We narrowed to 9,236 results for: mel
-
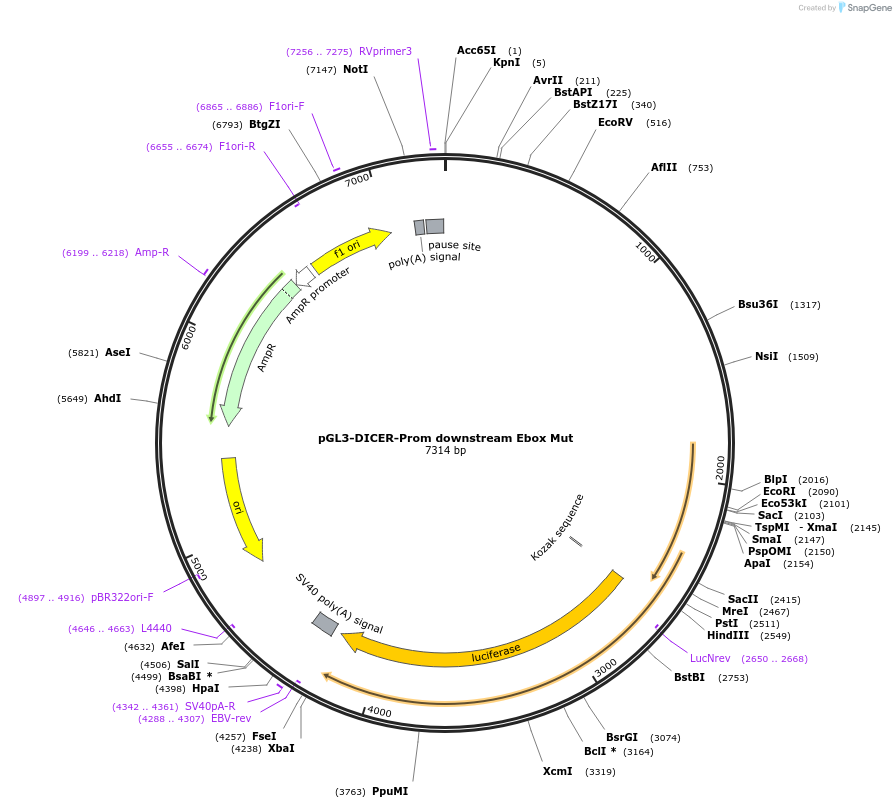
Plasmid#25853DepositorInsertDICER-Prom downstream Ebox Mut (DICER1 gene ID 23405, Human)
UseLuciferaseExpressionMammalianAvailable SinceAug. 25, 2010AvailabilityAcademic Institutions and Nonprofits only -
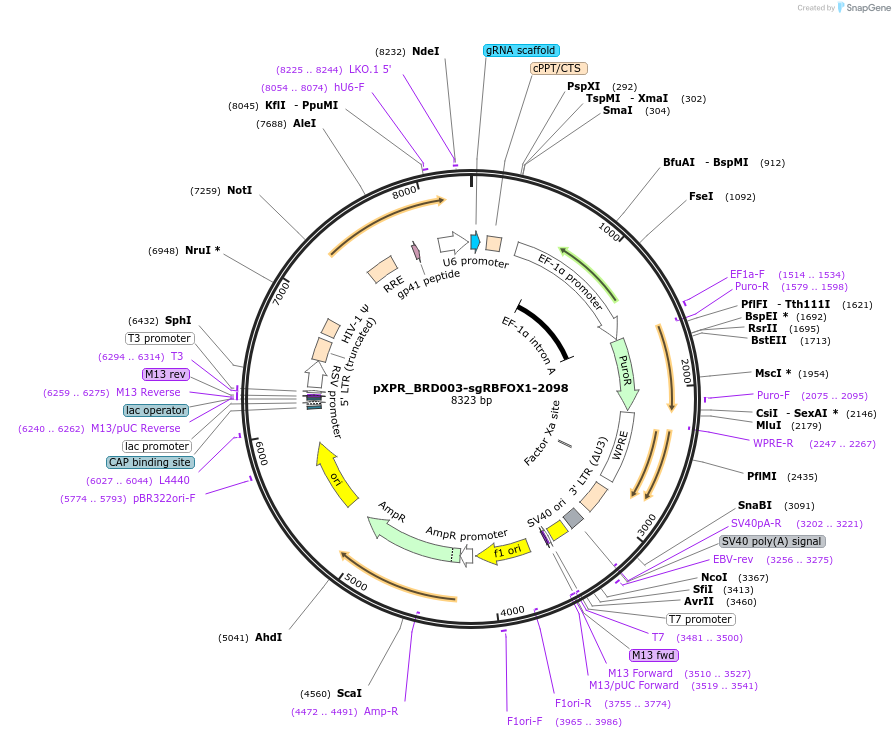
pXPR_BRD003-sgRBFOX1-2098
Plasmid#115452PurposeConstitutive lentiviral expressionDepositorAvailable SinceSept. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
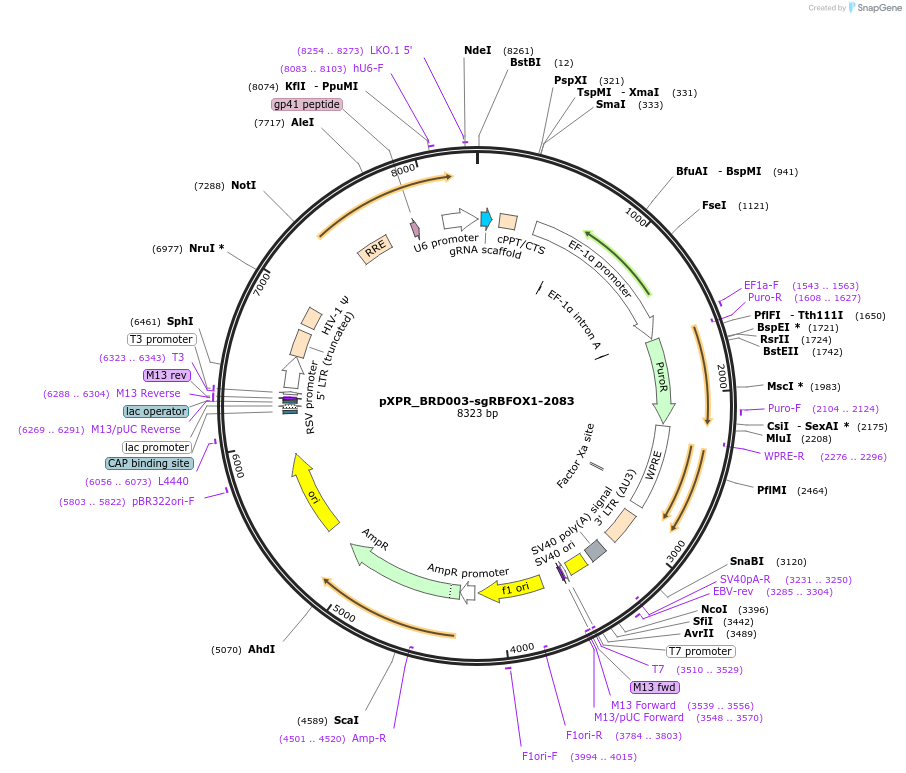
pXPR_BRD003-sgRBFOX1-2083
Plasmid#115451PurposeConstitutive lentiviral expressionDepositorAvailable SinceSept. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
RalA (G23V)( full )-pcw107-V5
Plasmid#64645Purpose(control) when used to produce lentivirus, express physiological levels of insertDepositorAvailable SinceDec. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
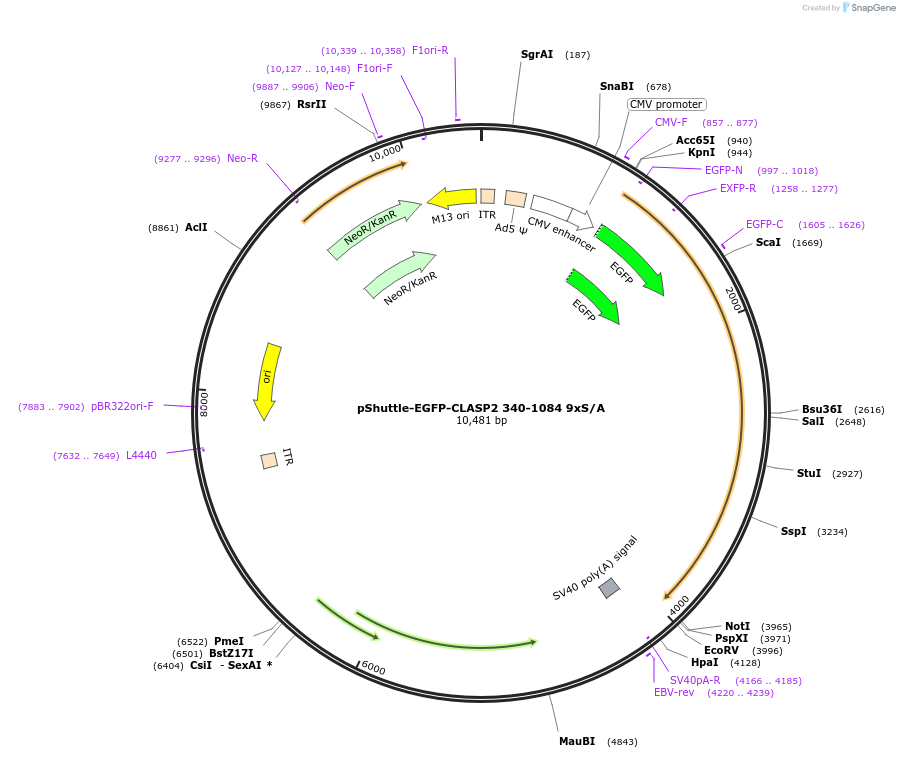
pShuttle-EGFP-CLASP2 340-1084 9xS/A
Plasmid#24384DepositorInsertCLASP2 (340-1084) 9xS/A (CLASP2 Human)
TagsEGFPExpressionMammalianMutationNonphosphorylatable CLASP2 deletion mutant. M…Available SinceJune 29, 2010AvailabilityAcademic Institutions and Nonprofits only -
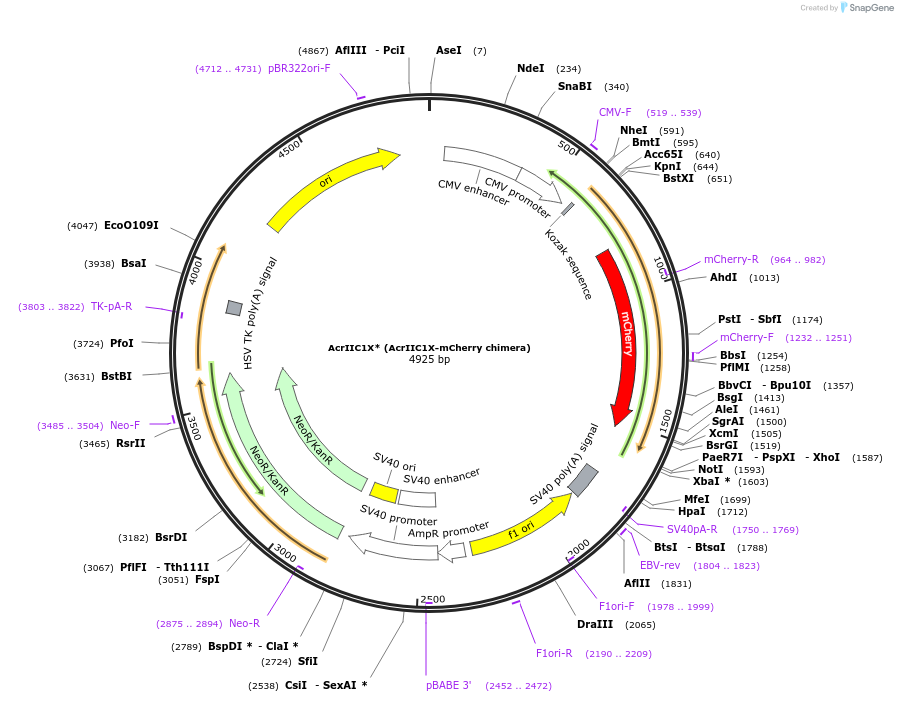
AcrIIC1X* (AcrIIC1X-mCherry chimera)
Plasmid#128114PurposeExpresses AcrIIC1(N3F/D15Q/A48I) fused to mCherry; mediates potent inhibition of both, N. meningitidis as well as S. aureus Cas9DepositorInsertAcrX* (AcrIIC1 N3F/D15Q/A48I fused to mCherry)
UseCRISPR and Synthetic BiologyExpressionMammalianPromoterCMVAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
pLenti6/V5-DEST-ACP5
Plasmid#31204DepositorAvailable SinceJuly 29, 2011AvailabilityAcademic Institutions and Nonprofits only -
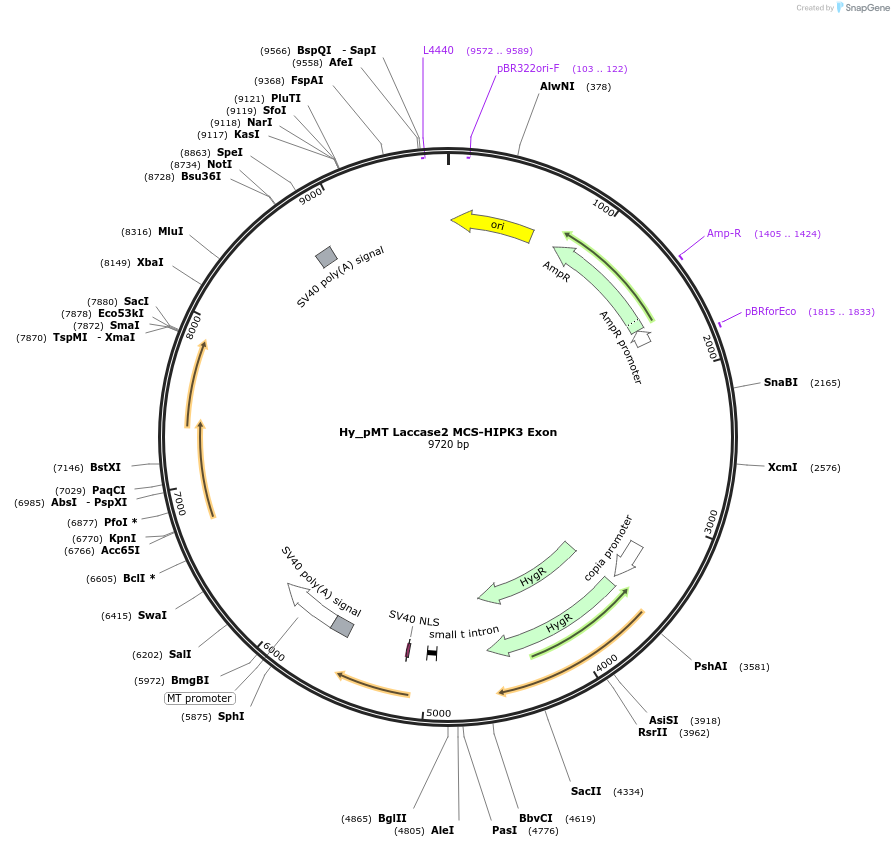
Hy_pMT Laccase2 MCS-HIPK3 Exon
Plasmid#69888PurposeExpresses the human HIPK3 exon 2 circular RNA in DrosophilaDepositorInsertHIPK3 (HIPK3 Fly, Human)
ExpressionInsectMutationExpresses aa1-194 of HIPK3PromoterMetallothionein Promoter (pMT)Available SinceNov. 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
pLenti6/V5-DEST-ITGB3BP
Plasmid#31211DepositorAvailable SinceJuly 29, 2011AvailabilityAcademic Institutions and Nonprofits only -
RalA (G23V)( full )-pcw107
Plasmid#64644Purposewhen used to produce lentivirus, express physiological levels of insertDepositorAvailable SinceDec. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCMV-Tag4A-MMP8 (G104R)
Plasmid#29548DepositorInsertMetalloproteinase protein 8 (MMP8 Human)
TagsFLAGExpressionBacterial and MammalianMutationG104RAvailable SinceJuly 22, 2011AvailabilityAcademic Institutions and Nonprofits only -
His-FLAG-Med20 pBacPAK8
Plasmid#15372DepositorAvailable SinceAug. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
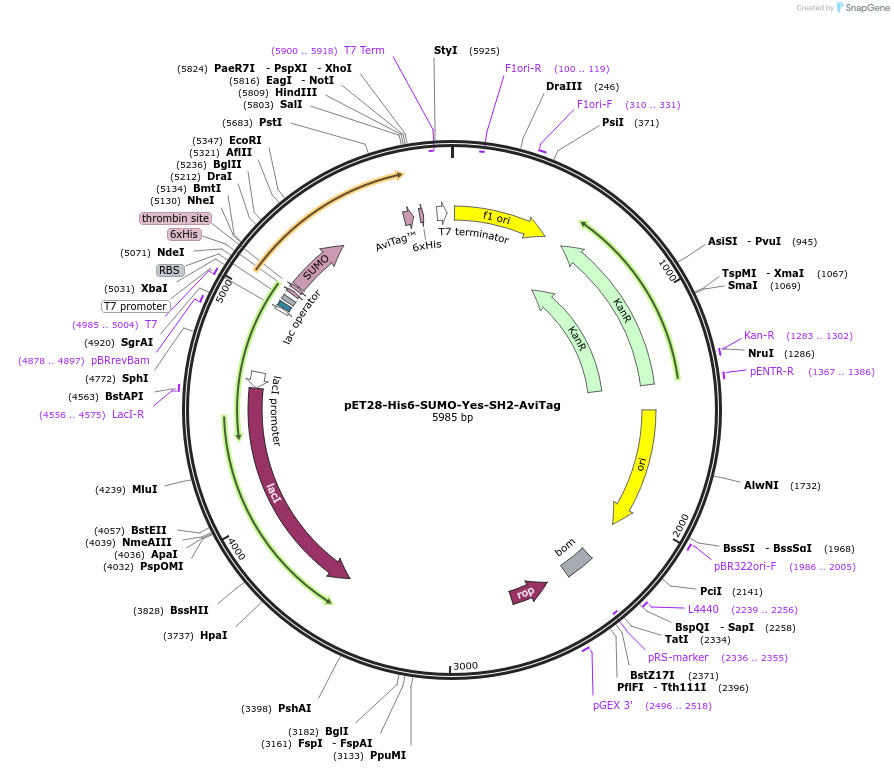
pET28-His6-SUMO-Yes-SH2-AviTag
Plasmid#214208PurposeBacterial expression of SH2 domain of the Yes kinase with N-terminal Thrombin-cleavable 6xHis tag and SUMO and C-terminal AviTag for Streptavidin bead functionalization for binder selectionsDepositorInsertYes kinase SH2 domain (YES1 Human)
Tags6xHis, AviTag, and SUMOExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
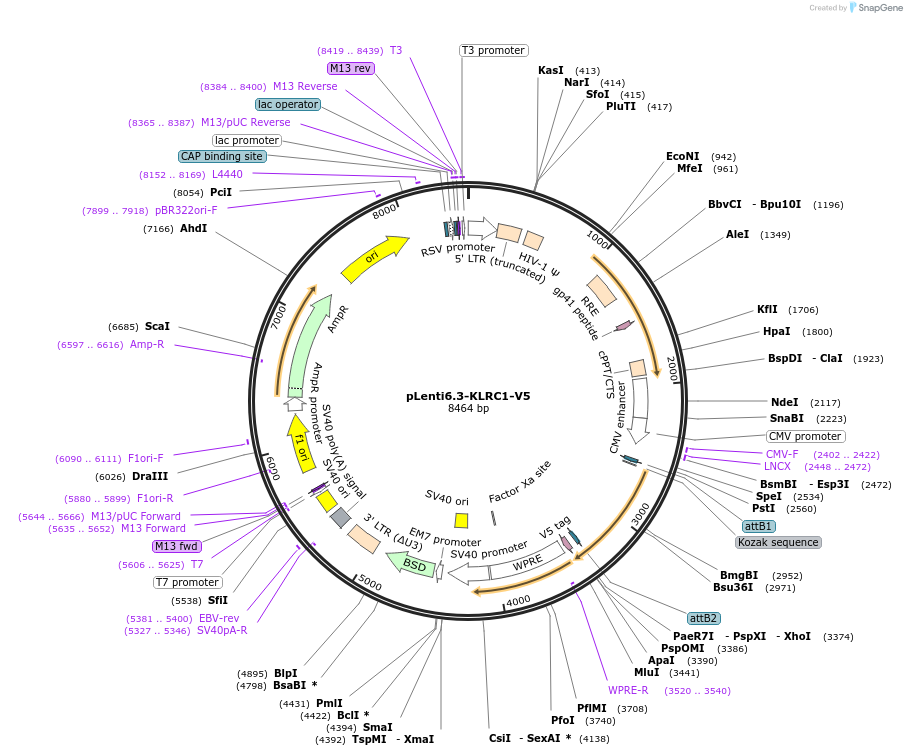
pLenti6.3-KLRC1-V5
Plasmid#124093PurposeKLRC1-V5 tagged expression vectorDepositorAvailable SinceOct. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
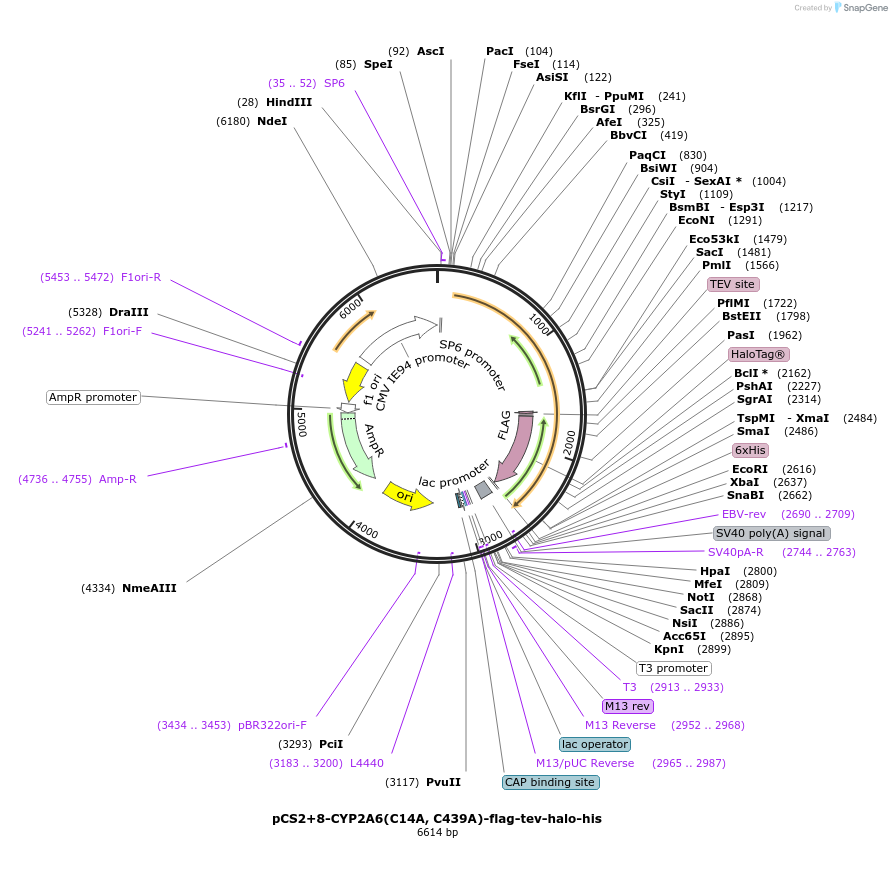
pCS2+8-CYP2A6(C14A, C439A)-flag-tev-halo-his
Plasmid#227162PurposeFor T-REX experiments of CYP2A6 C14A and C439A double mutant. Less sensing ability to reactive electrophilic species (RES).DepositorInsertCYP2A6 (CYP2A6 Human)
TagsFlag, HaloTag, and His6ExpressionMammalianMutationchanged cysteine 14 and cysteine 439 to alaninePromoterCMV and SP6Available SinceAug. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
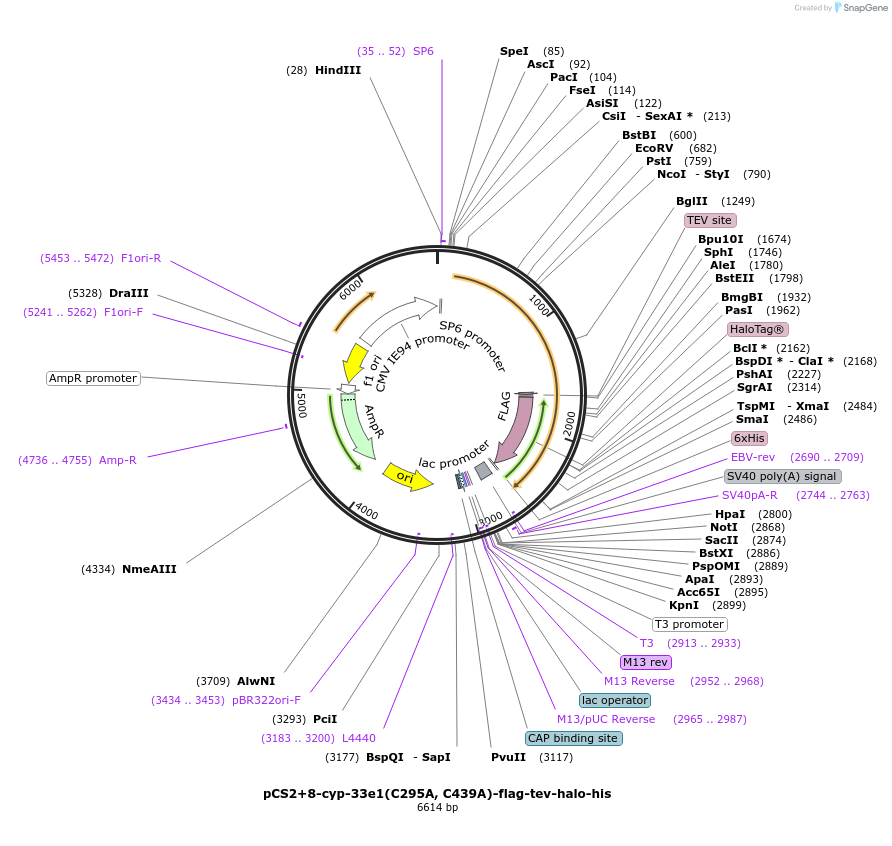
pCS2+8-cyp-33e1(C295A, C439A)-flag-tev-halo-his
Plasmid#227158PurposeFor T-REX experiments of cyp-33e1 C295A and C439A double mutant. Almost no sensing ability to reactive electrophilic species (RES).DepositorInsertcyp-33e1 (cyp-33E1 Nematode)
TagsFlag, HaloTag, and His6ExpressionMammalianMutationchanged cysteine 295 and cysteine 439 to alaninePromoterCMV and SP6Available SinceAug. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
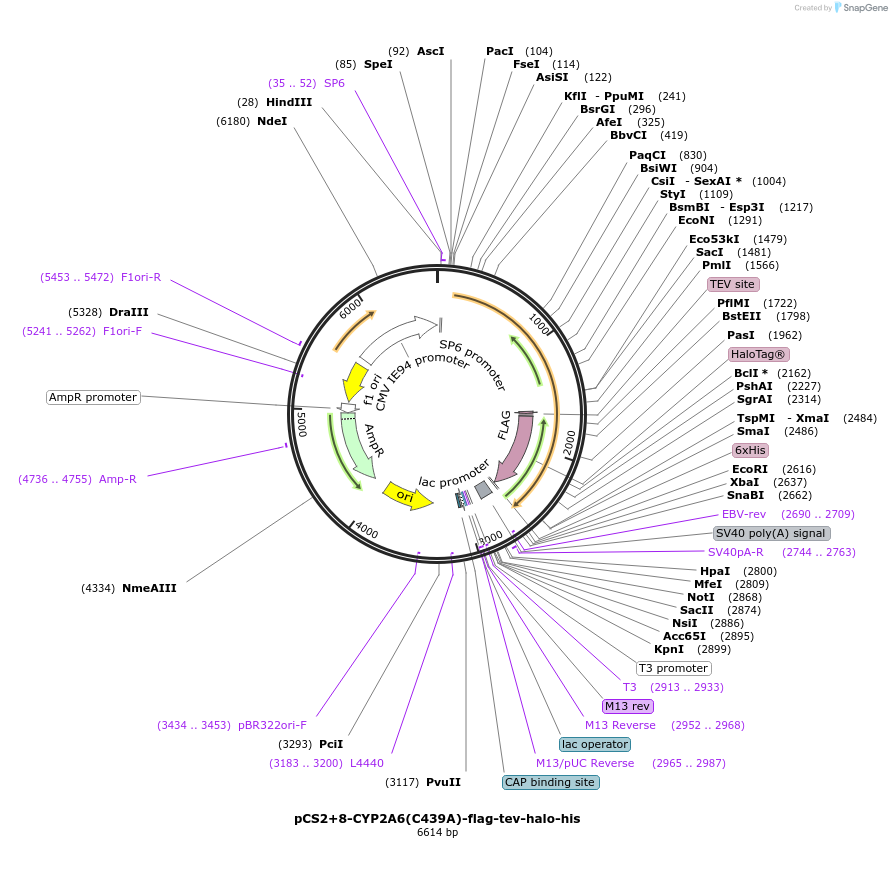
pCS2+8-CYP2A6(C439A)-flag-tev-halo-his
Plasmid#227161PurposeFor T-REX experiments of CYP2A6 C439A mutant. Less sensing ability to reactive electrophilic species (RES).DepositorInsertCYP2A6 (CYP2A6 Human)
TagsFlag, HaloTag, and His6ExpressionMammalianMutationchanged cysteine 439 to alaninePromoterCMV and SP6Available SinceAug. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
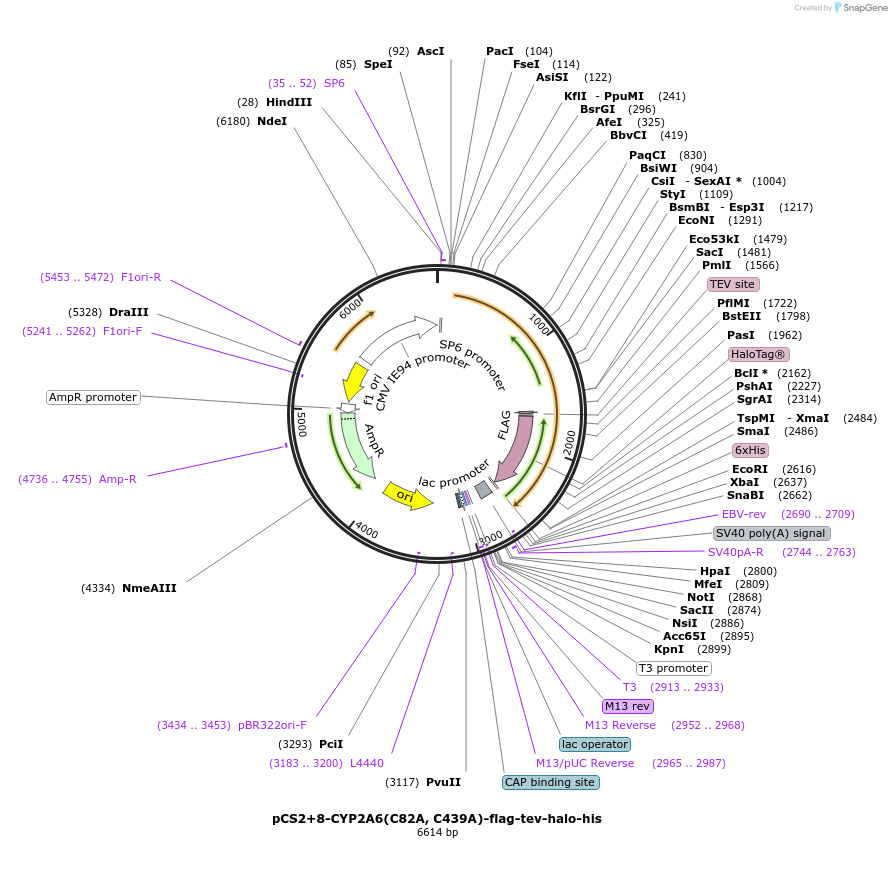
pCS2+8-CYP2A6(C82A, C439A)-flag-tev-halo-his
Plasmid#227163PurposeFor T-REX experiments of CYP2A6 C82A and C439A double mutant. Almost no sensing ability to reactive electrophilic species (RES).DepositorInsertCYP2A6 (CYP2A6 Human)
TagsFlag, HaloTag, and His6ExpressionMammalianMutationchanged cysteine 82 and cysteine 439 to alaninePromoterCMV and SP6Available SinceAug. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
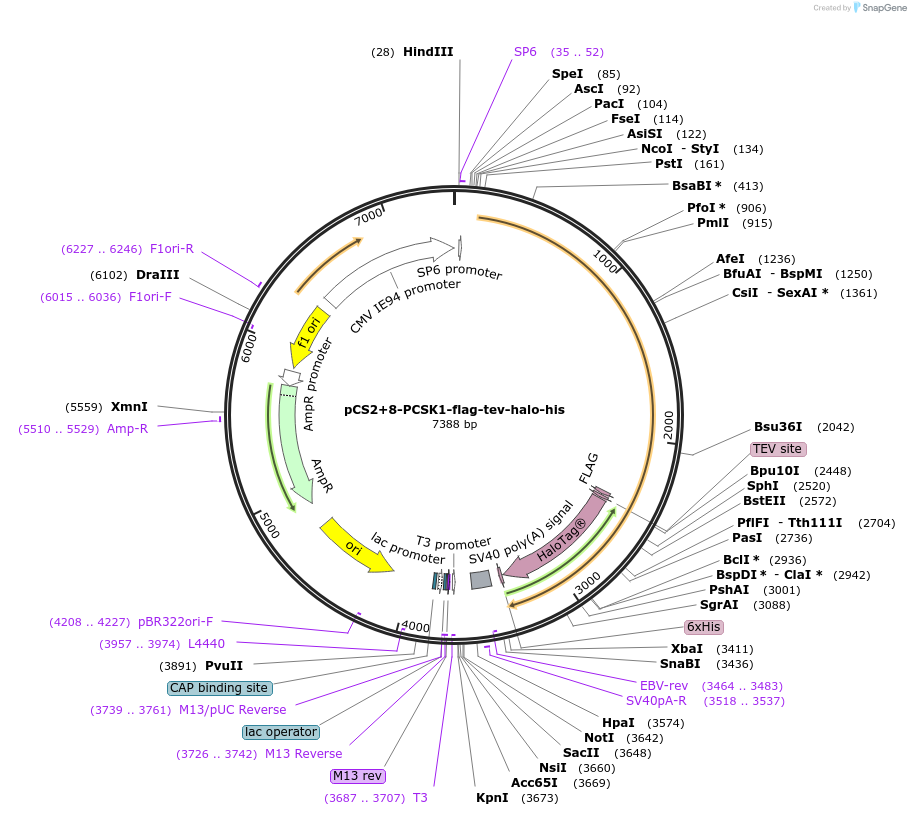
pCS2+8-PCSK1-flag-tev-halo-his
Plasmid#227155PurposeFor T-REX expriments of PCSK1.DepositorAvailable SinceAug. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
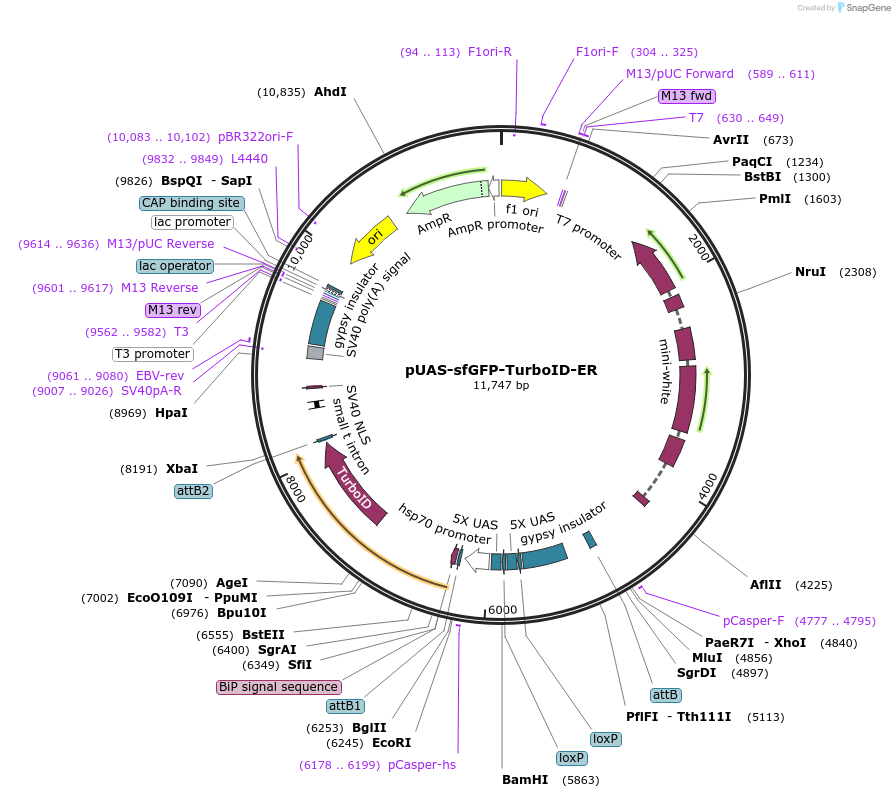
pUAS-sfGFP-TurboID-ER
Plasmid#240232PurposeExpression of ER-localized sfGFP-TurboID under control of UAS promoter. Can be used to generate transgenic fly lines by white+ fly eye selection and phiC31 integration.DepositorInsertBIP-sfGFP-TurboID-KDEL
ExpressionInsectPromoterUASAvailable SinceJuly 11, 2025AvailabilityAcademic Institutions and Nonprofits only