We narrowed to 8,782 results for: set
-
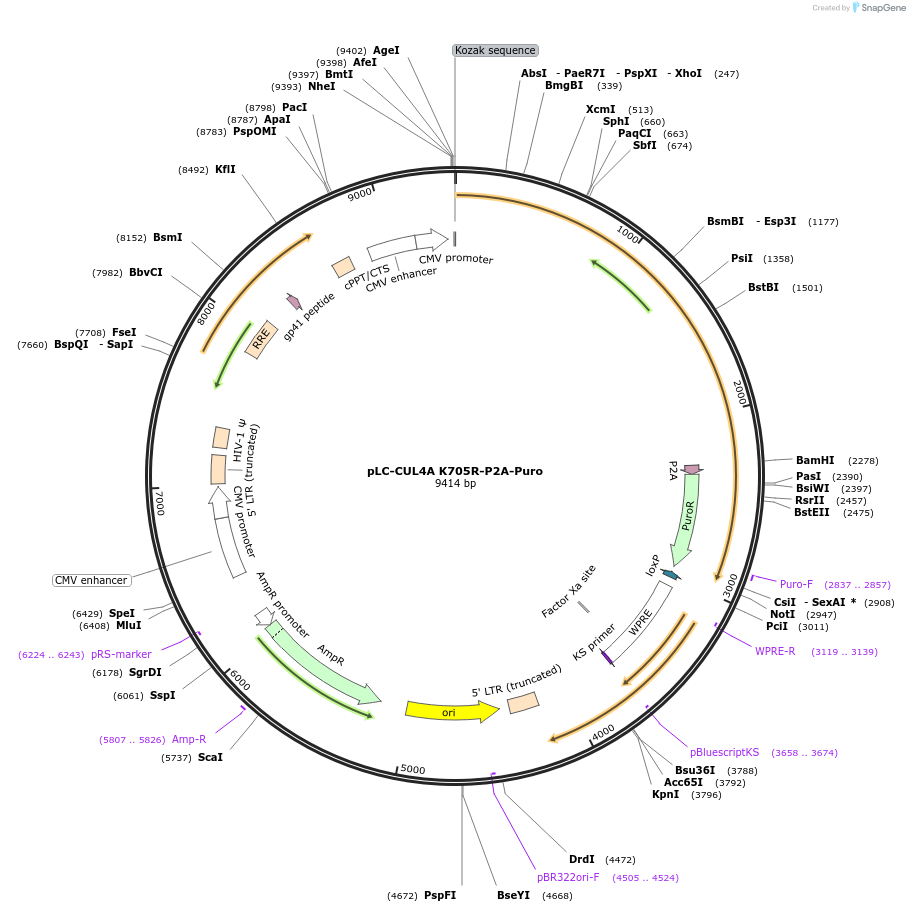
Plasmid#124305PurposeLentiviral vector for expression of Nedd8 attachment mutant K705R CUL4A-P2A-Puro casette from a CMV promoterDepositorInsertCUL4A (CUL4A Human)
UseLentiviralExpressionMammalianMutationchanged Lysine 705 to ArgininePromoterCMVAvailable SinceJuly 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
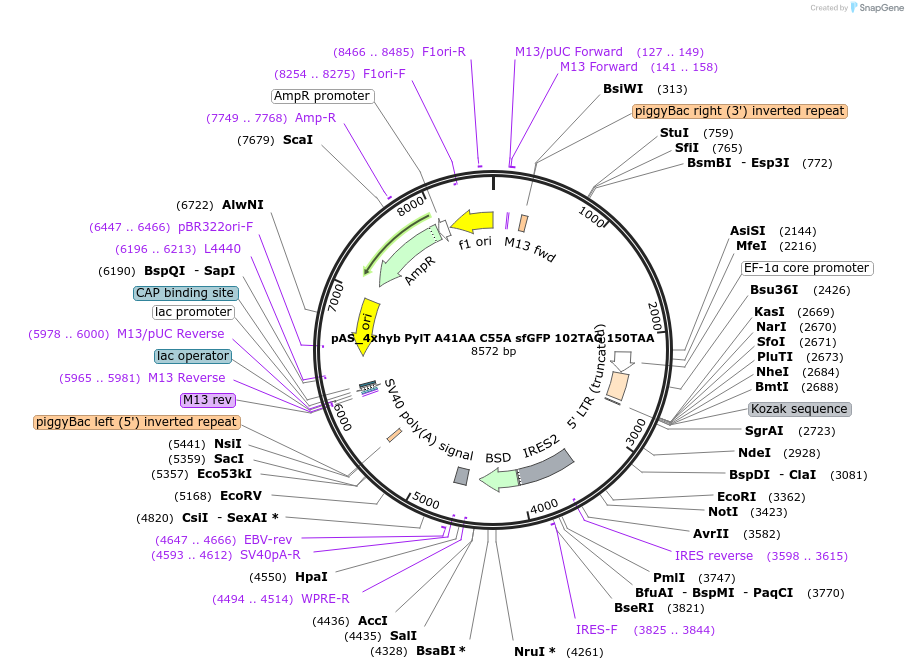
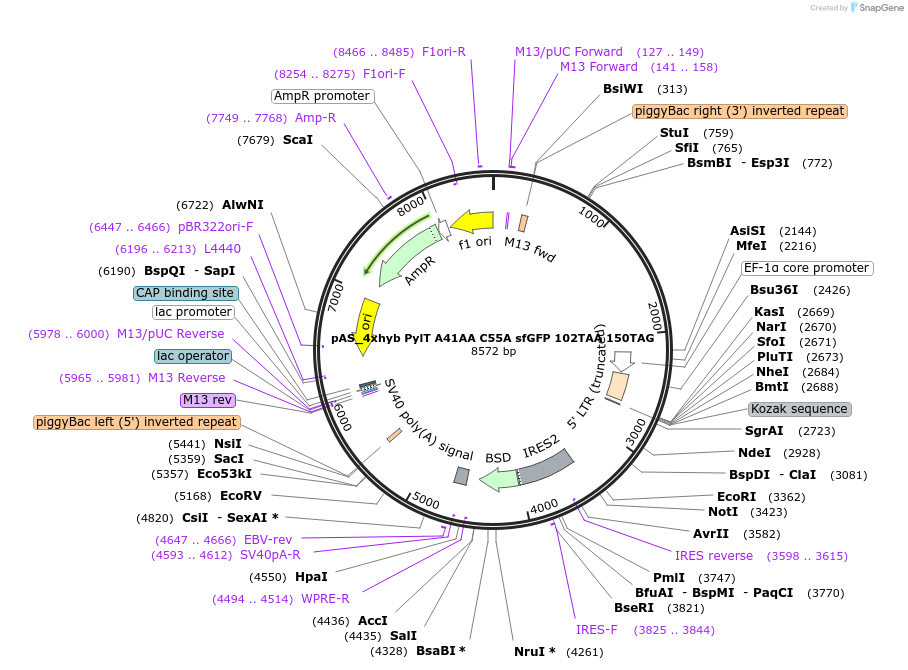
pAS_4xhyb PylT A41AA C55A sfGFP 102TAG 150TAA
Plasmid#154776Purposeplasmid with 4xhybrid PylT cassette (mutant A41AA C55A) and dual suppression reporter sfGFP 102TAG 150 TAA stop, for transient transfection or stable, piggybac-mediated, integrationDepositorInsertsfGFP
ExpressionMammalianMutation102TAG 150TAA in GFP reporter, hybrid PylT with A…PromoterEF1Available SinceOct. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
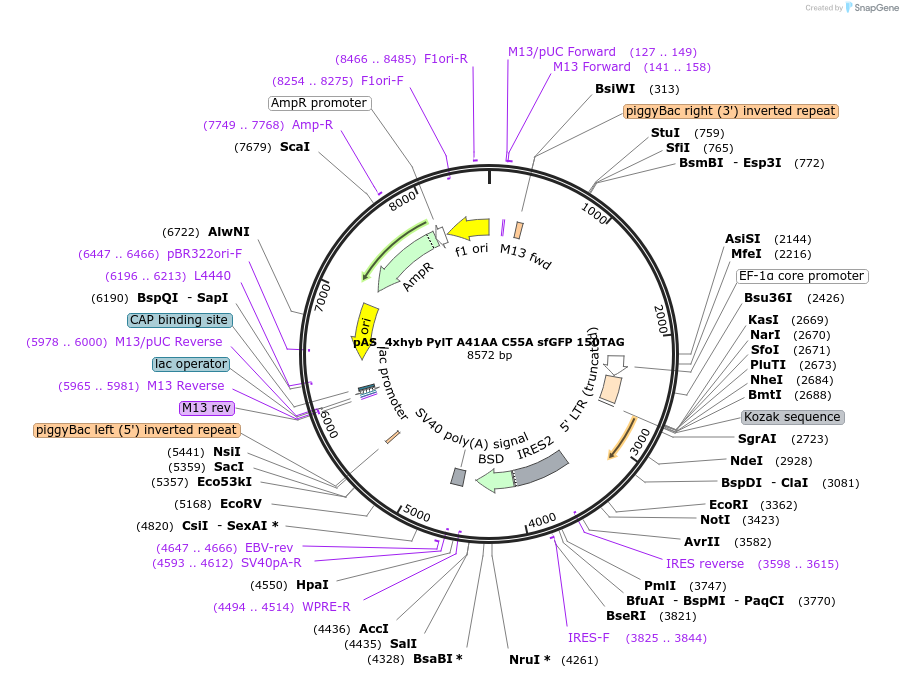
pAS_4xhyb PylT A41AA C55A sfGFP 150TAG
Plasmid#154772Purposeplasmid with 4xhybrid PylT cassette (Mx1201 G1 PylT hybrid mutant A41AA C55A) and amber suppression reporter sfGFP 150 TAG stop, for transient transfection or stable, piggybac-mediated, integrationDepositorInsertsfGFP
ExpressionMammalianMutation150TAG in GFP reporter, hybrid PylT with A41AA an…PromoterEF1Available SinceOct. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
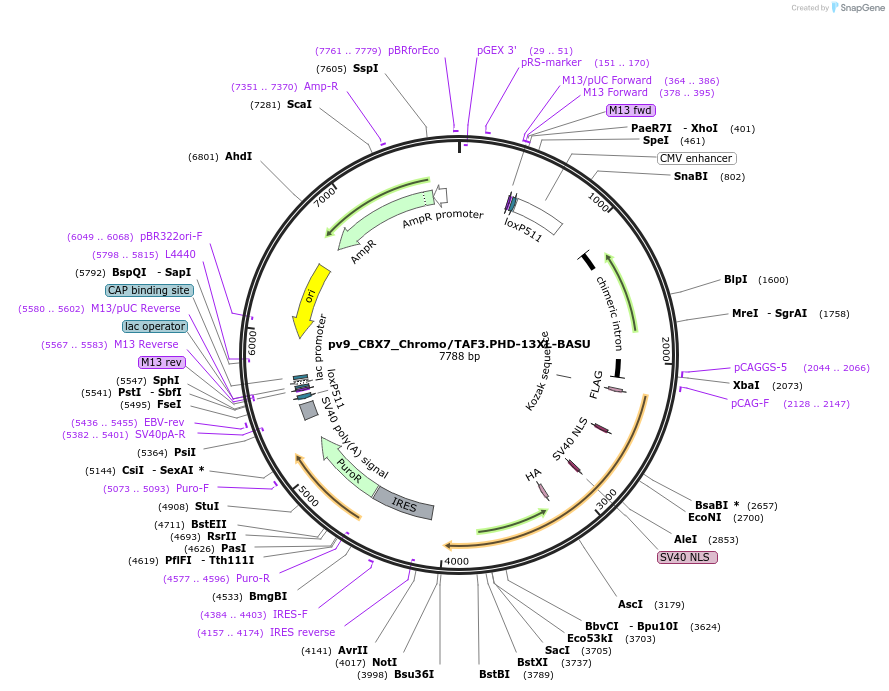
pv9_CBX7_Chromo/TAF3.PHD-13XL-BASU
Plasmid#179415PurposeCell line generation via recombination-mediated cassette exchange (RMCE) and stable expression of CBX7.Chromo/TAF3.PHD-13XL-BASUDepositorInsertCBX7.Chromo/TAF3.PHD-13XL-BASU
ExpressionMammalianPromoterCAGGSAvailable SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
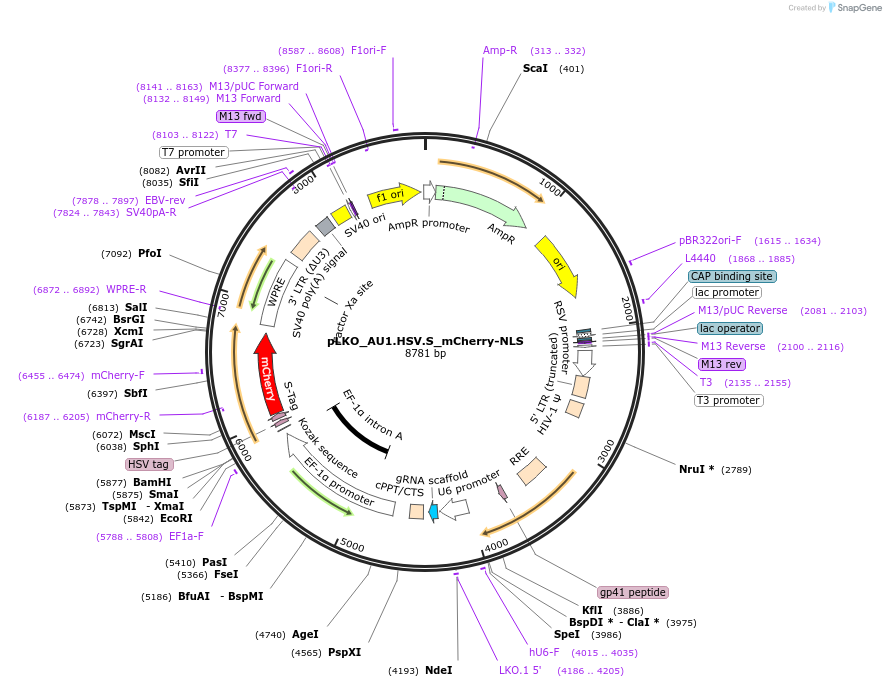
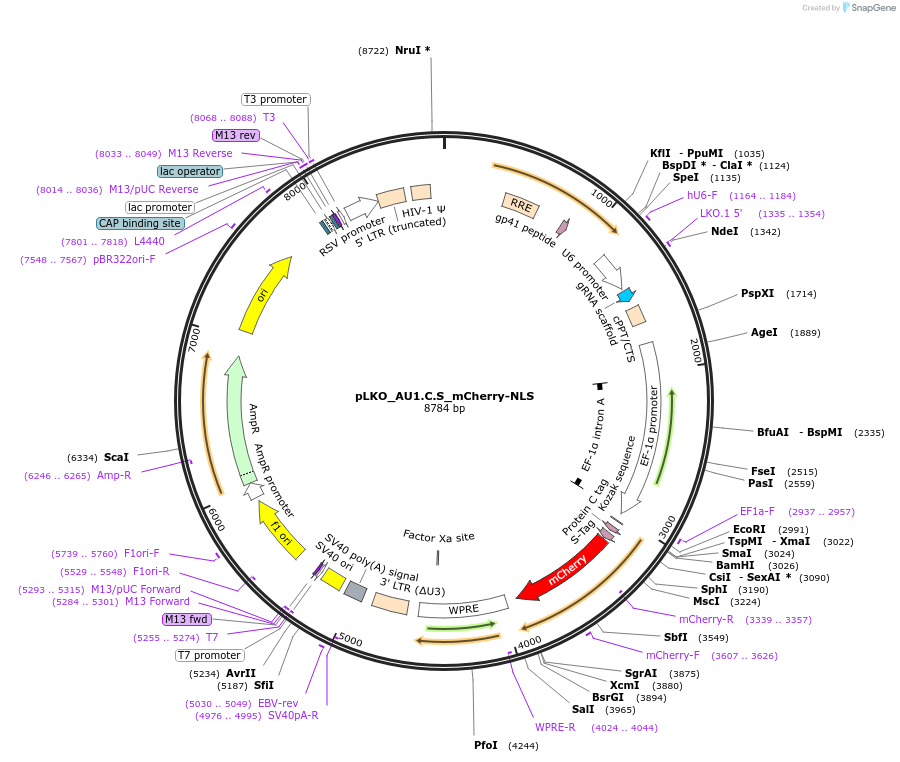
pLKO_AU1.HSV.S_mCherry-NLS
Plasmid#178224PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.HSV.S and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
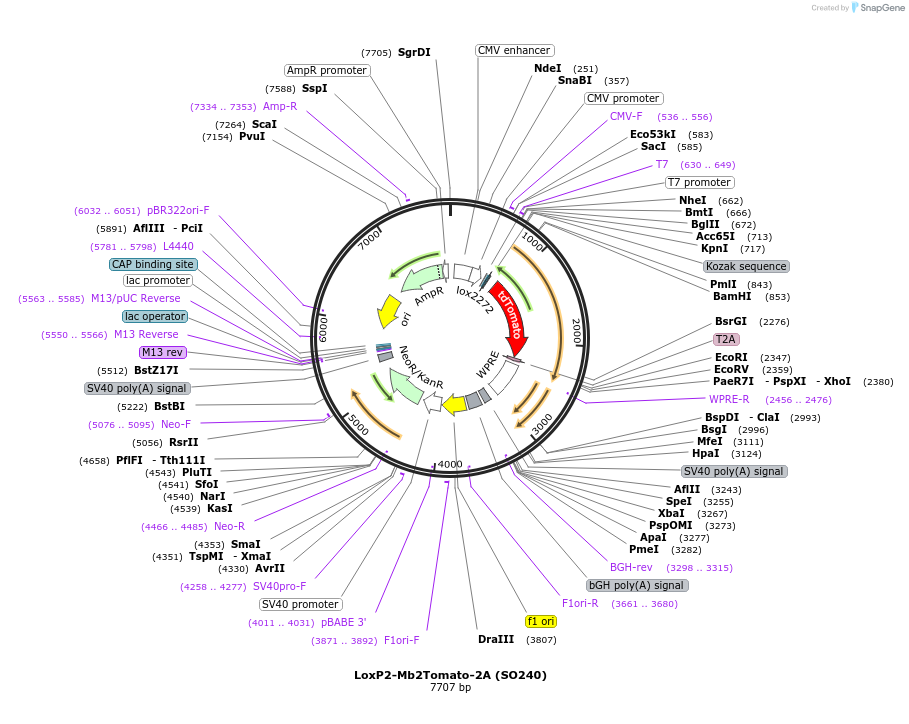
LoxP2-Mb2Tomato-2A (SO240)
Plasmid#99614PurposeTo clone gene of interest downstream of LoxP2-Mb2Tomato-2A cassetteDepositorInsertMb2Tomato
TagsT2AExpressionMammalianAvailable SinceSept. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
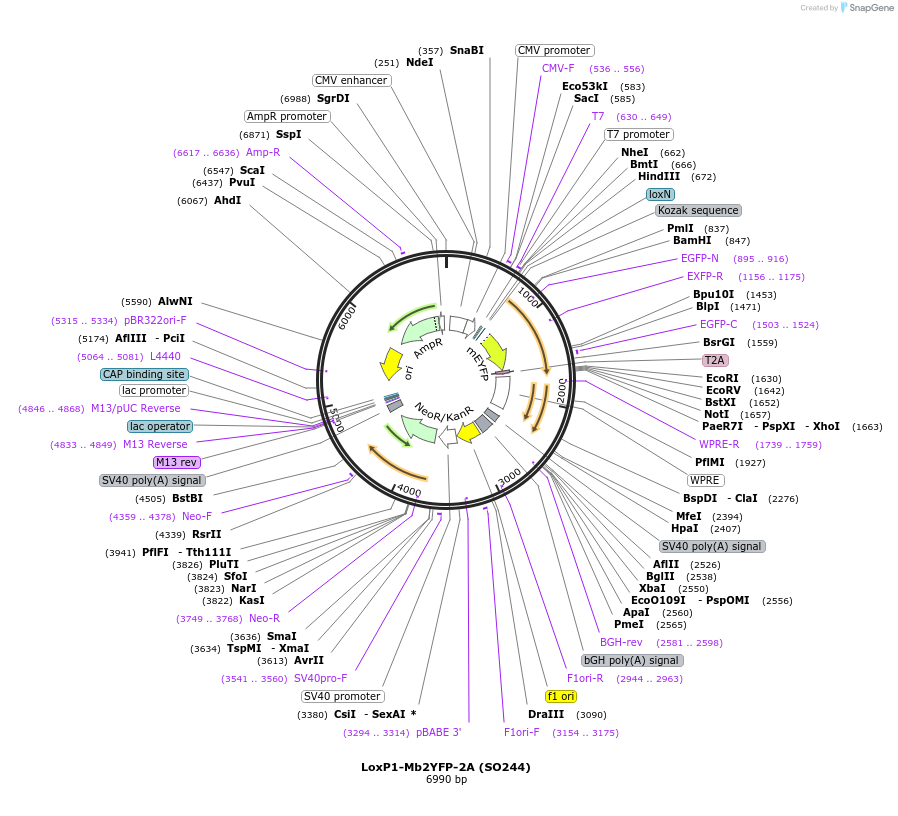
LoxP1-Mb2YFP-2A (SO244)
Plasmid#99613PurposeTo clone gene of interest downstream of LoxP1-Mb2YFP-2A cassetteDepositorInsertMb2EYFP
TagsT2AExpressionMammalianAvailable SinceSept. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
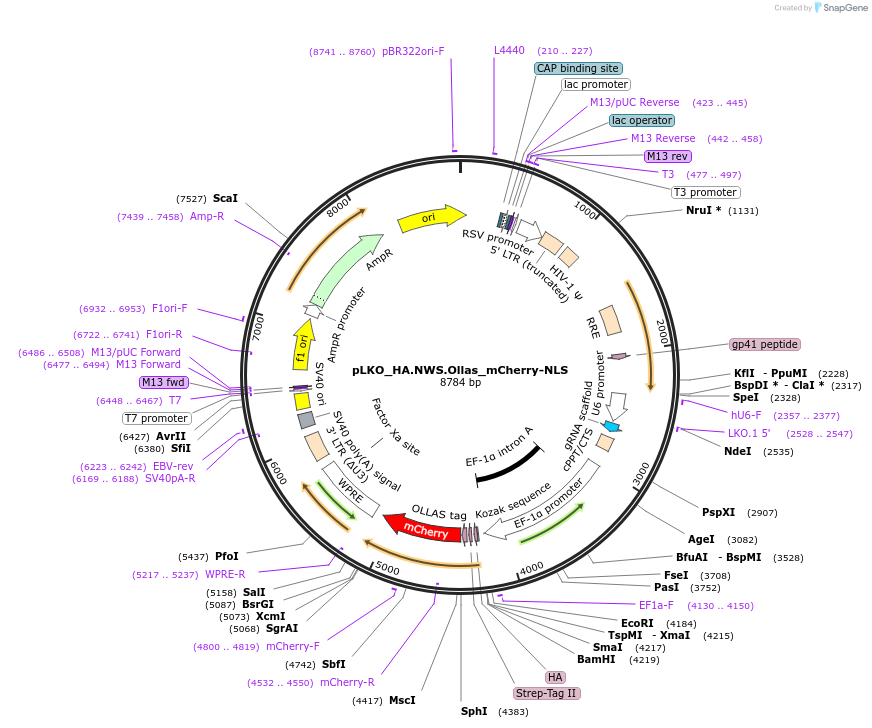
pLKO_HA.NWS.Ollas_mCherry-NLS
Plasmid#178263PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsHA.NWS.Ollas and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
pAS_4xhyb PylT A41AA C55A sfGFP 102TAA 150TAG
Plasmid#154777Purposeplasmid with 4xhybrid PylT cassette (mutant A41AA C55A) and dual suppression reporter sfGFP 102TAA 150 TAG stop, for transient transfection or stable, piggybac-mediated, integrationDepositorInsertsfGFP
ExpressionMammalianMutation102TAA 150TAG in GFP reporter, hybrid PylT with …PromoterEF1Available SinceOct. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
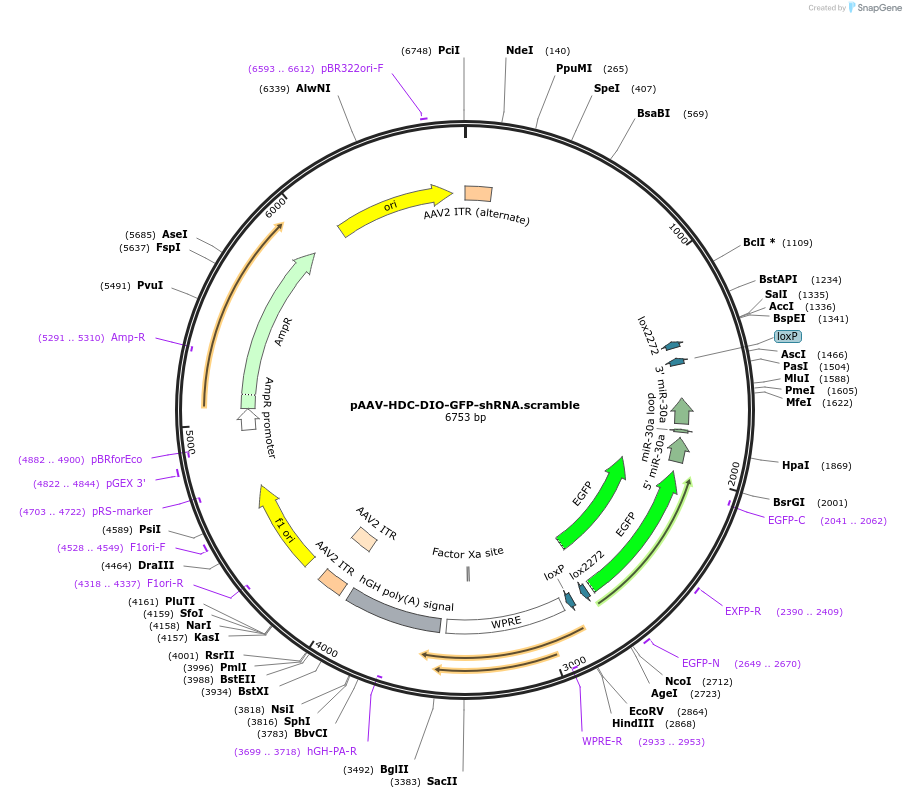
pAAV-HDC-DIO-GFP-shRNA.scramble
Plasmid#184331PurposeExpresses scramble shRNA for control expts. Flexed (DIO) cassette driven by hdc promoter fragment for pan neuronal expressionDepositorInserthdc pan neuronal gene promoter, flex switch, GFP, scramble shRNA
UseAAV, Cre/Lox, Mouse Targeting, and RNAiExpressionMammalianPromoterhistidine decarboxylase promoter fragment that g…Available SinceJune 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
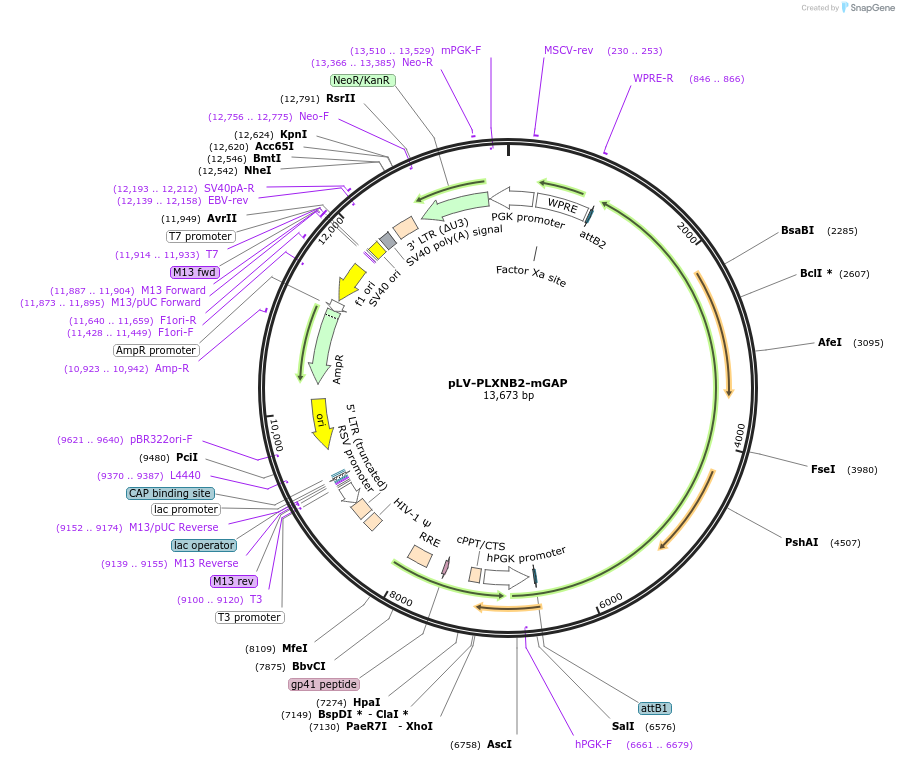
pLV-PLXNB2-mGAP
Plasmid#86241PurposeLentivirus for expression of human Plexin-B2 with mutant GAP domainDepositorInsertPlexin-B2 (PLXNB2 Human)
UseLentiviralExpressionMammalianMutationPLXNB2 with point mutations of GAP domainPromoterhPGKAvailable SinceJan. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
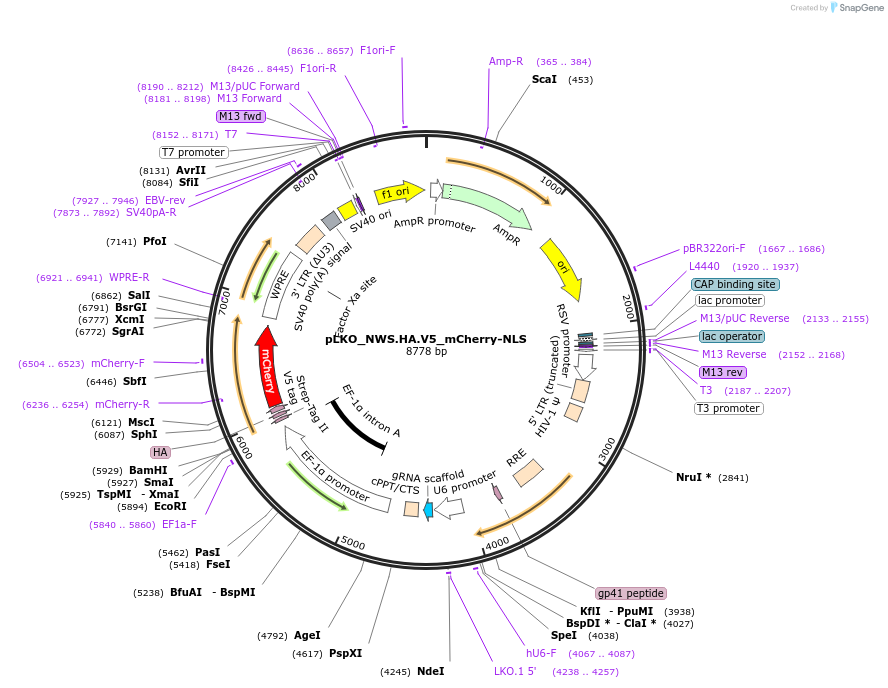
pLKO_NWS.HA.V5_mCherry-NLS
Plasmid#178283PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsNWS.HA.V5 and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
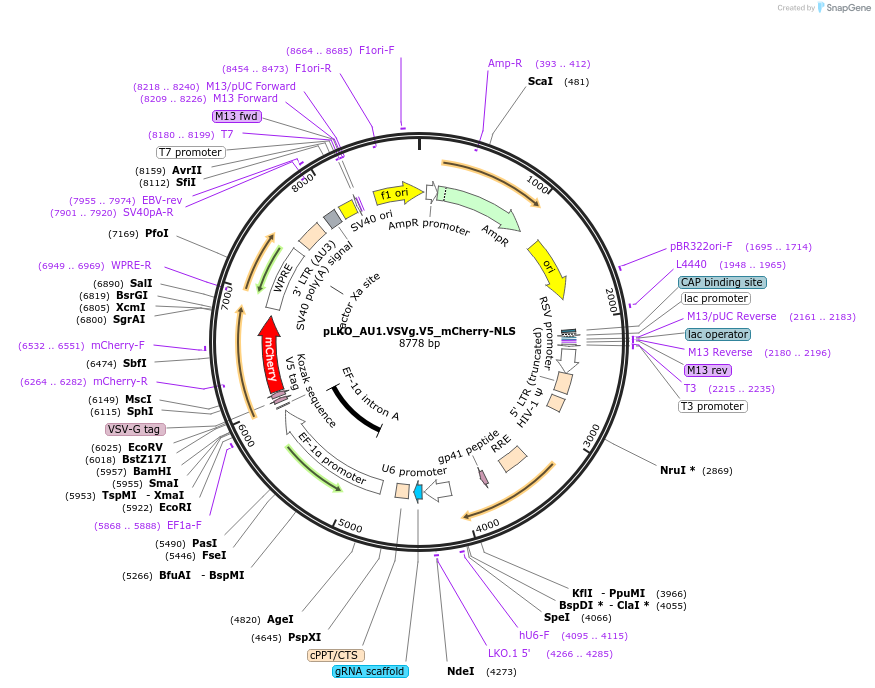
pLKO_AU1.VSVg.V5_mCherry-NLS
Plasmid#178236PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.VSVg.V5 and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
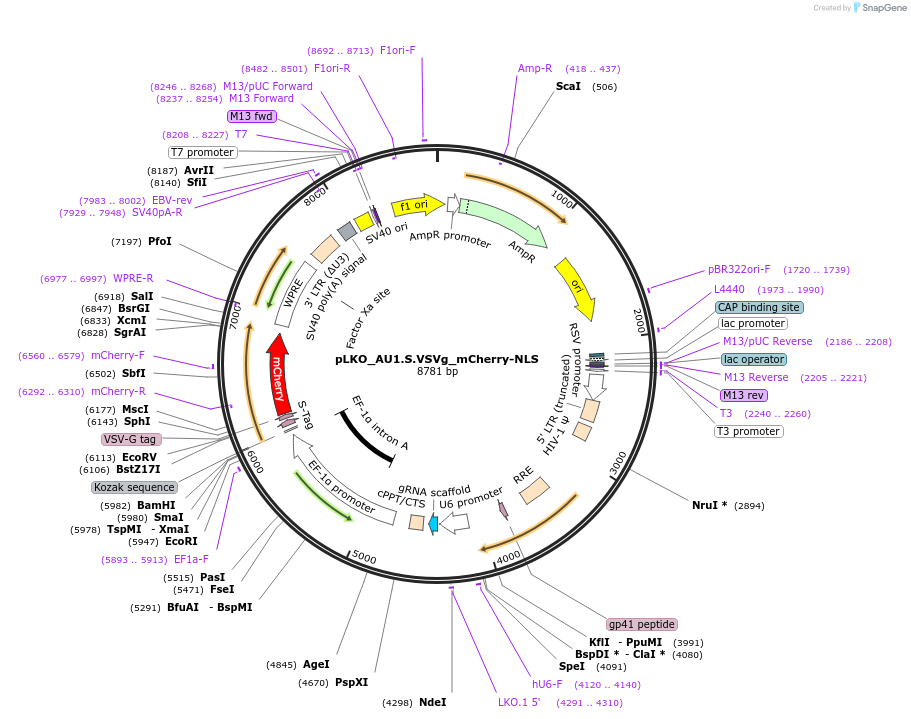
pLKO_AU1.S.VSVg_mCherry-NLS
Plasmid#178234PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.S.VSVg and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
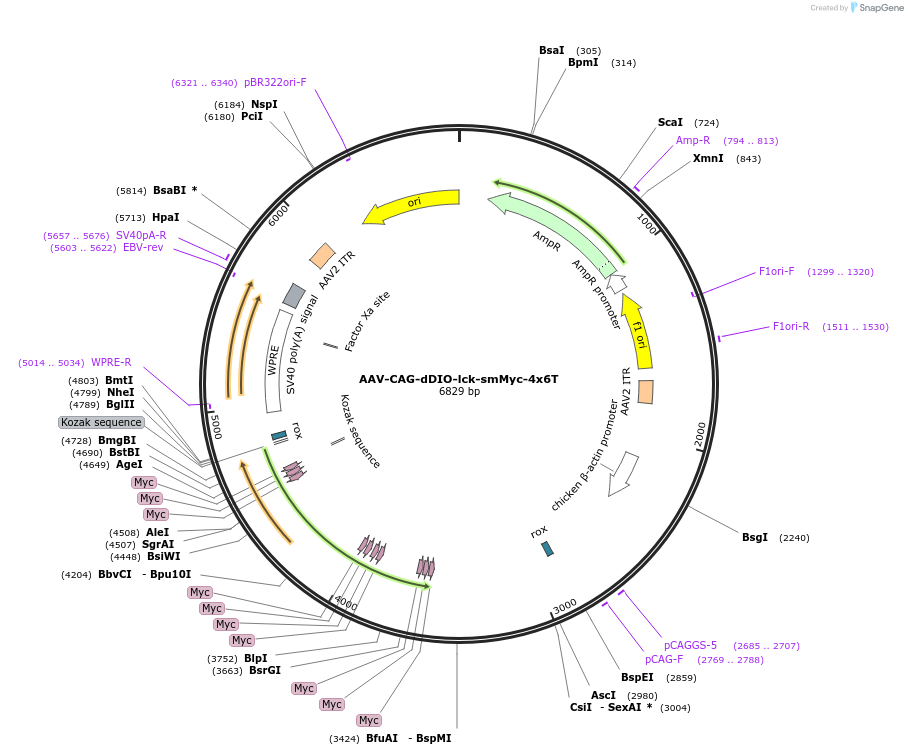
AAV-CAG-dDIO-lck-smMyc-4x6T
Plasmid#196421PurposeDre-dependent AAV expression of membrane-targeted Myc spaghetti monster reporter preferentially in astrocytes; astrocyte selectivity generated with 4x6T miRNA targeting cassetteDepositorInsertLck-smMyc
UseAAV; Dre/rox; astrocyte-selectiveTagsLckExpressionMammalianPromoterCAGAvailable SinceFeb. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
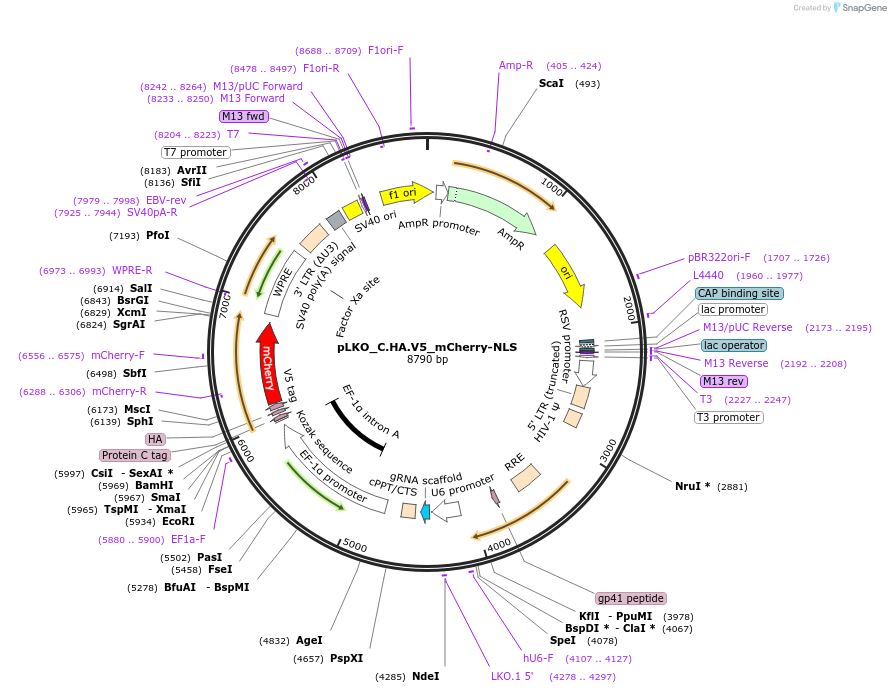
pLKO_C.HA.V5_mCherry-NLS
Plasmid#178242PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsC.HA.V5 and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
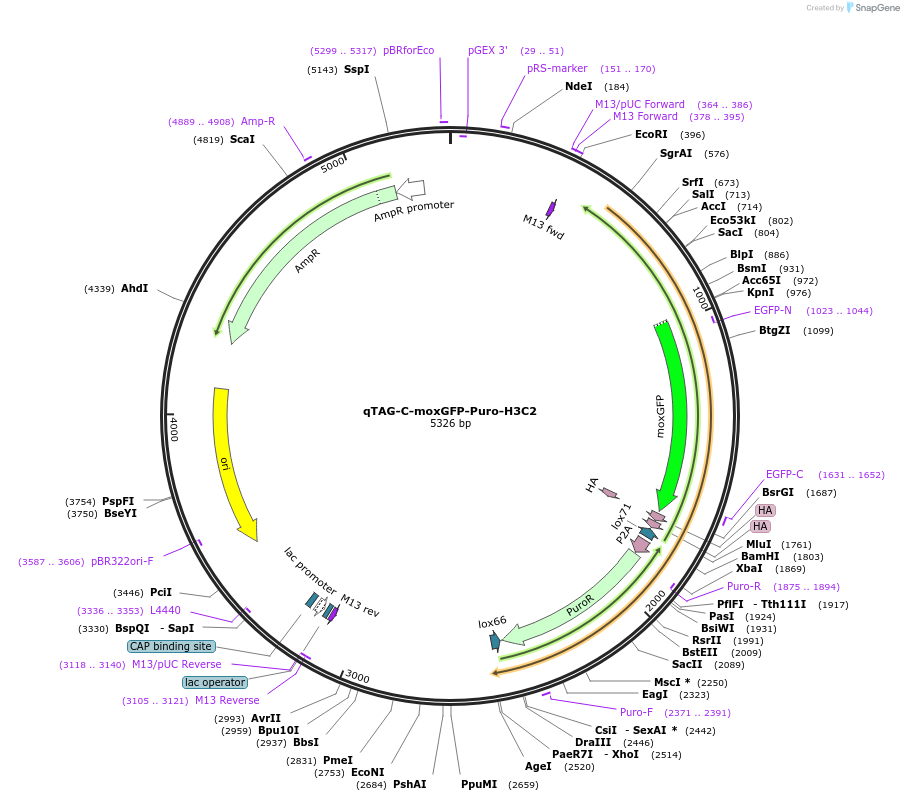
qTAG-C-moxGFP-Puro-H3C2
Plasmid#207783PurposeDonor template for moxGFP-2A-Puro insertion into the C-terminus of the H3C2 locus for nuclei visualization. To be co-transfected with sgRNA plasmid px330-PITCh-H3C2 Addgene #207780DepositorInsertH3C2 Homology Arms flanking a moxGFP-Puro Cassette (H3C2 Human)
UseCRISPR; Donor templateExpressionMammalianAvailable SinceNov. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
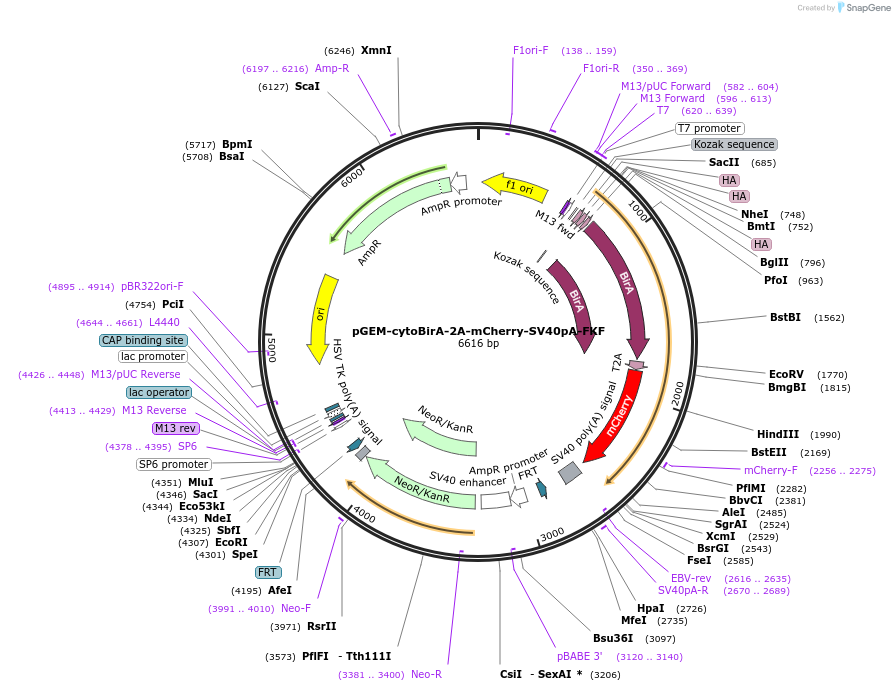
pGEM-cytoBirA-2A-mCherry-SV40pA-FKF
Plasmid#79887PurposeDonor cassette containing HA-tagged biotin ligase (BirA), a Thosea asigna virus peptide (2A), mCherry protein and SV40 polyA tail, followed by FRT site-flanked Kanamycin selection geneDepositorInsertHA-tagged BirA, 2A, mCherry protein and SV40 polyA, followed by FRT site-flanked Kanamycin selection gene
ExpressionBacterialAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
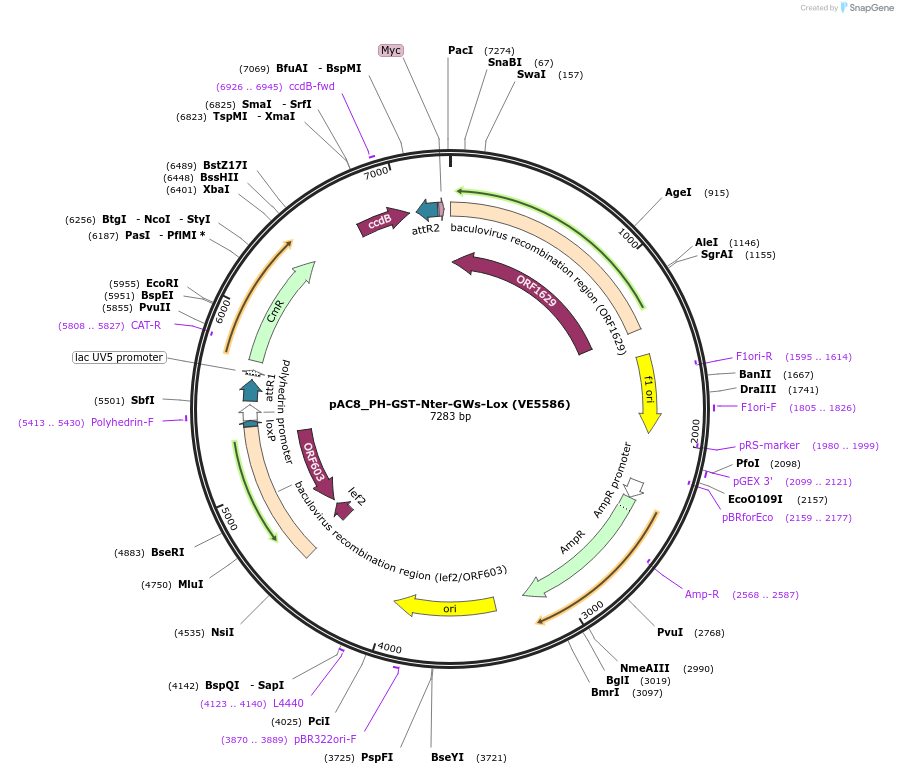
pAC8_PH-GST-Nter-GWs-Lox (VE5586)
Plasmid#163766PurposeTransfer vector for gene expression to generate recombinant baculoviruses by homologous recombination. Contains expression cassette with an N-ter GST tag under the pH promoter.DepositorInsertN-terminal GST tag
TagsGST TagExpressionInsectPromoterPHAvailable SinceDec. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
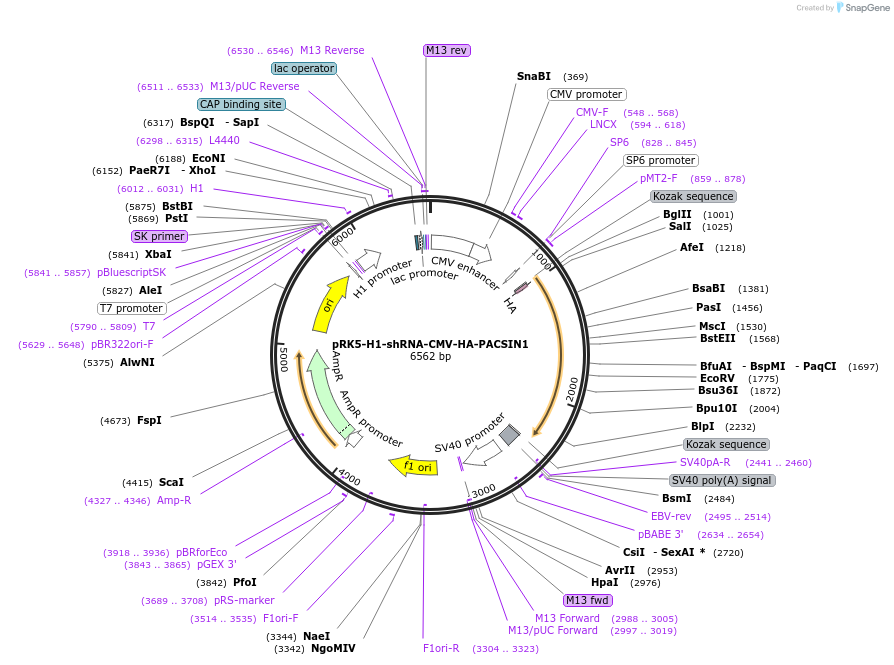
pRK5-H1-shRNA-CMV-HA-PACSIN1
Plasmid#72577PurposeBicistronic construct expressing PACSIN1 shRNA and HA-PACSIN1-rescue constructDepositorInsertsUseRNAiTagsHAExpressionMammalianMutation5 silent mutations around shRNA recognition seque…PromoterCMV and H1Available SinceMay 10, 2017AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.HA.VSVg_mCherry-NLS
Plasmid#178220PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.HA.VSVg and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
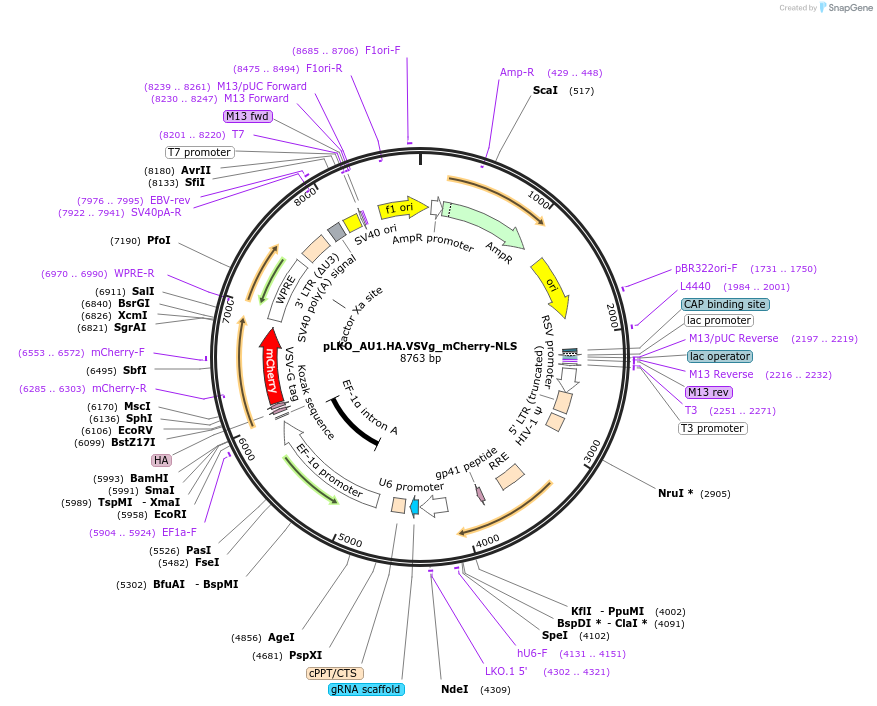
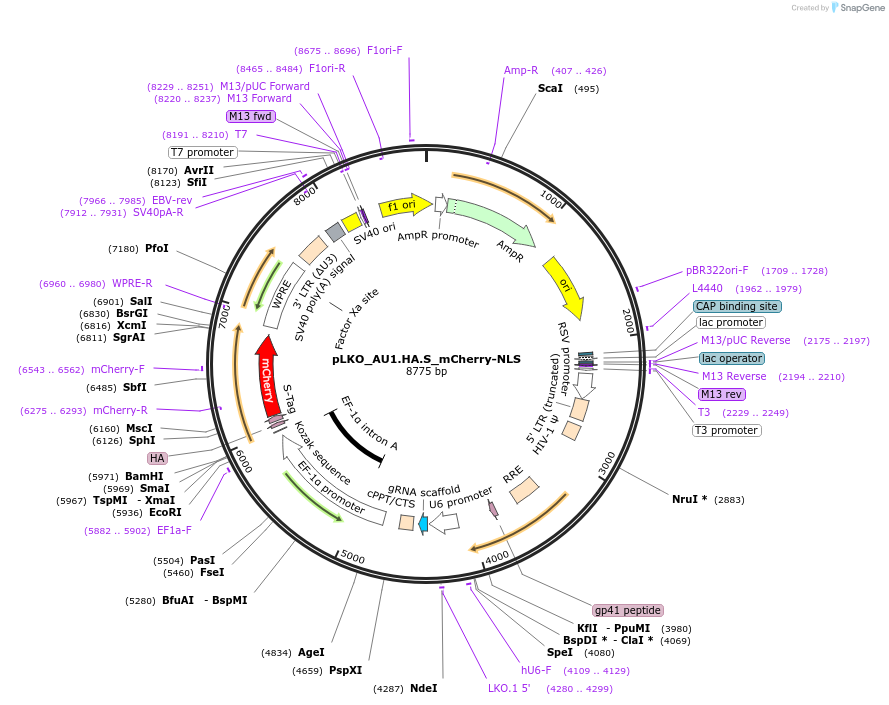
pLKO_AU1.HA.S_mCherry-NLS
Plasmid#178219PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.HA.S and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
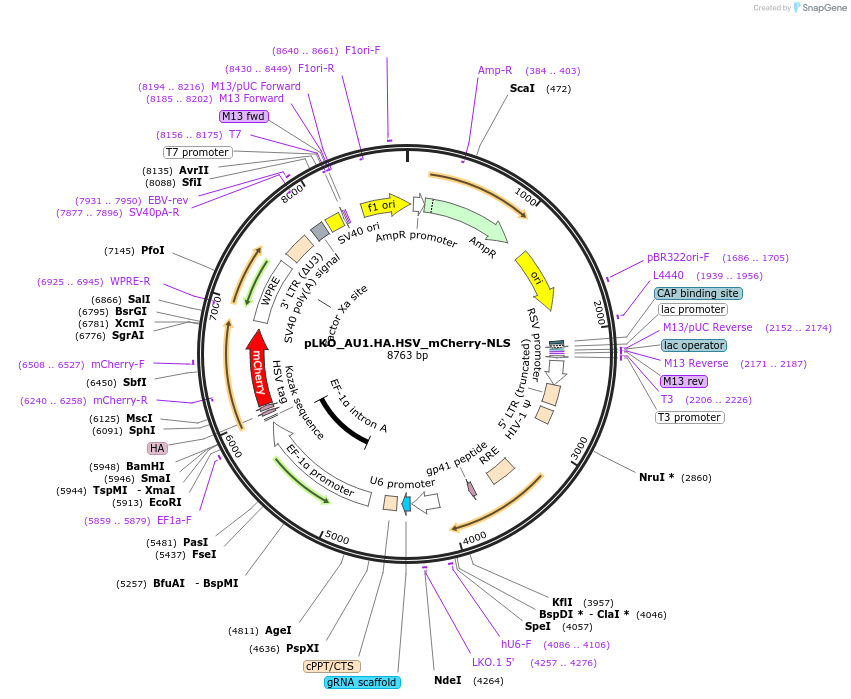
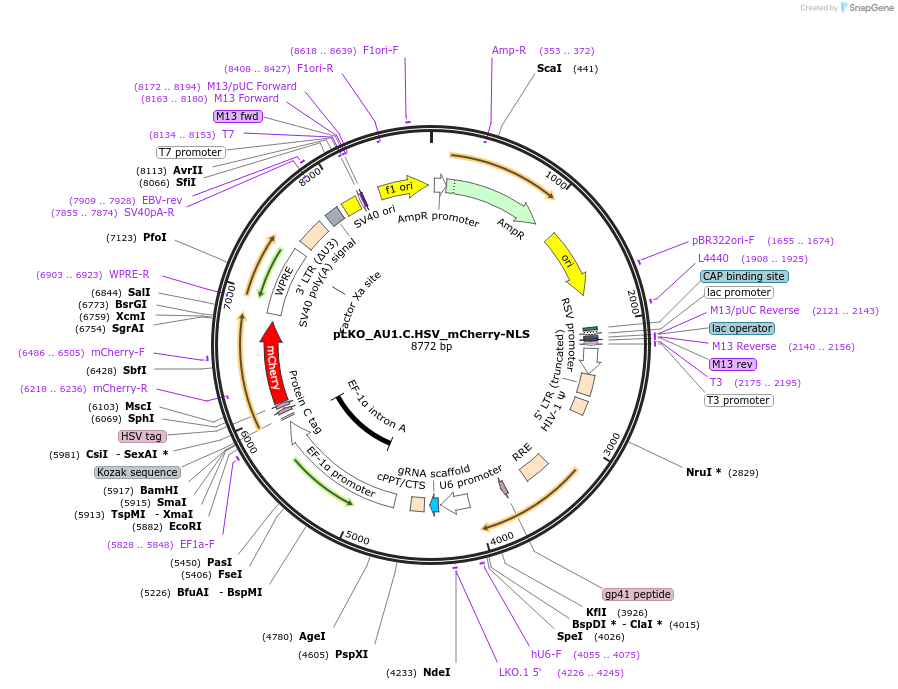
pLKO_AU1.HA.HSV_mCherry-NLS
Plasmid#178216PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.HA.HSV and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
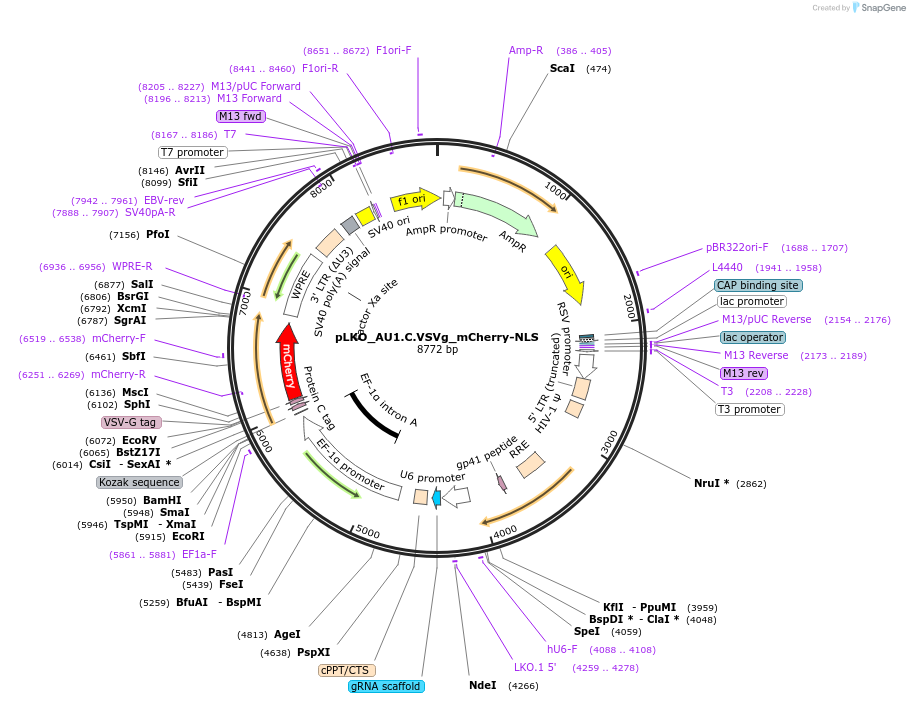
pLKO_AU1.C.VSVg_mCherry-NLS
Plasmid#178214PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.C.VSVg and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.C.S_mCherry-NLS
Plasmid#178213PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.C.S and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLKO_AU1.C.HSV_mCherry-NLS
Plasmid#178210PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsAU1.C.HSV and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
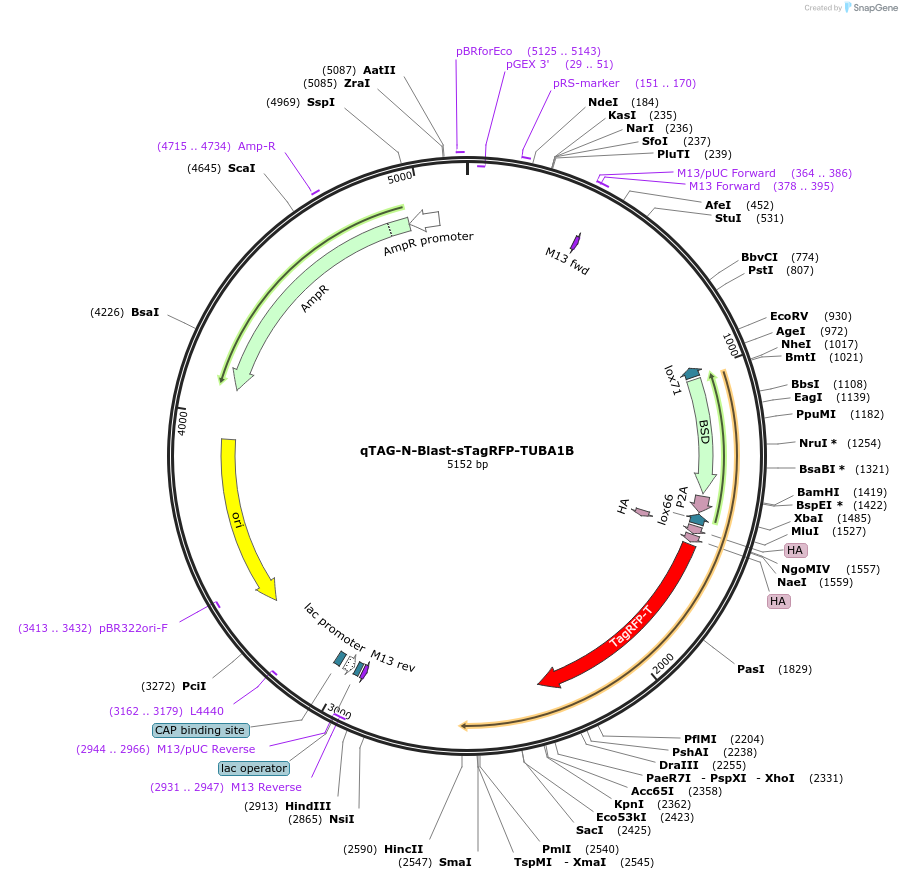
qTAG-N-Blast-sTagRFP-TUBA1B
Plasmid#207768PurposeDonor template for Blast-2A-sTagRFP insertion into the N-terminus of the TUBA1B locus for tubulin visualization. To be co-transfected with sgRNA plasmid px330-PITCh-TUBA1B Addgene #207763DepositorInsertTUBA1B Homology Arms flanking a Blast-sTagRFP Cassette (TUBA1B Human)
UseCRISPR; Donor templateExpressionMammalianAvailable SinceNov. 15, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
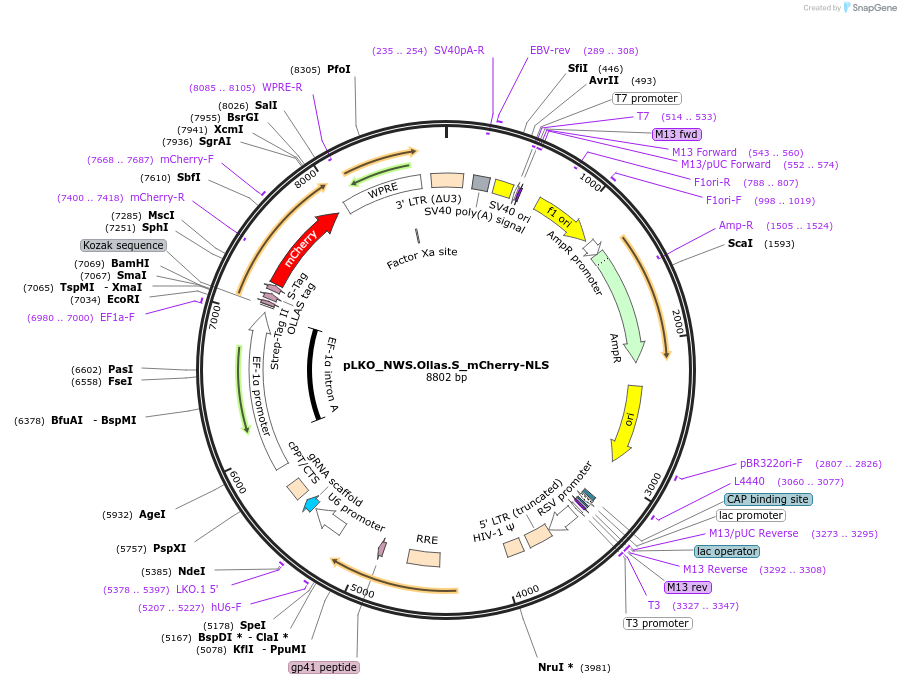
pLKO_NWS.Ollas.S_mCherry-NLS
Plasmid#178279PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsNWS.Ollas.S and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
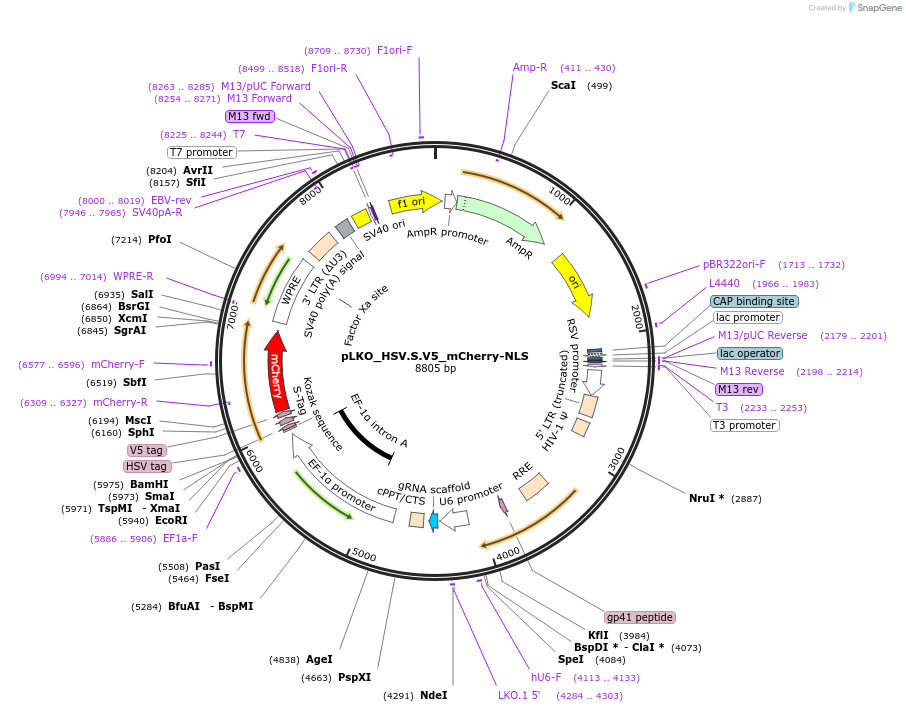
pLKO_HSV.S.V5_mCherry-NLS
Plasmid#178278PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsHSV.S.V5 and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
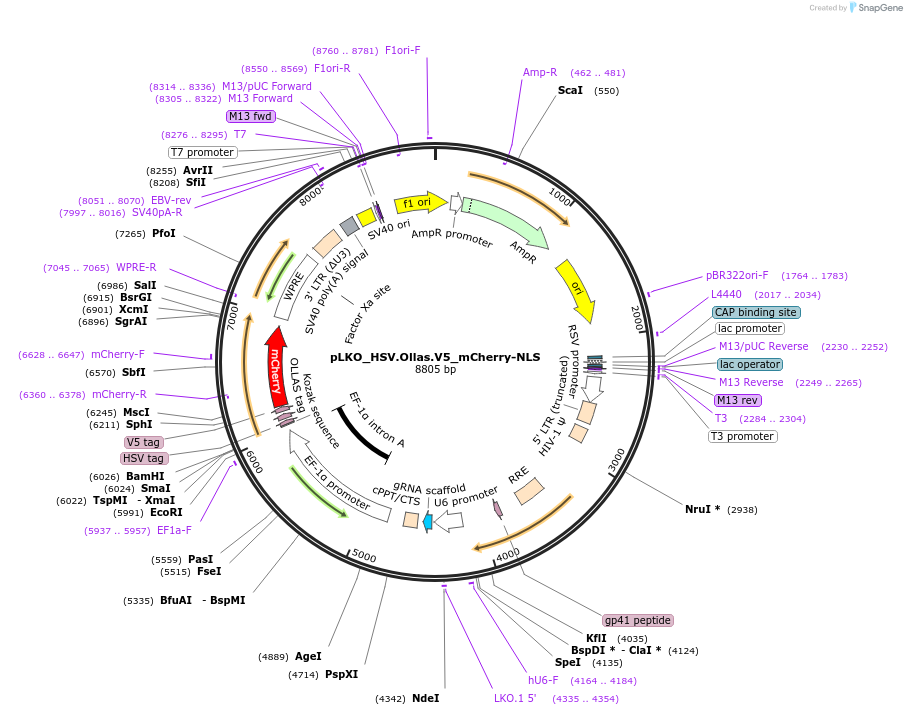
pLKO_HSV.Ollas.V5_mCherry-NLS
Plasmid#178277PurposeNuclear Protein Barcode (nPC) for vector/cell tracking. Contains U6 trRNA cassette for sgRNA cloning. Enables spatial detection of CRISPR screens.DepositorInsertnPC Tagged mCherry-NLS
UseLentiviralTagsHSV.Ollas.V5 and Nuclear Localization SignalPromotereF1aAvailable SinceJan. 27, 2022AvailabilityAcademic Institutions and Nonprofits only