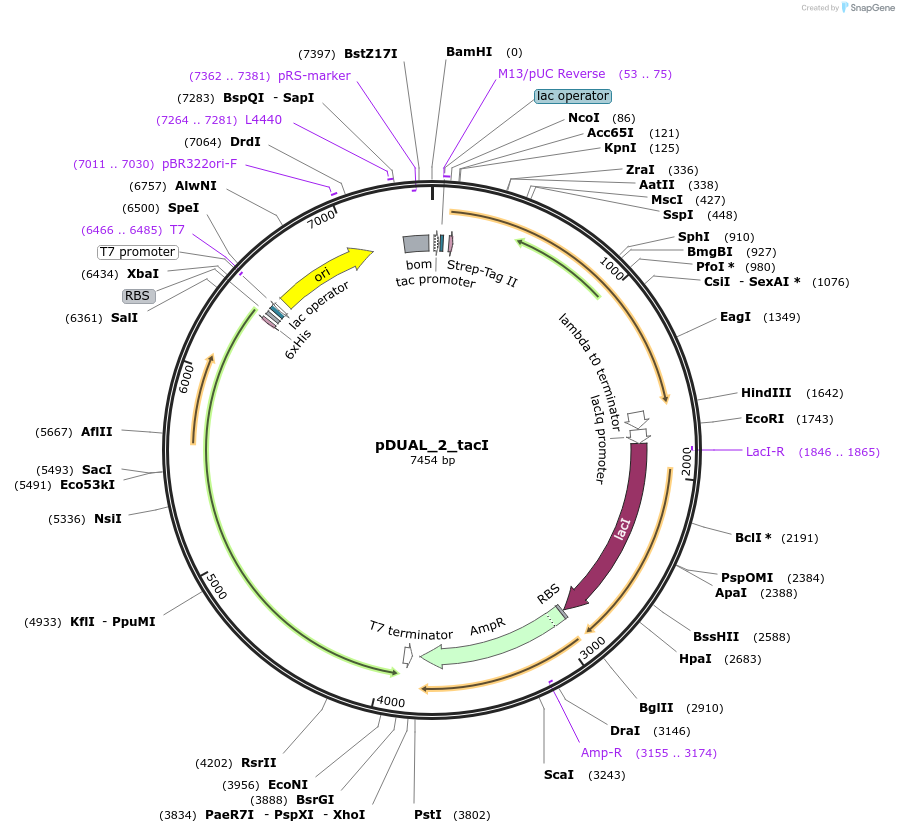
pDUAL_2_tacI
Plasmid#155174Purposesee pDUAL_tacI: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
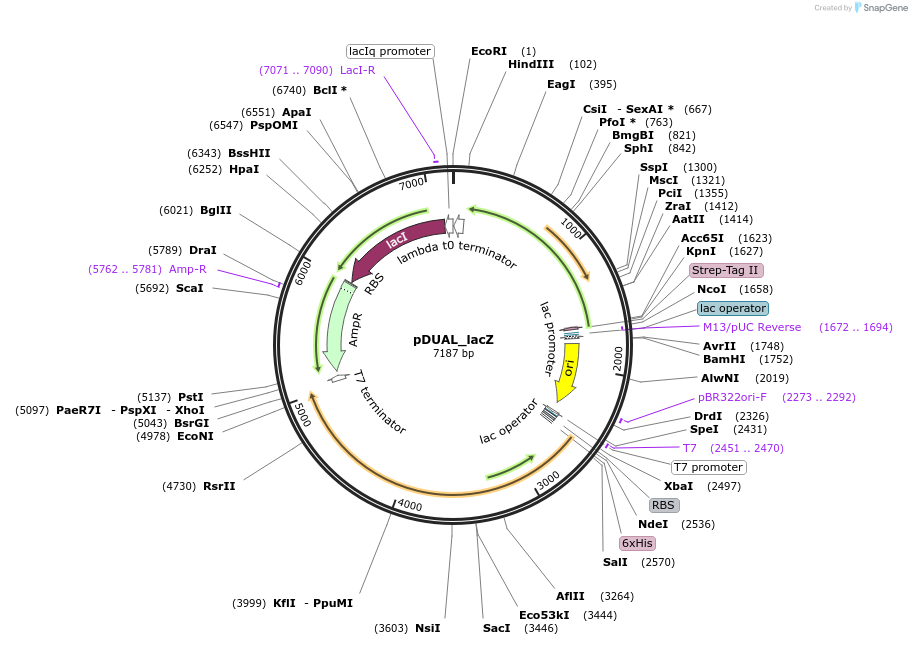
pDUAL_lacZ
Plasmid#155169PurposeBaSP and CuCBP co-expression: lacZ promoter controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
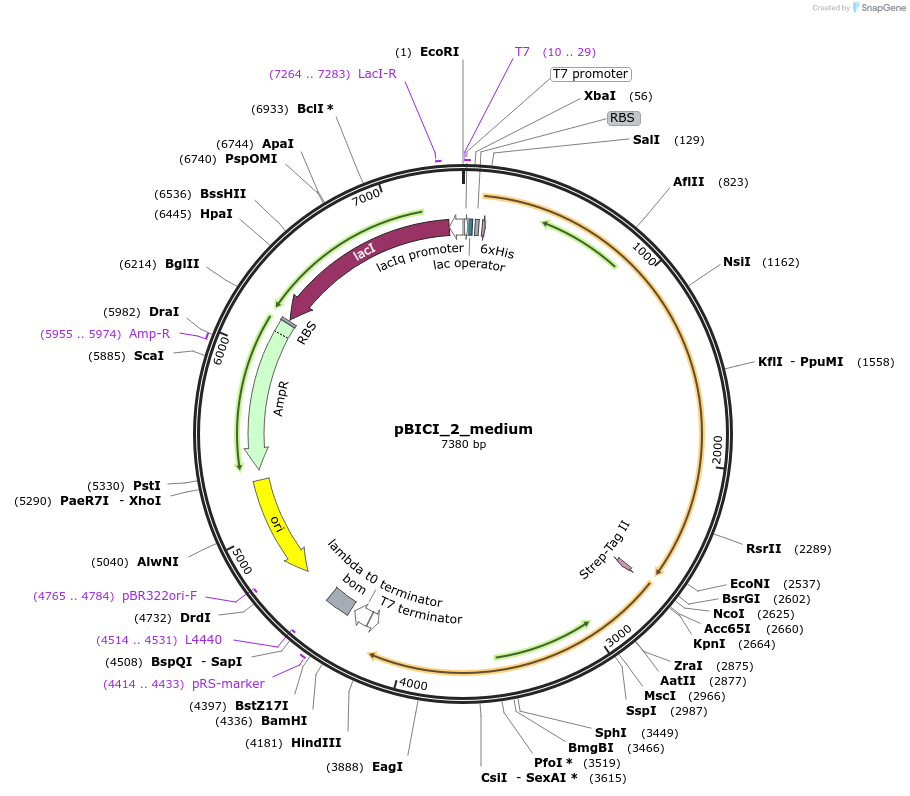
pBICI_2_medium
Plasmid#155167Purposesee pBICI_medium: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
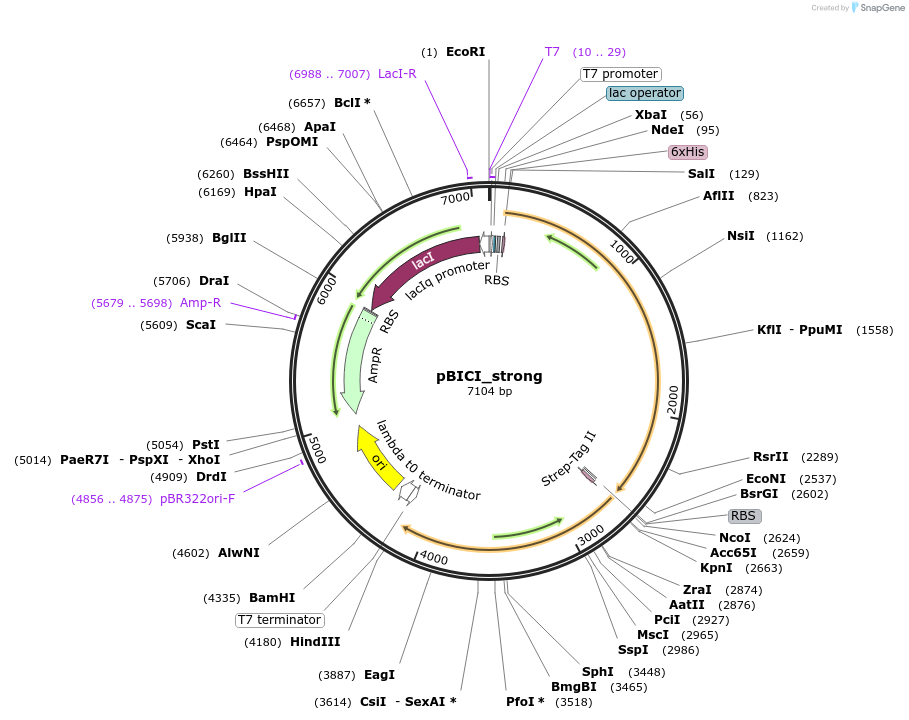
pBICI_strong
Plasmid#155165PurposeBaSP and CuCBP co-expression: strong RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
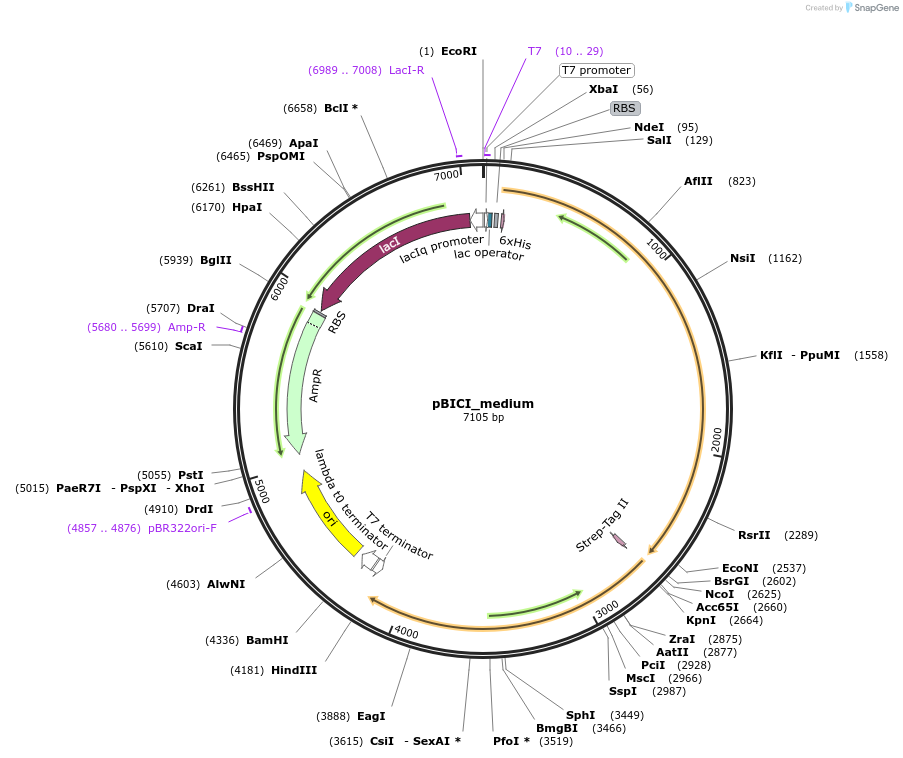
pBICI_medium
Plasmid#155164PurposeBaSP and CuCBP co-expression: medium RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
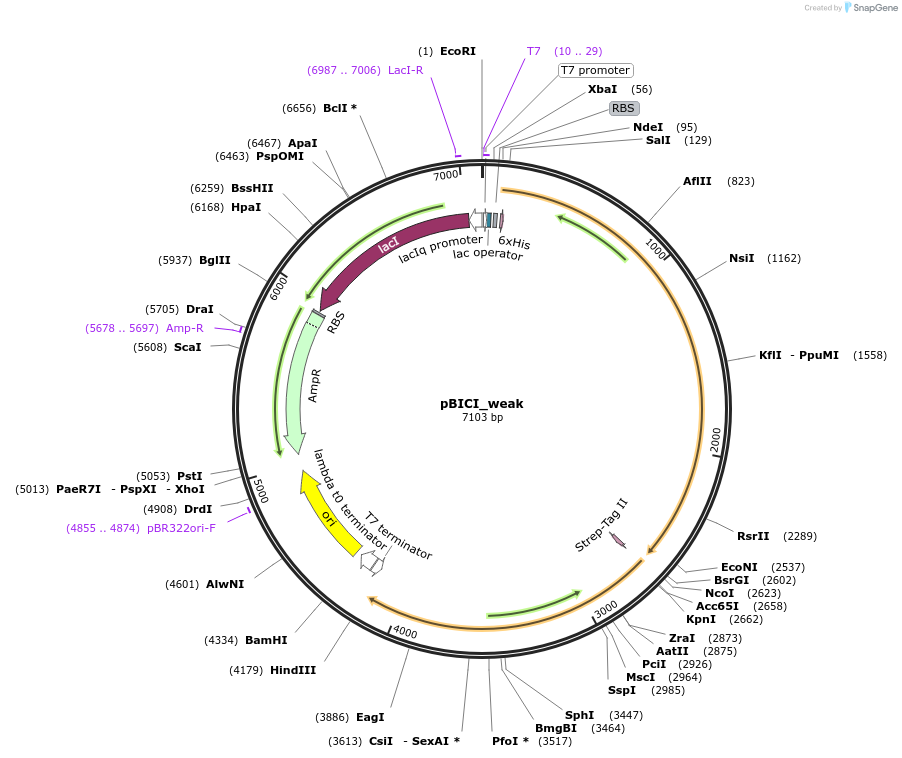
pBICI_weak
Plasmid#155163PurposeBaSP and CuCBP co-expression: weak RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
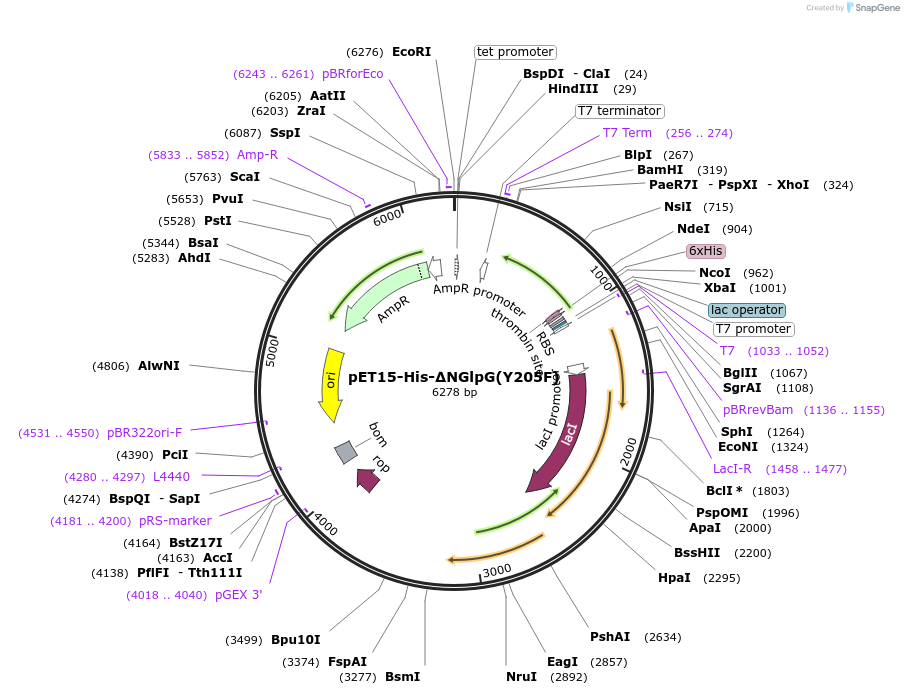
pET15-His-∆NGlpG(Y205F)
Plasmid#155132PurposeExpresses N-terminally His-tagged E. coli GlpG Y205F variant (residues 87-276) from pET15bDepositorInsertGlpG (glpG Escherichia coli)
TagsHisExpressionBacterialMutationY205F, insert codes for GlpG residues 87-276PromoterT7Available SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
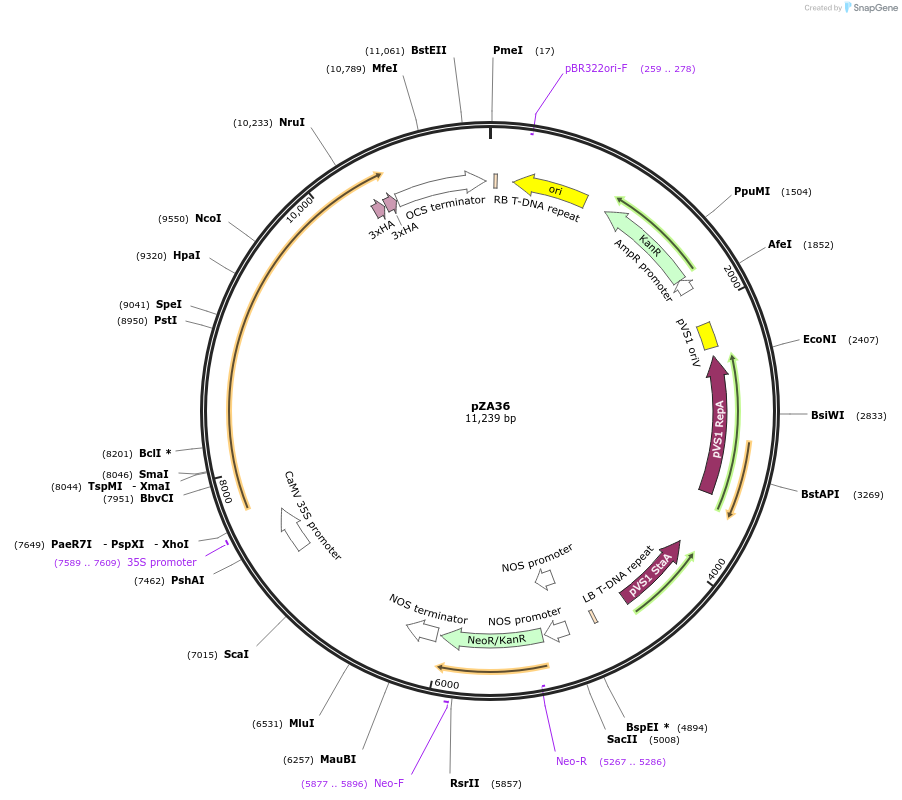
pZA36
Plasmid#158514PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V/L17E-6xHA. Km plant selection marker.DepositorAvailable SinceSept. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
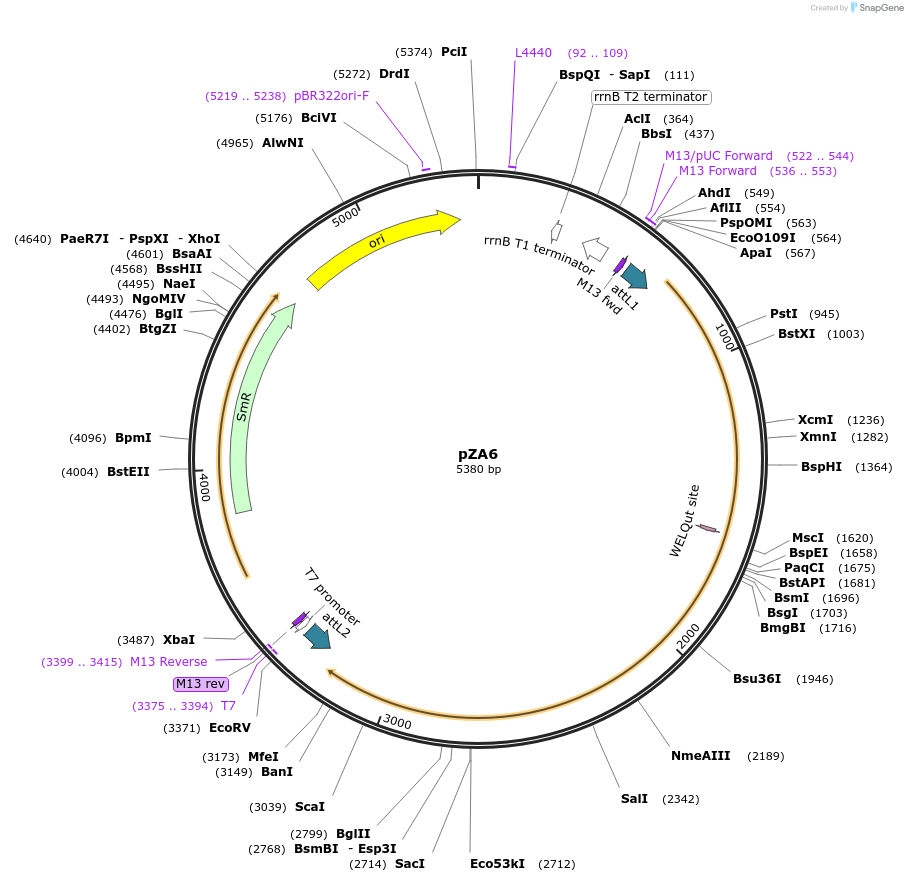
pZA6
Plasmid#158484PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, STOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
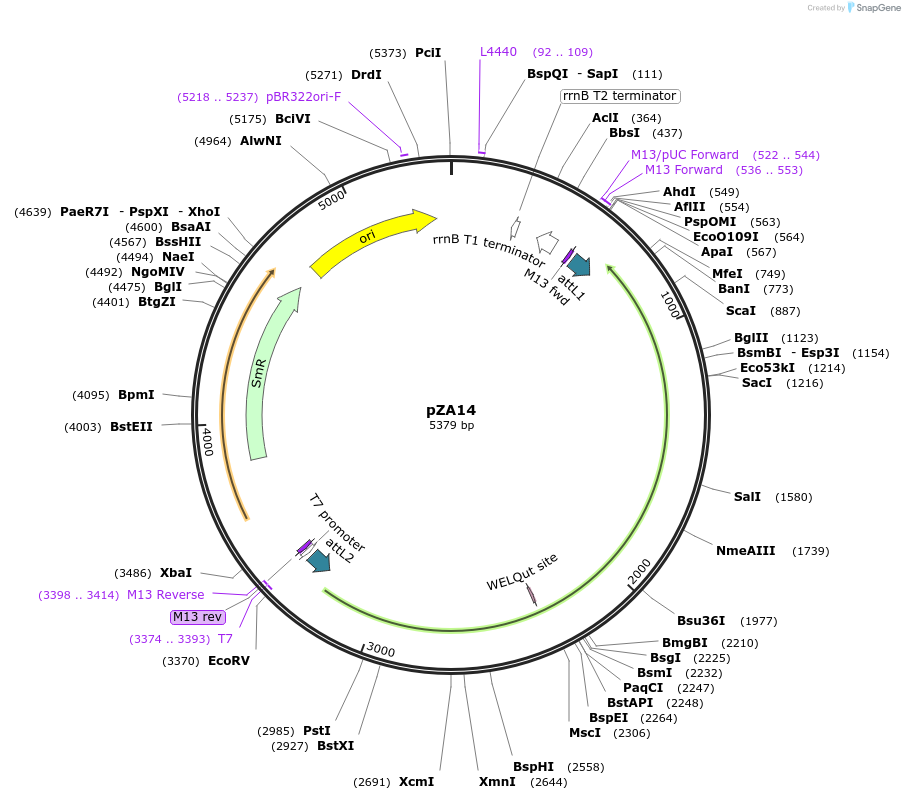
pZA14
Plasmid#158492PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 D481V without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationD481V, no STOP codonAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
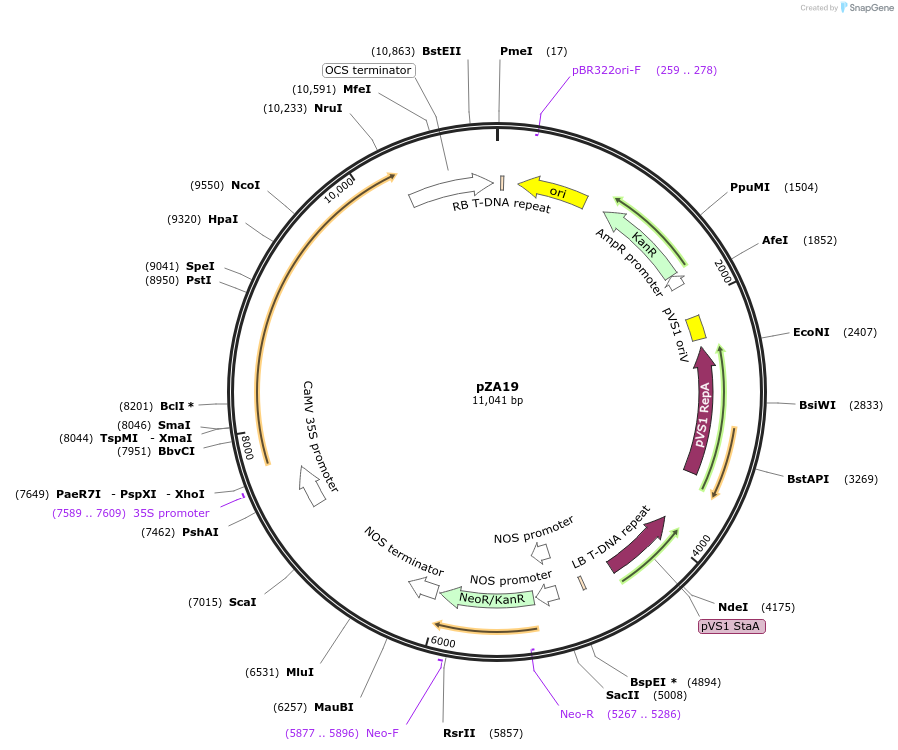
pZA19
Plasmid#158497PurposeIn planta gene expression of Golden gate compatlible A. thaliana ZAR1 L17E. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
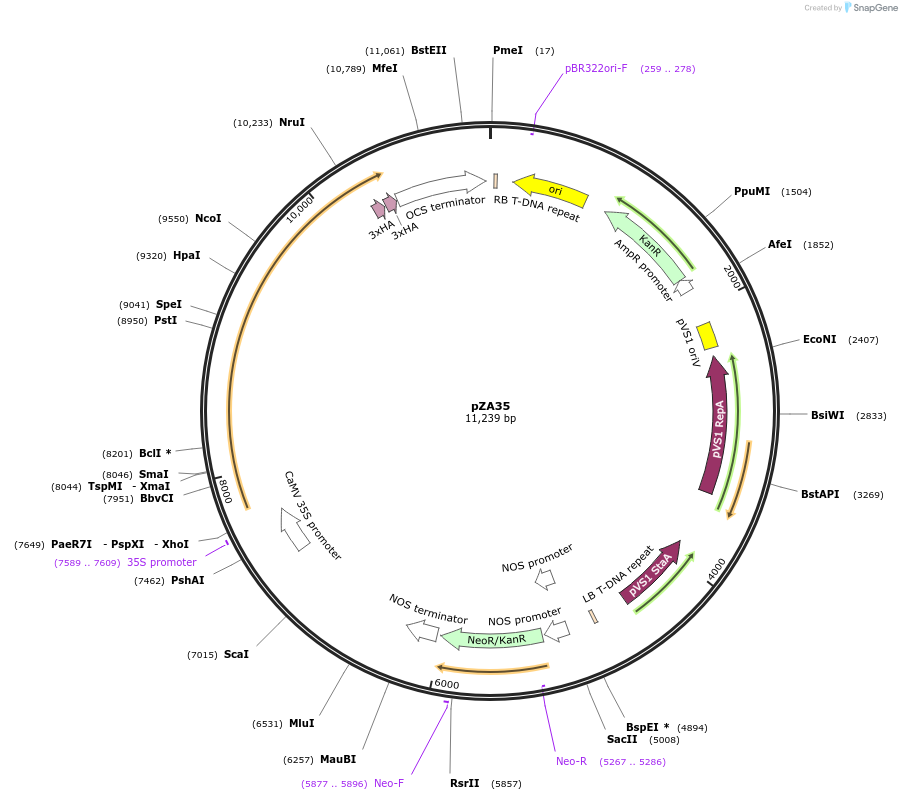
pZA35
Plasmid#158513PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-6xHA. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
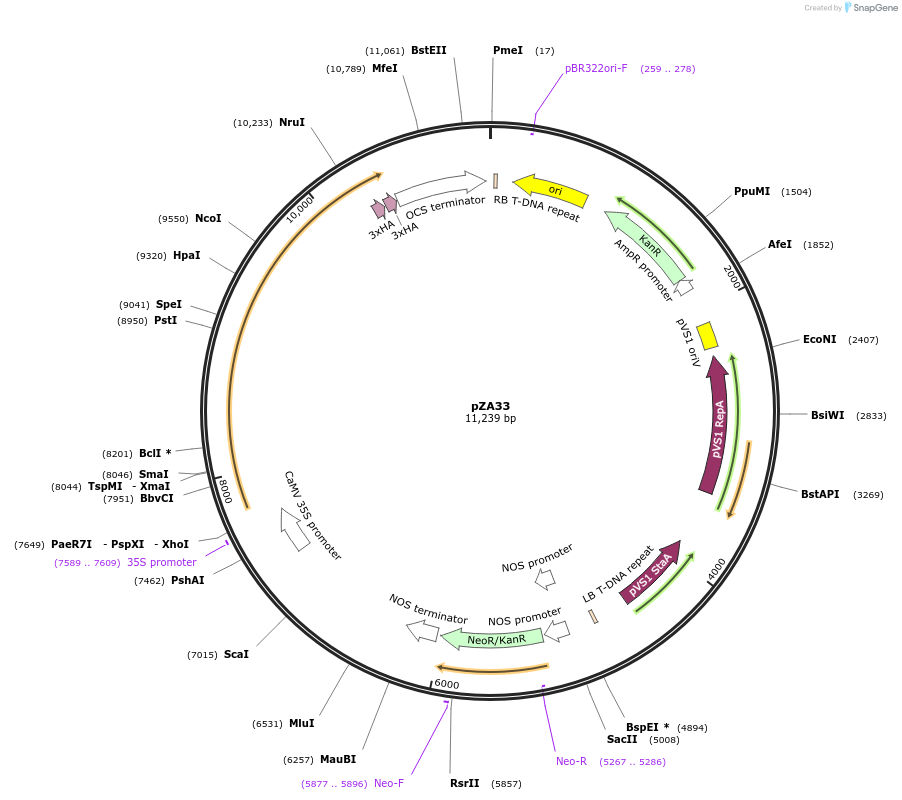
pZA33
Plasmid#158511PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1-6xHA. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
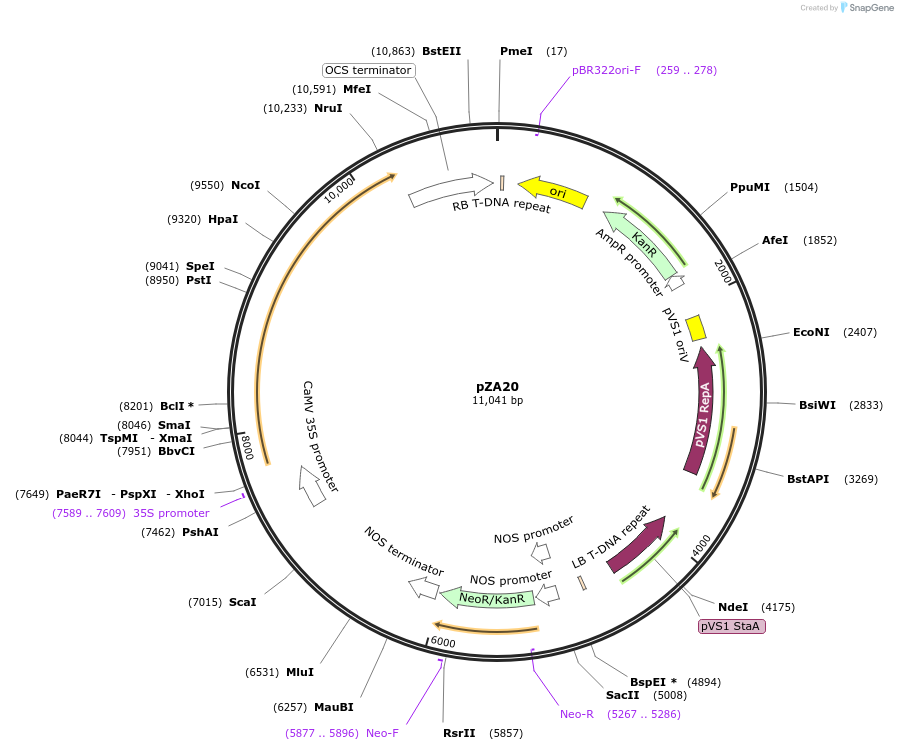
pZA20
Plasmid#158498PurposeIn planta gene expression of Golden gate complatible A. thaliana ZAR1 D498V/L17E. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
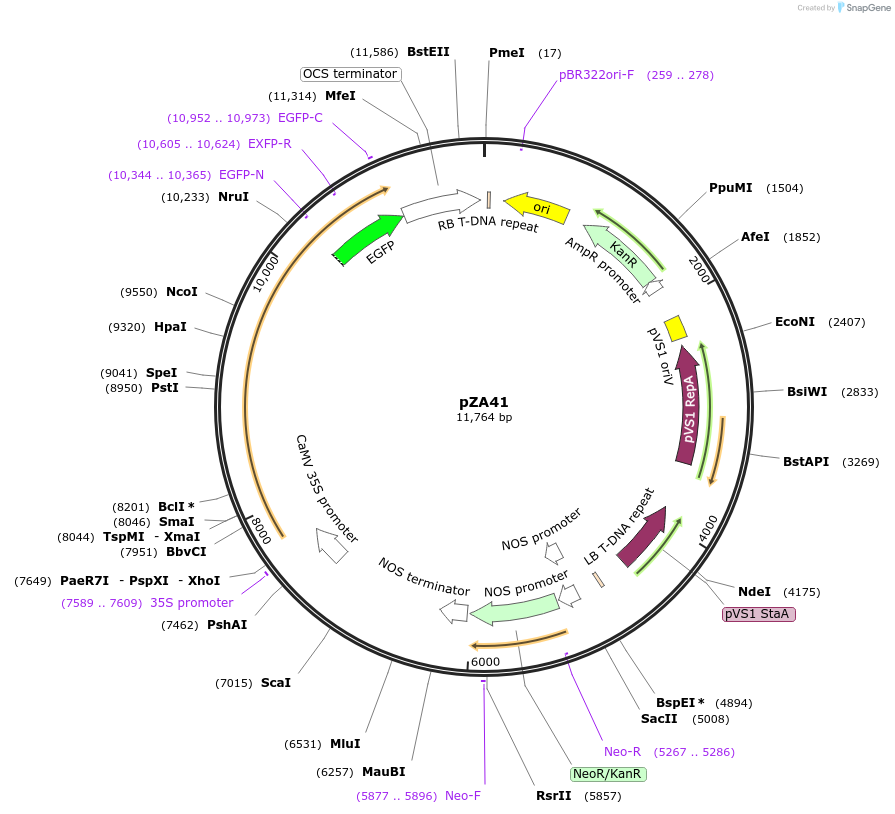
pZA41
Plasmid#158519PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
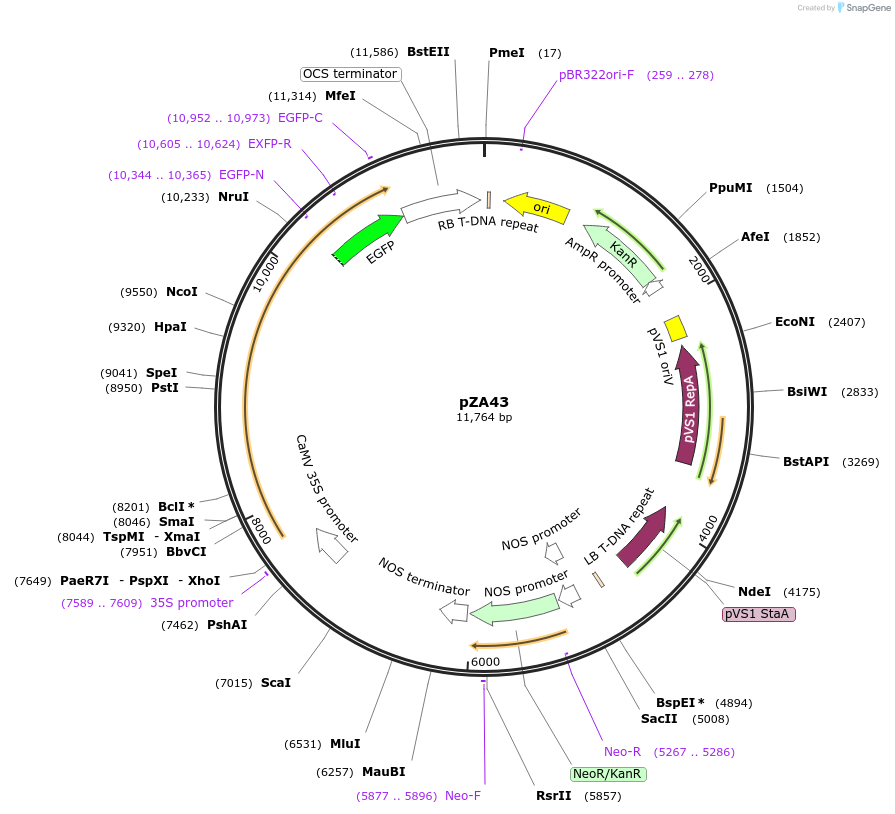
pZA43
Plasmid#158521PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
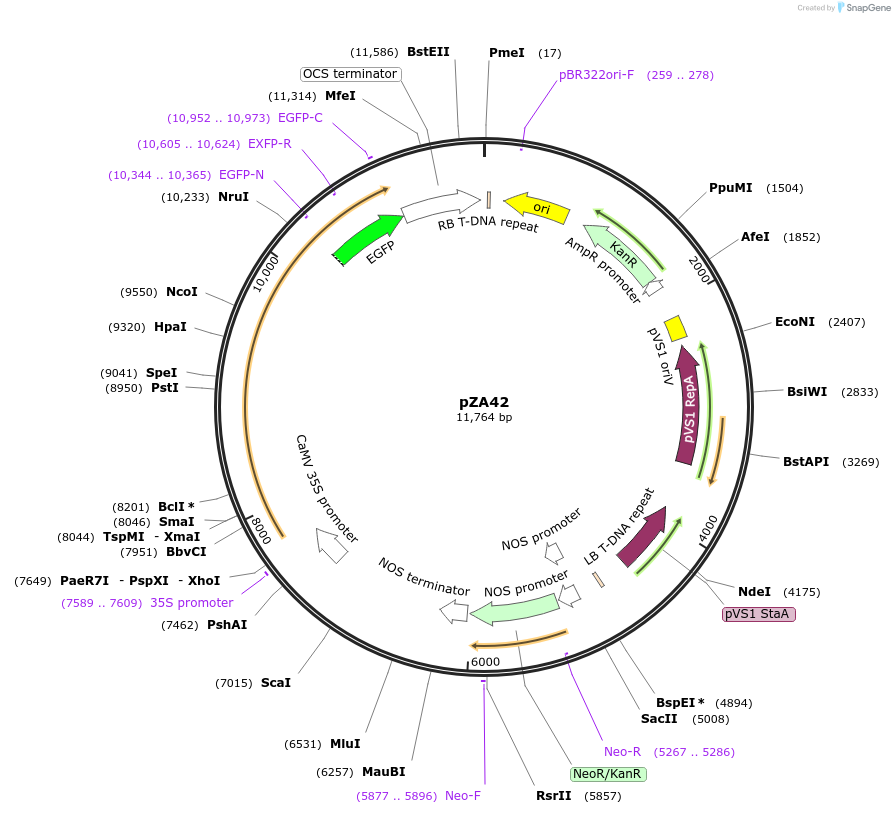
pZA42
Plasmid#158520PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V-eGFP. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only