We narrowed to 6,821 results for: sas
-
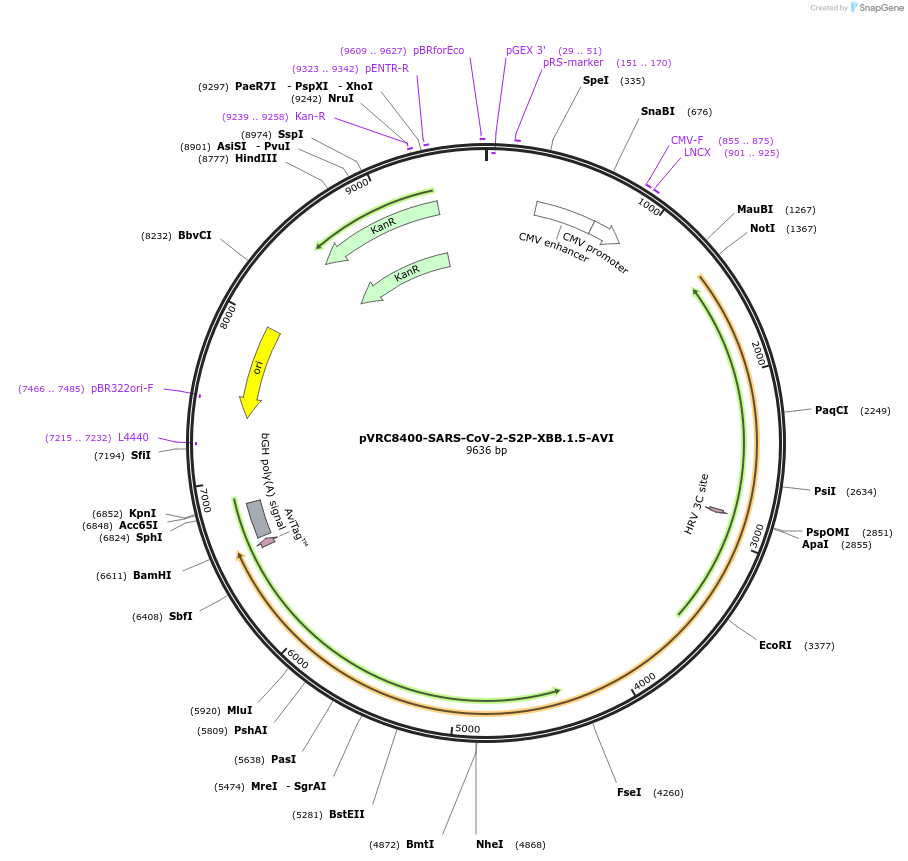
Plasmid#198465PurposeExpresses SARS-CoV-2 S2P protein from the XBB.1.5 variant with single chain Fc, HRV3C protease cleavage site, and AVI tagDepositorInsertSARS-CoV-2-S2P-XBB.1.5 (S )
TagsAVI, HRV3C, and scFcExpressionMammalianMutationT19I, L24-, P25-, P26-, A27S, V83A, G142D, Y144-,…Available SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
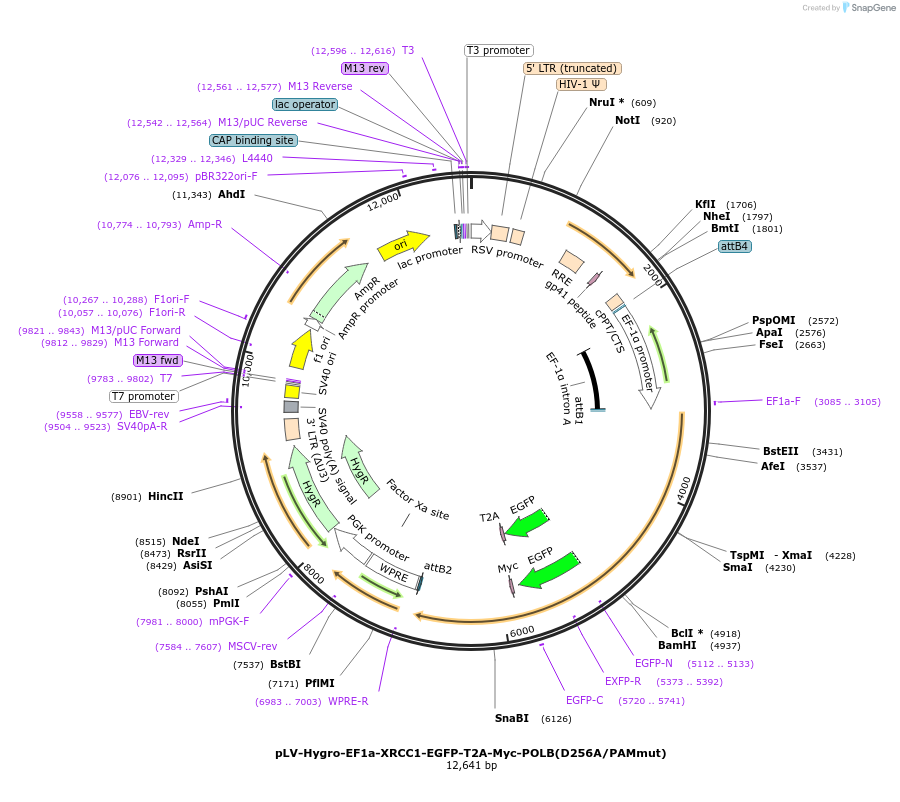
pLV-Hygro-EF1a-XRCC1-EGFP-T2A-Myc-POLB(D256A/PAMmut)
Plasmid#176141PurposeEGFP fused to the C-terminus of XRCC1, linked by T2A to N-terminus Myc-tagged POLB with the mutation Asp256Ala, a mutation in the PAM site used by POLBKO gRNA1 & a hygromycin resistance cassetteDepositorUseLentiviralTagsEGFP and MYCExpressionMammalianMutationMyc-tagged PolB with mutation in Asp256 to Ala, a…PromoterEF1AAvailable SinceJan. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
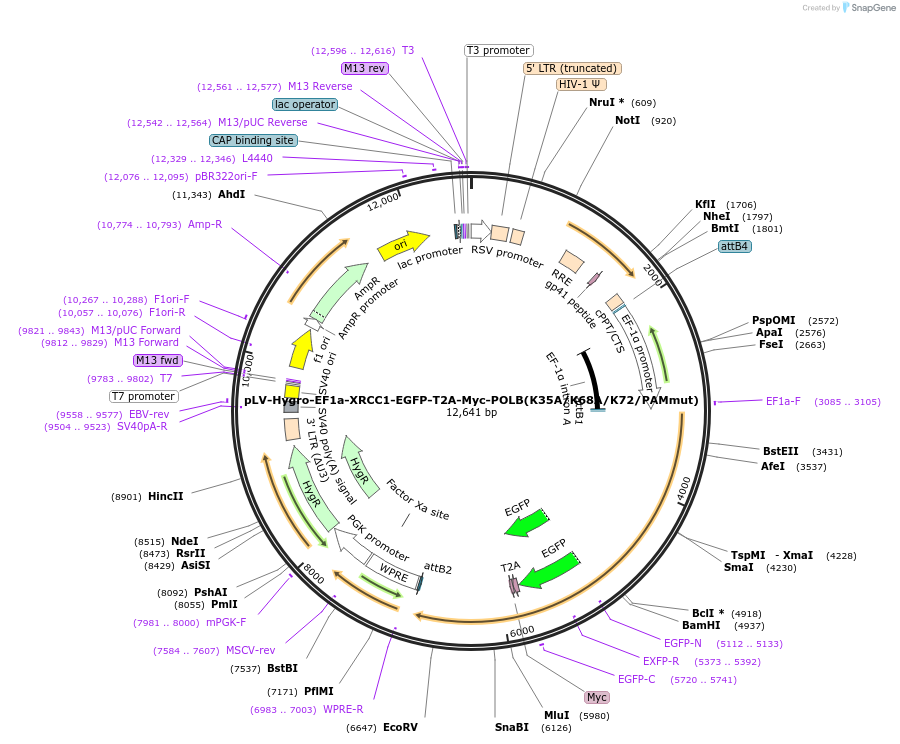
pLV-Hygro-EF1a-XRCC1-EGFP-T2A-Myc-POLB(K35A/K68A/K72/PAMmut)
Plasmid#176140PurposeEGFP fused to the C-terminus of XRCC1, linked by T2A to N-terminus Myc-tagged POLB with the mutations Lys35Ala, Lys68Ala, Lys72Ala, a mutation in the PAM site used by POLBKO gRNA1 & a hygromycin resisDepositorUseLentiviralTagsEGFP and MYCExpressionMammalianMutationMyc-tagged PolB with mutations Lys35 to Ala, Lys6…PromoterEF1AAvailable SinceJan. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
ML24
Bacterial Strain#61915PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG asnA genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML25
Bacterial Strain#61916PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG asnA asnB genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML42
Bacterial Strain#61921PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG argH metA lysA thrC asnB genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML26
Bacterial Strain#61917PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG argH genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML36
Bacterial Strain#61919PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG metA genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML31
Bacterial Strain#61918PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG argH metA genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
Target Accelerator Lung Cancer Mutant Collection
Plasmid Kit#1000000104PurposePanel of lung adenocarcinoma alleles that have undergone basic, functional screening; full-length human clones (180 mutant and 45 corresponding wild-type) and 82 wild-type experssion controlsDepositorAvailable SinceApril 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
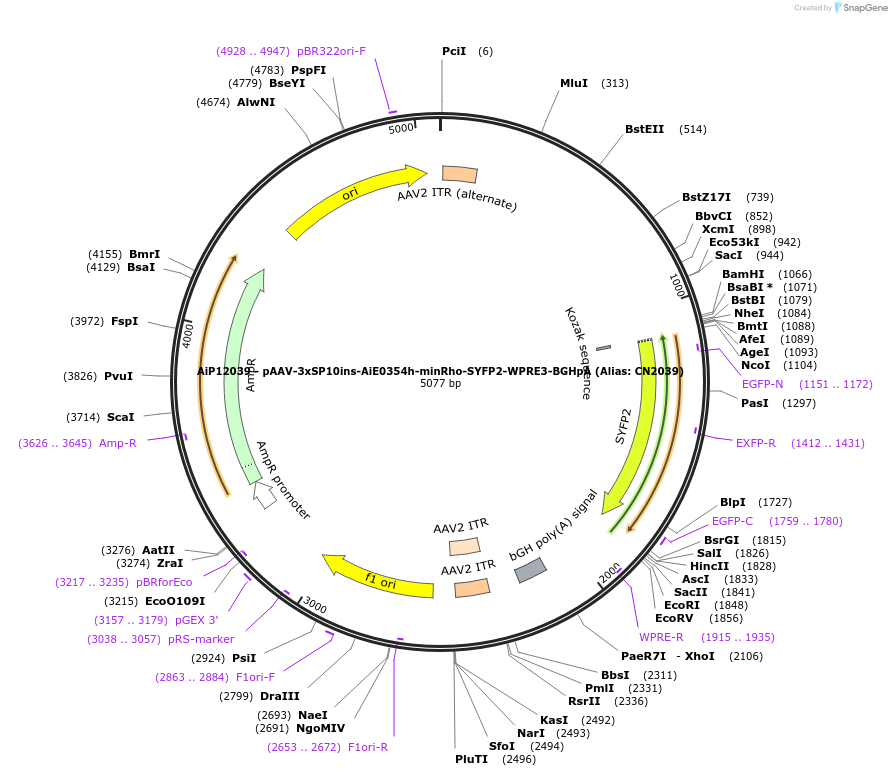
AiP12039 - pAAV-3xSP10ins-AiE0354h-minRho-SYFP2-WPRE3-BGHpA (Alias: CN2039)
Plasmid#208401PurposeAiE0354h is an enhancer sequence designed to drive AAV-mediated transgene expression in Vip+ cortical interneuronsDepositorInsertSYFP2
UseAAV and Synthetic BiologyExpressionMammalianPromotereHGT_354h (enhancer)Available SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
ML43
Bacterial Strain#61922PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG argH metA lysA thrC asnA asnB genes deleted.DepositorBacterial ResistanceNoneAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML40K1
Bacterial Strain#61920PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. cyo ilvE avtA aspC hisG argH metA lysA genes. deleted. Contains pACYC plasmid.DepositorBacterial ResistanceChloramphenicolAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML12
Bacterial Strain#61910PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. ilvE avtA aspC genes deleted.DepositorBacterial ResistanceKanamycinAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
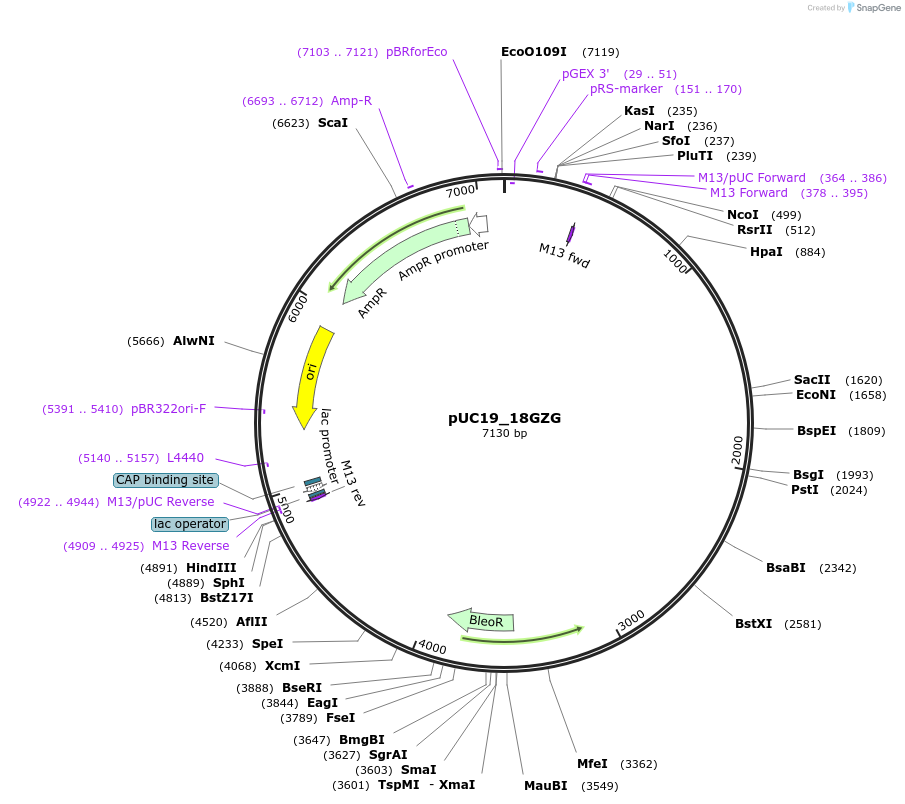
pUC19_18GZG
Plasmid#117228Purposeshble flanked by an endogenous GAPDH promoter and terminator systemDepositorInsertshble
UseThraustochytrid expressionPromoterGAPDHAvailable SinceOct. 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
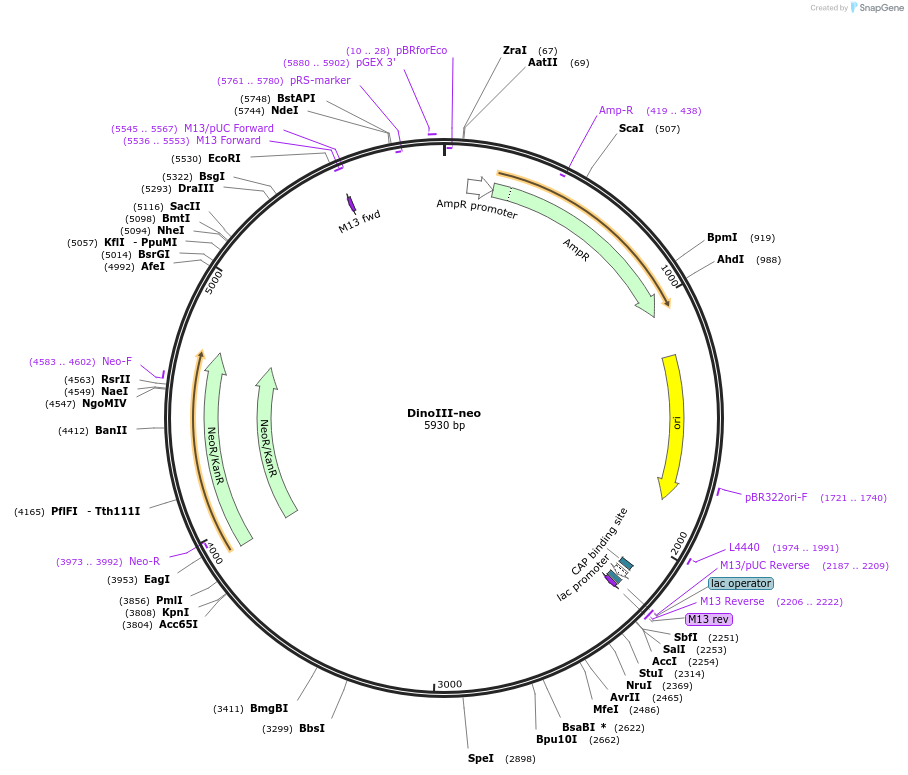
DinoIII-neo
Plasmid#155026PurposeDinoflagelate backbone for amino 3'-glycosyl phosphotransferase (neo) geneDepositorInsertamino 3'-glycosyl phosphotransferase
ExpressionBacterialAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
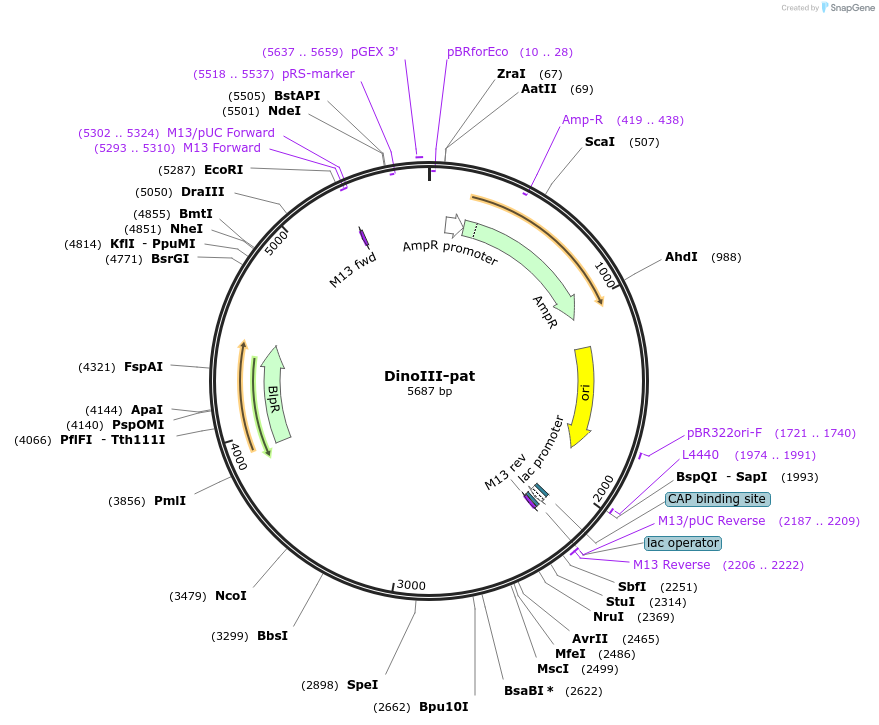
DinoIII-pat
Plasmid#155028PurposeDinoflagelate backbone for Phosphinothricin N-acetyltransferase, pat, geneDepositorInsertPhosphinothricin N-acetyltransferase
ExpressionBacterialAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
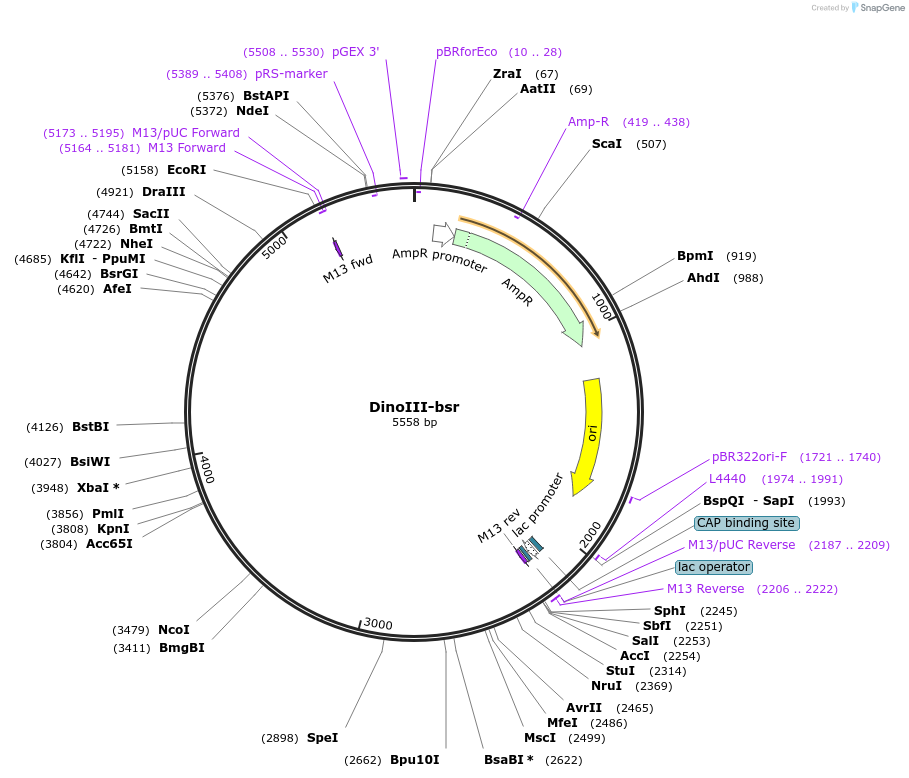
DinoIII-bsr
Plasmid#155027PurposeDinoflagelate backbone for blasticidin S-resistance gene (bsr)DepositorInsertBlasticidin-S deaminase
ExpressionBacterialAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
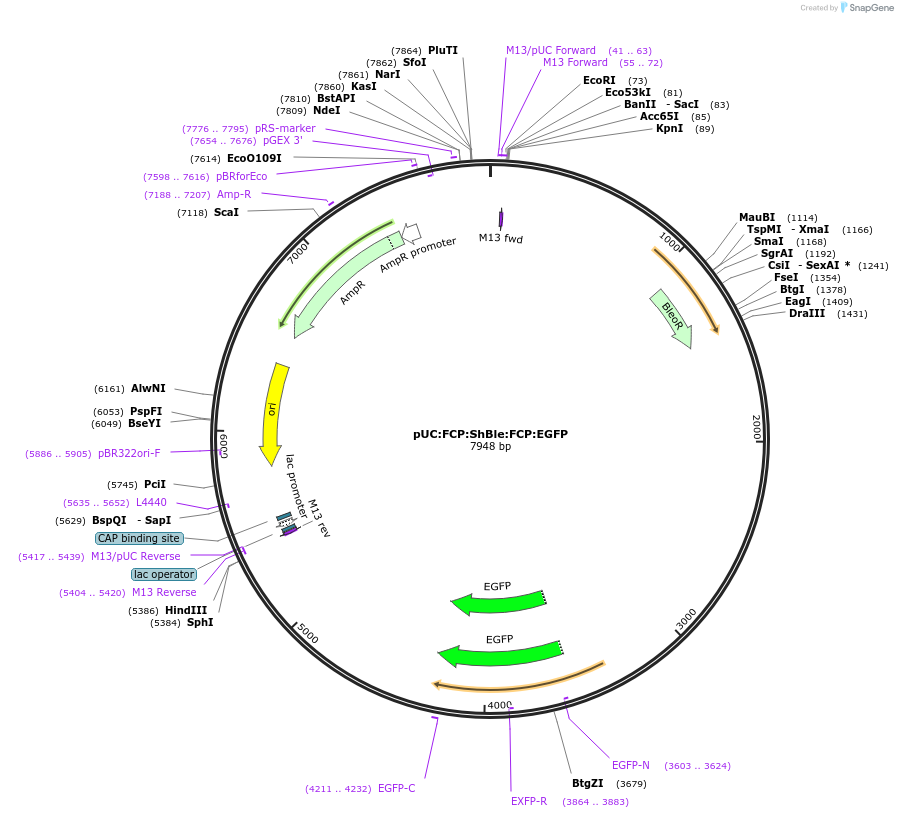
pUC:FCP:ShBle:FCP:EGFP
Plasmid#85987PurposeExpresses ShBle and EGFP with FCP promoters/terminators for F. cylindrus transformation.DepositorInsertsShBle
EGFP
PromoterFCPAvailable SinceJan. 18, 2017AvailabilityAcademic Institutions and Nonprofits only -
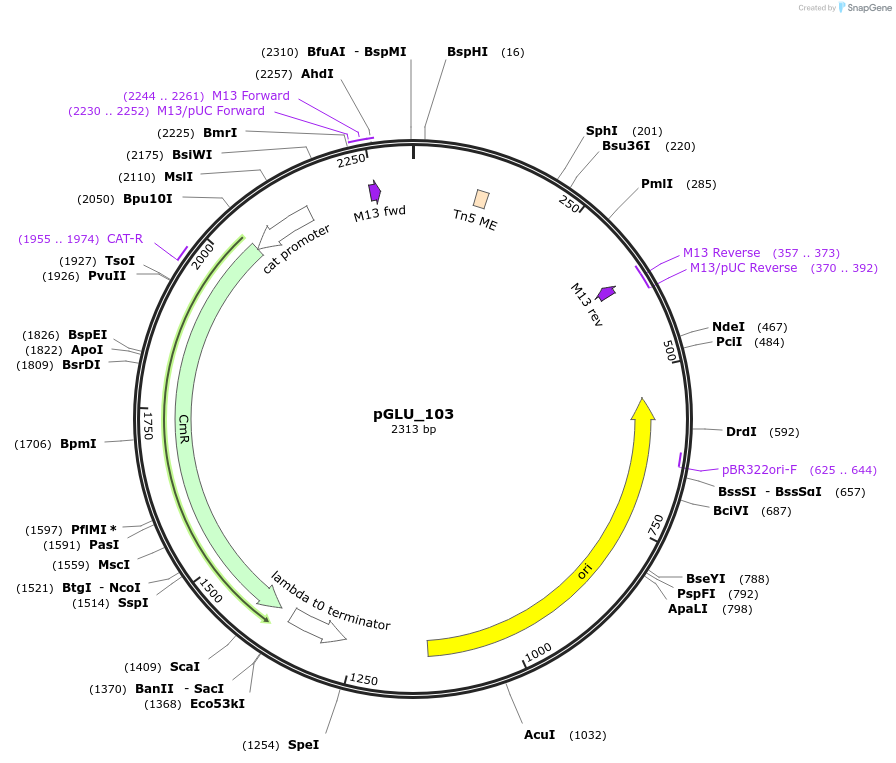
pGLU_103
Plasmid#225785PurposeThis is the promoter region taken from upstream of Tn5 transposase in pBAM1. It can be considered the "default" promoter to use.DepositorInsertPtn5_[Tn5]
UseSynthetic BiologyMutationN/AAvailable SinceSept. 27, 2024AvailabilityAcademic Institutions and Nonprofits only