We narrowed to 4,565 results for: golden gate
-
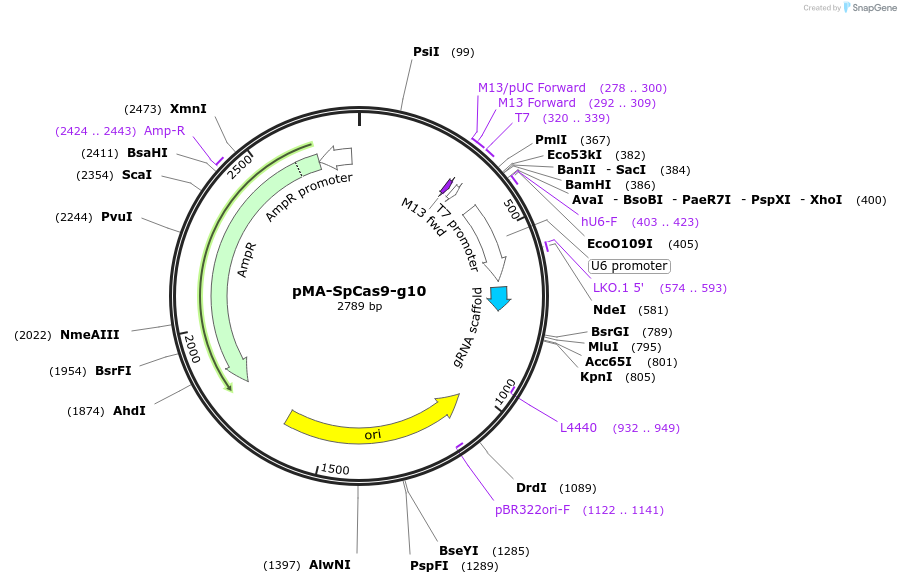
Plasmid#80793PurposeBsaI-based cloning of SpCas9 gRNA guide sequence, position 10/20/30 in the arrayDepositorInsertCRISPR gRNA expression cassette (for SpCas9)
UseCRISPRTagsnaExpressionMammalianPromoterU6Available SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
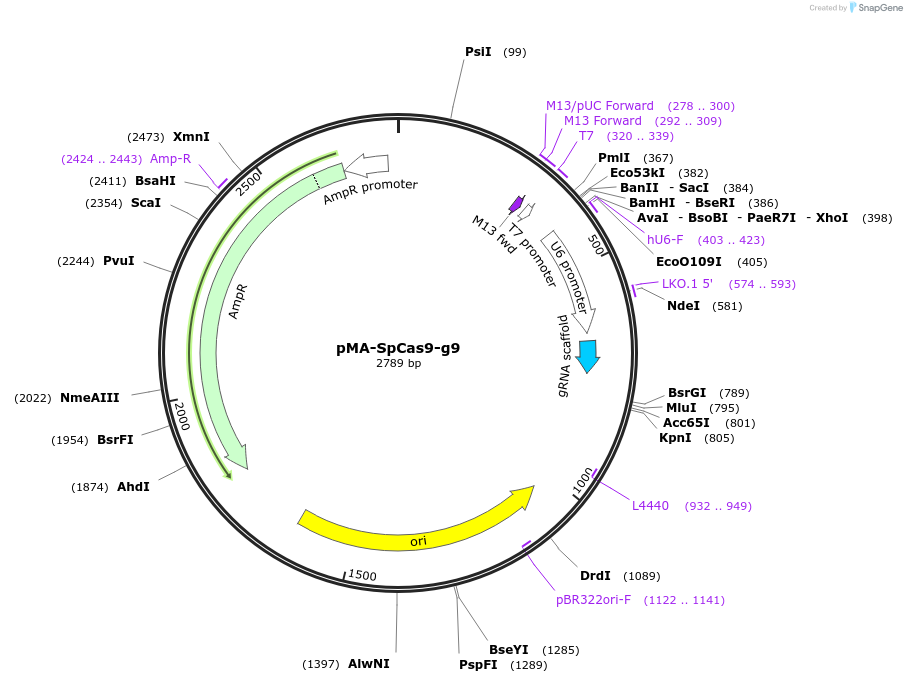
pMA-SpCas9-g9
Plasmid#80792PurposeBsaI-based cloning of SpCas9 gRNA guide sequence, position 9/19/29 in the arrayDepositorInsertCRISPR gRNA expression cassette (for SpCas9)
UseCRISPRTagsnaExpressionMammalianPromoterU6Available SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
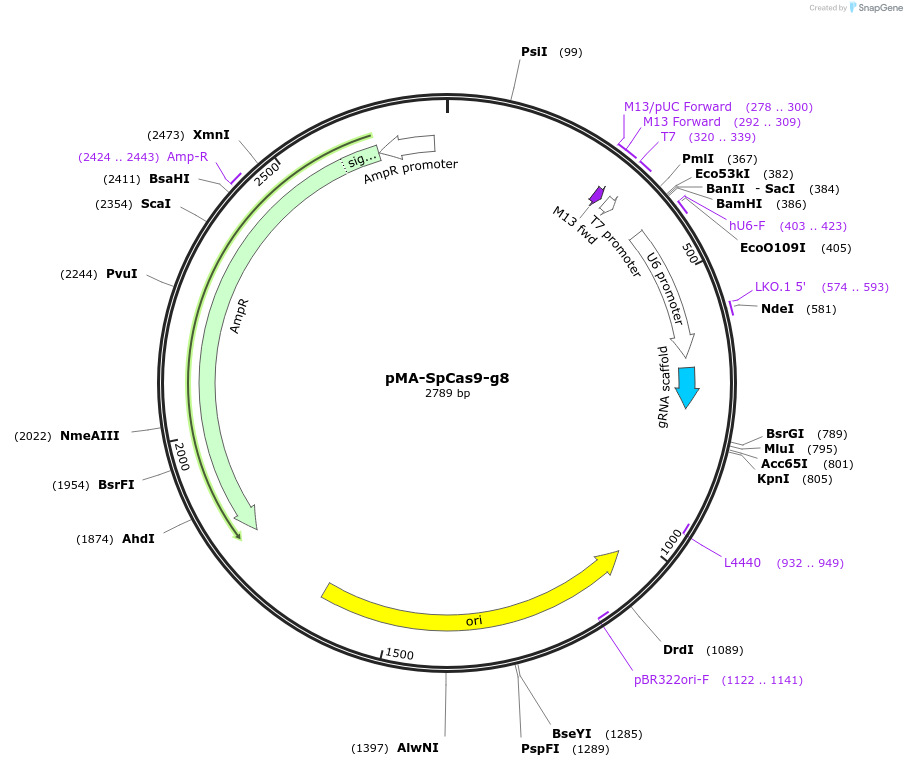
pMA-SpCas9-g8
Plasmid#80791PurposeBsaI-based cloning of SpCas9 gRNA guide sequence, position 8/18/28 in the arrayDepositorInsertCRISPR gRNA expression cassette (for SpCas9)
UseCRISPRTagsnaExpressionMammalianPromoterU6Available SinceSept. 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
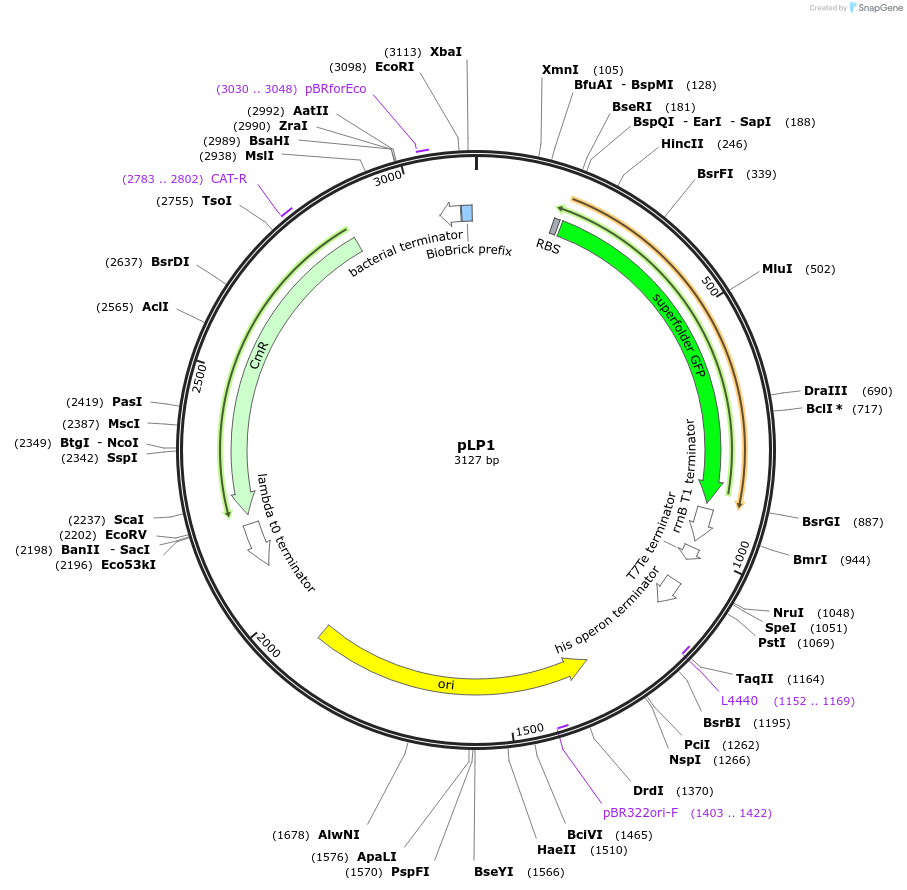
pLP1
Plasmid#209988PurposeEntry vector to store Level 0 partsDepositorTypeEmpty backboneExpressionBacterialAvailable SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
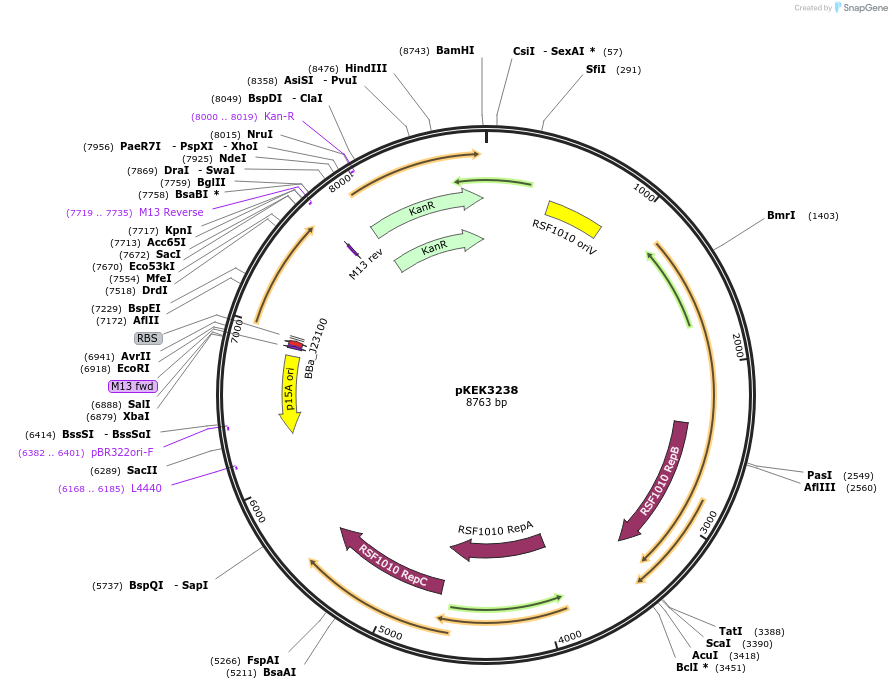
pKEK3238
Plasmid#234092PurposeCCP expression in diverse gram-negative bacteria, with pre-cloned Golden Gate Compatible (BsaI) restriction sites to facilitate promoter swapping.DepositorInsertsfCherry2
ExpressionBacterialMutationE117Q/T127I/G219APromoterpJ23100-BsaIAvailable SinceApril 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
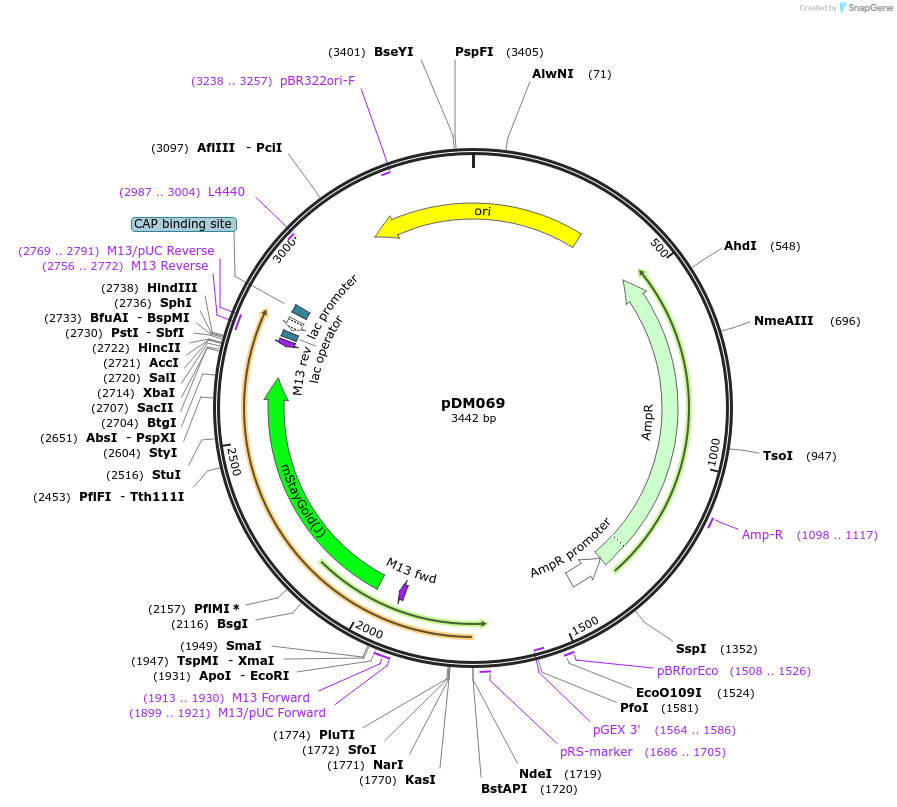
pDM069
Plasmid#216821PurposeAspergillus nidulans codon-adjusted mStayGold2 fluorescent protein, includes linker for N-terminal tagging.DepositorInsertmStayGold2
TagsRIPGLIN and VinnyLinkerExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
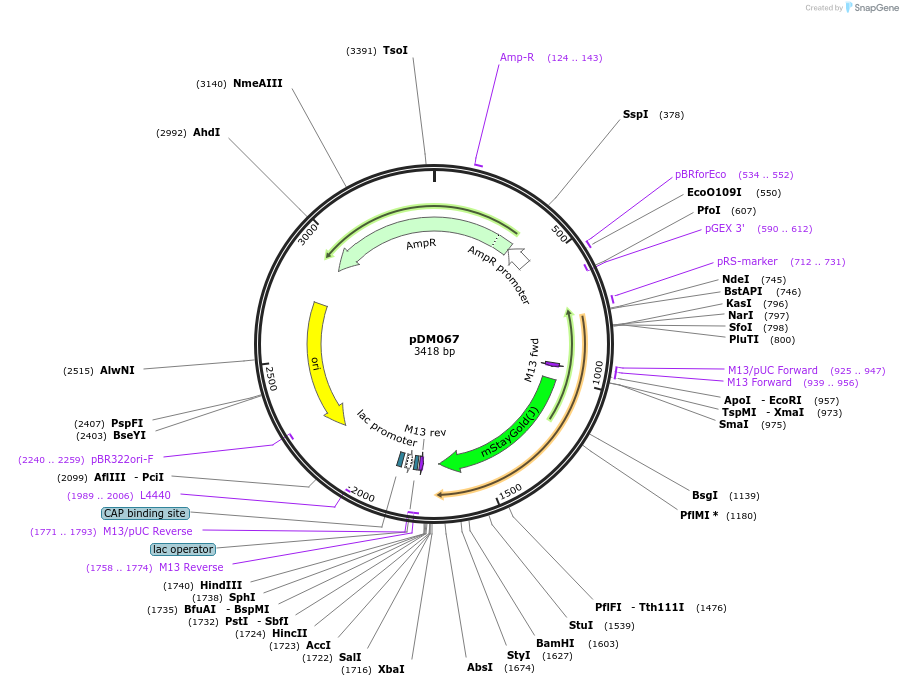
pDM067
Plasmid#216820PurposeAspergillus nidulans codon-adjusted mStayGold2 fluorescent protein, includes linker for C-terminal tagging.DepositorInsertmStayGold2
TagsVinnyLinkerExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
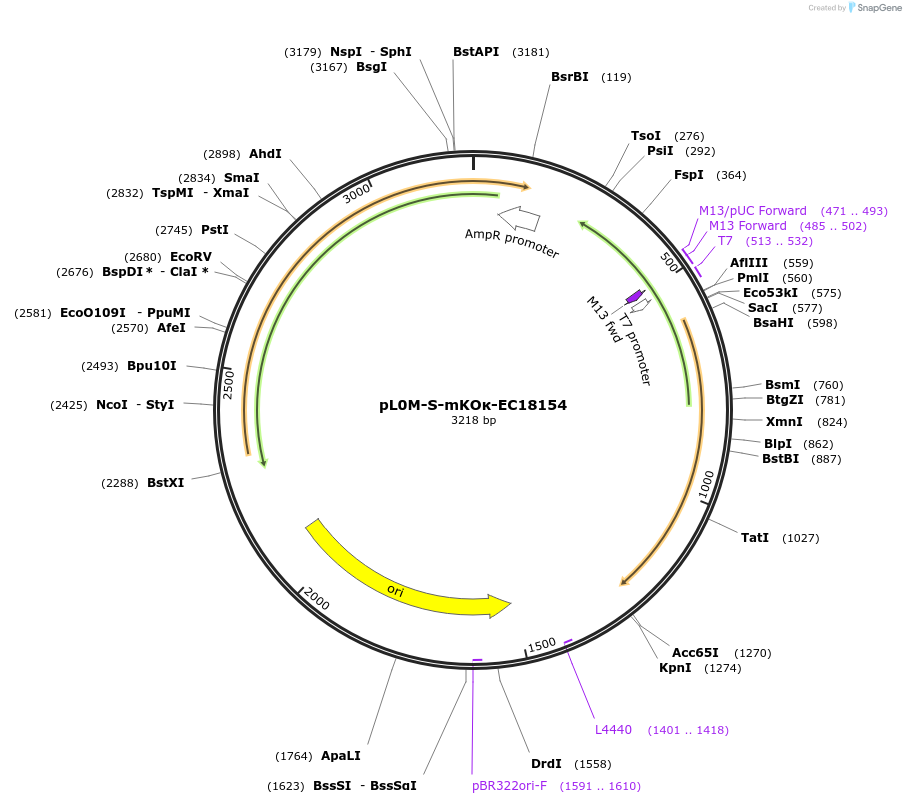
pL0M-S-mKOκ-EC18154
Plasmid#137077PurposeGolden Gate Level 0 S module for N-terminal fluorescent protein taggingDepositorInsertmKOκ DNA synthesis product with 5' and 3' extensions for Golden Gate cloning
UseSynthetic BiologyExpressionBacterialAvailable SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
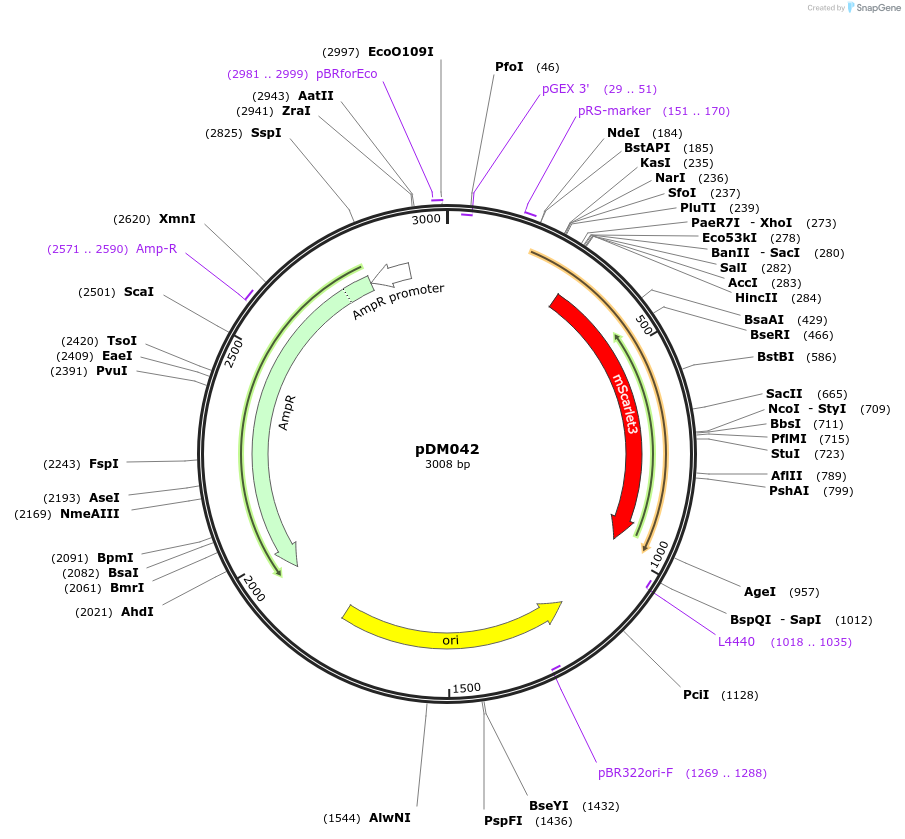
pDM042
Plasmid#216812PurposeAspergillus nidulans codon-adjusted mScarlet3 fluorescent protein, includes linker for C-terminal tagging.DepositorInsertmScarlet3
TagsVinnyLinkerExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
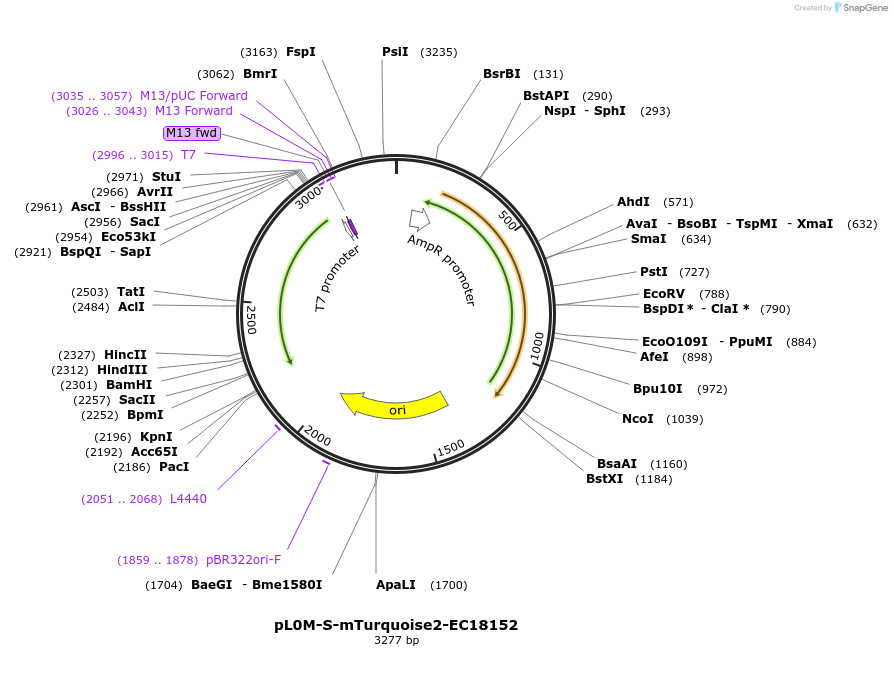
pL0M-S-mTurquoise2-EC18152
Plasmid#137074PurposeGolden Gate Level 0 S module for N-terminal fluorescent protein taggingDepositorInsertmTurquoise2 DNA synthesis product with 5' and 3' extensions for Golden Gate cloning
UseSynthetic BiologyExpressionBacterialAvailable SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
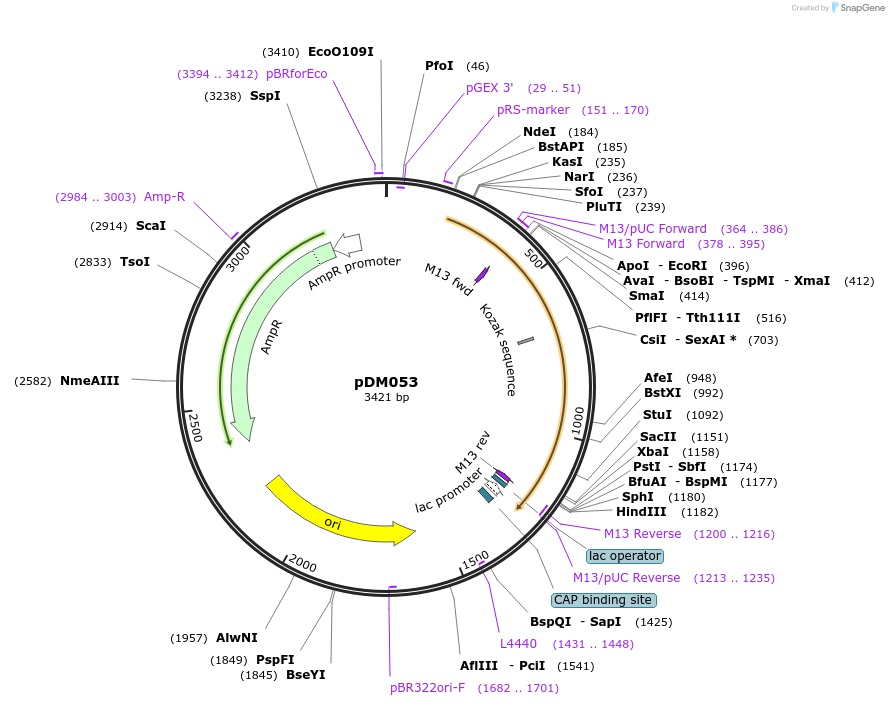
pDM053
Plasmid#216815PurposeAspergillus nidulans codon-adjusted mNeonGreen fluorescent protein, includes linker for N-terminal tagging.DepositorInsertmNeonGreen
TagsRIPGLINExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
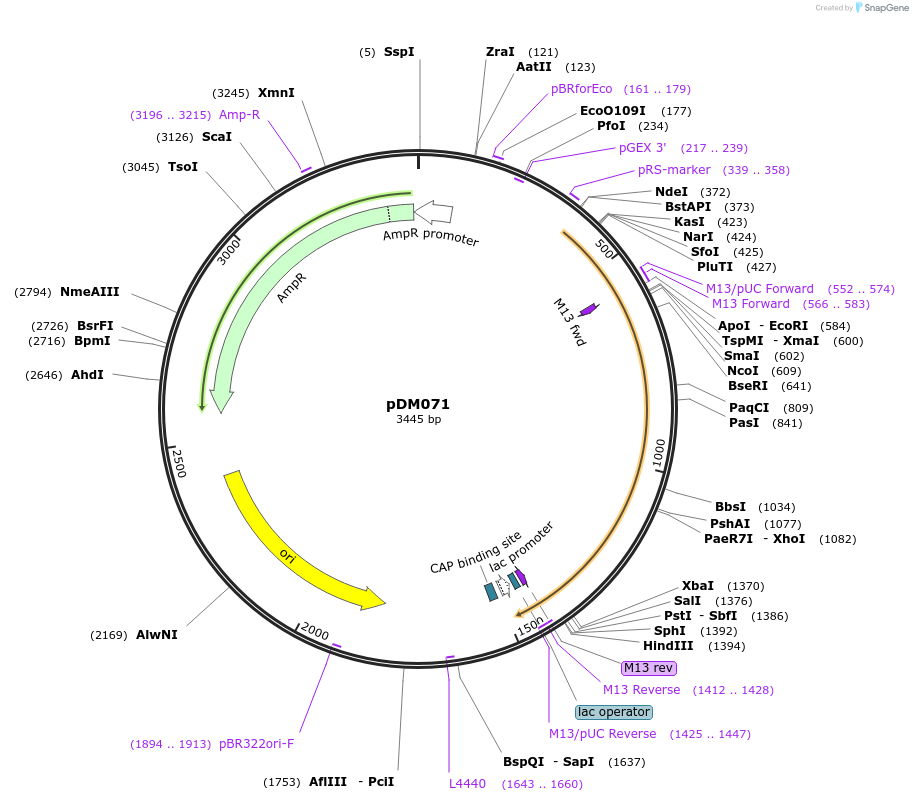
pDM071
Plasmid#216817PurposeAspergillus nidulans codon-adjusted mCardinal fluorescent protein, includes linker for N-terminal tagging.DepositorInsertmCardinal
TagsRIPGLINExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
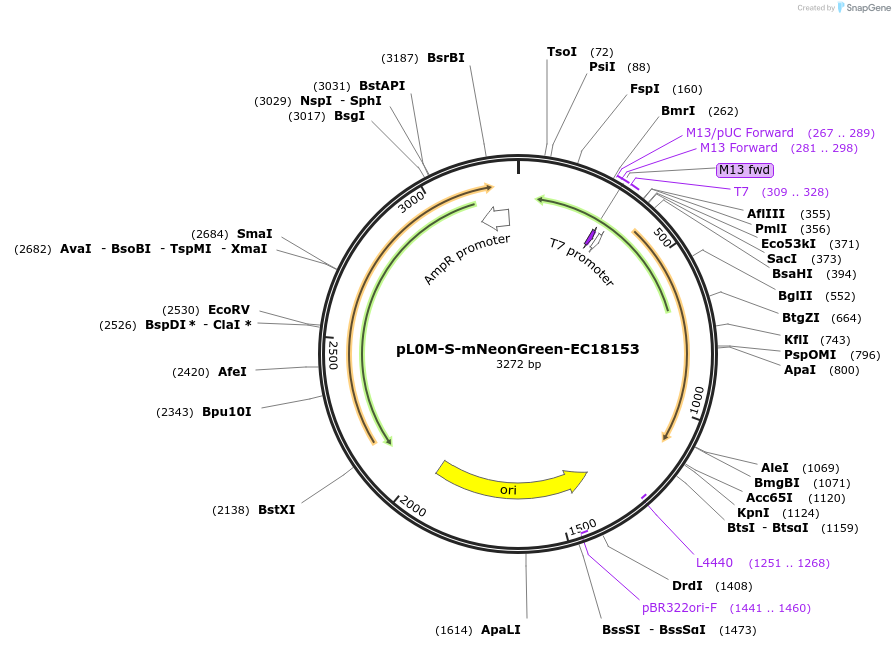
pL0M-S-mNeonGreen-EC18153
Plasmid#137075PurposeGolden Gate Level 0 S module for N-terminal fluorescent protein taggingDepositorInsertmNeonGreen DNA synthesis product with 5' and 3' extensions for Golden Gate cloning
UseSynthetic BiologyExpressionBacterialAvailable SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
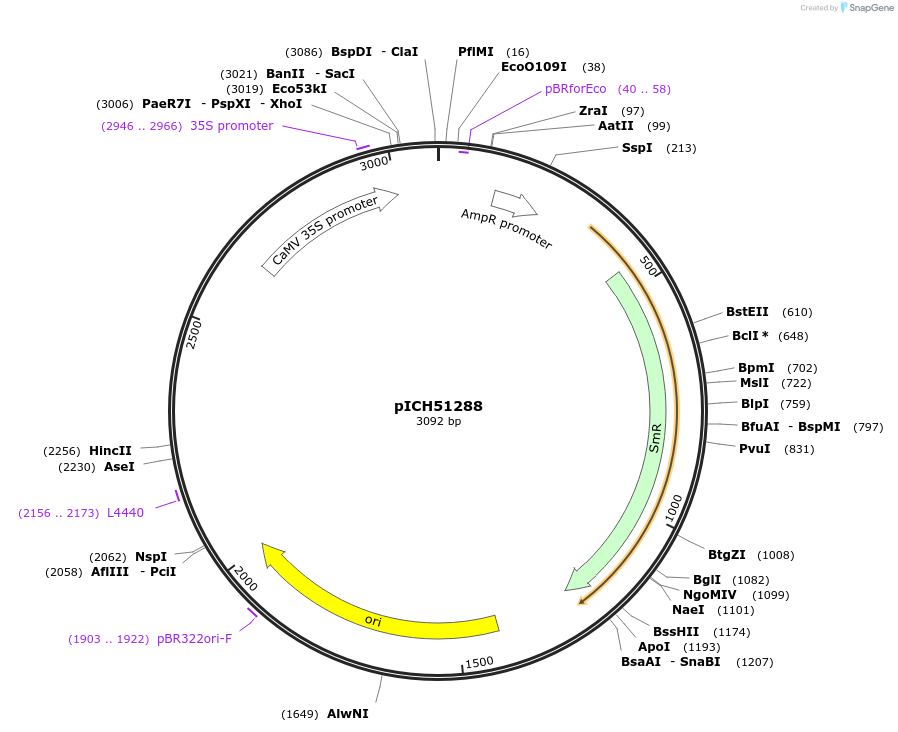
pICH51288
Plasmid#50269DepositorInsertpromoter (double), 35s (Cauliflower Mosaic Virus) + 5'UTR, omega (Tobacco Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
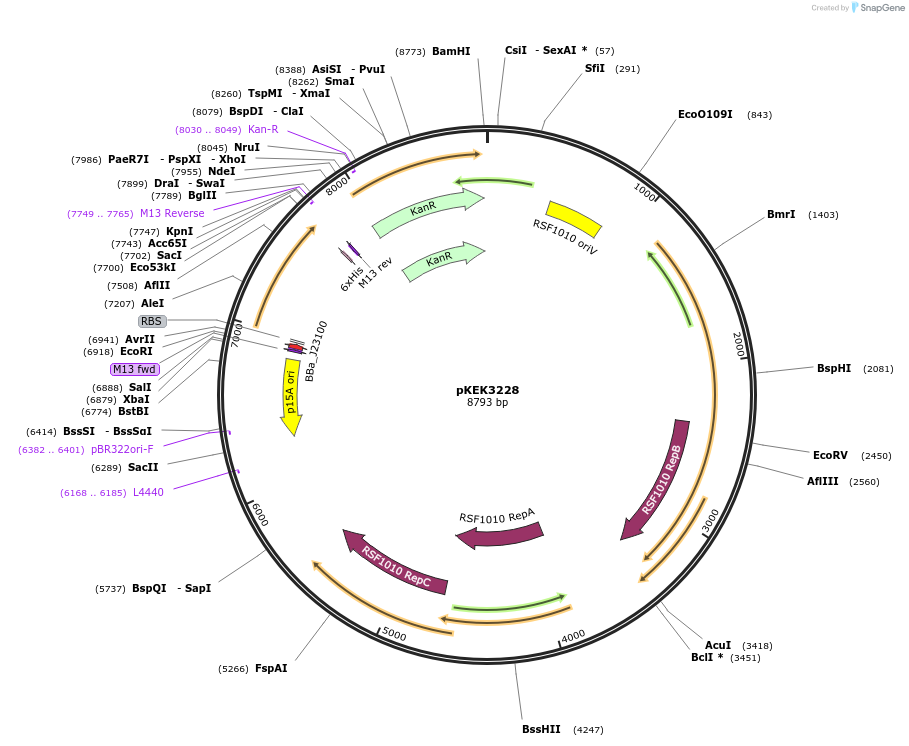
pKEK3228
Plasmid#234090PurposeCCP expression in diverse gram-negative bacteria, with pre-cloned Golden Gate Compatible (BsaI) restriction sites to facilitate promoter swapping.DepositorInsertaeBlue
ExpressionBacterialPromoterpJ23100-BsaIAvailable SinceApril 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
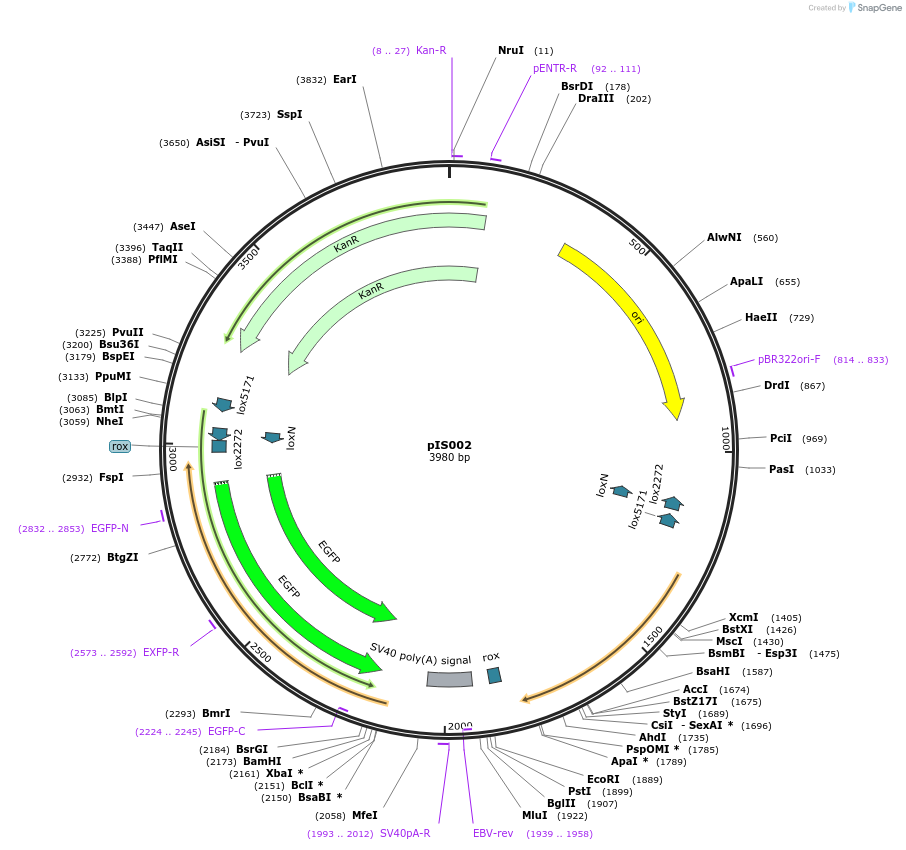
pIS002
Plasmid#194013PurposeThis plasmid is designed for use in a procedure to synthesize single-stranded DNA containing only the insert.DepositorInsertC16 Ex3 XTR f1
ExpressionBacterialAvailable SinceJan. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
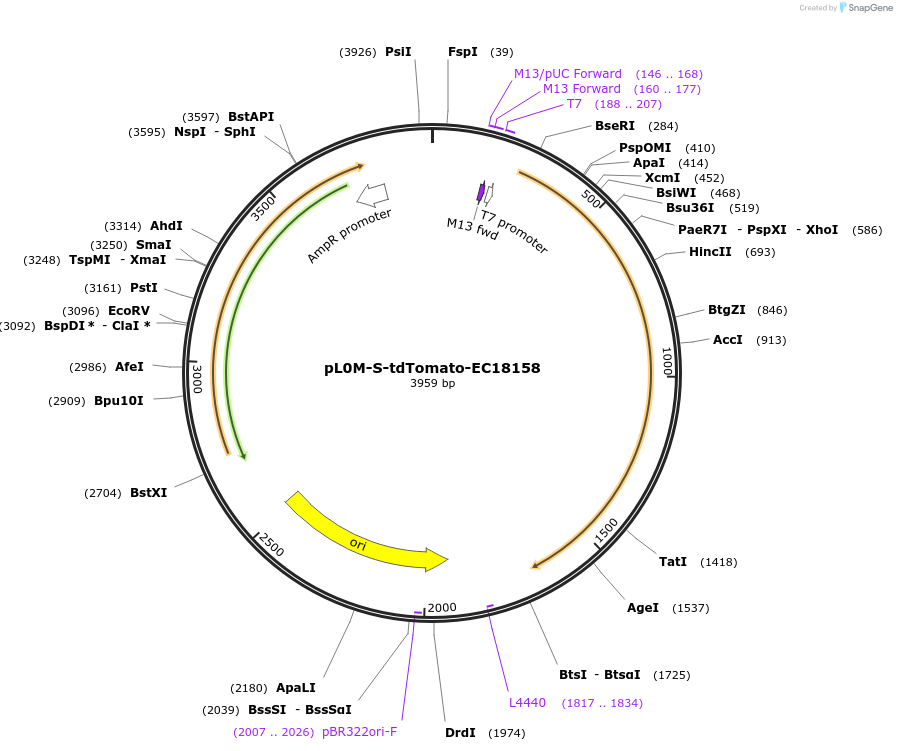
pL0M-S-tdTomato-EC18158
Plasmid#137078PurposeGolden Gate Level 0 S module for N-terminal fluorescent protein taggingDepositorInserttdTomato DNA synthesis product with 5' and 3' extensions for Golden Gate cloning
UseSynthetic BiologyExpressionBacterialAvailable SinceJune 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
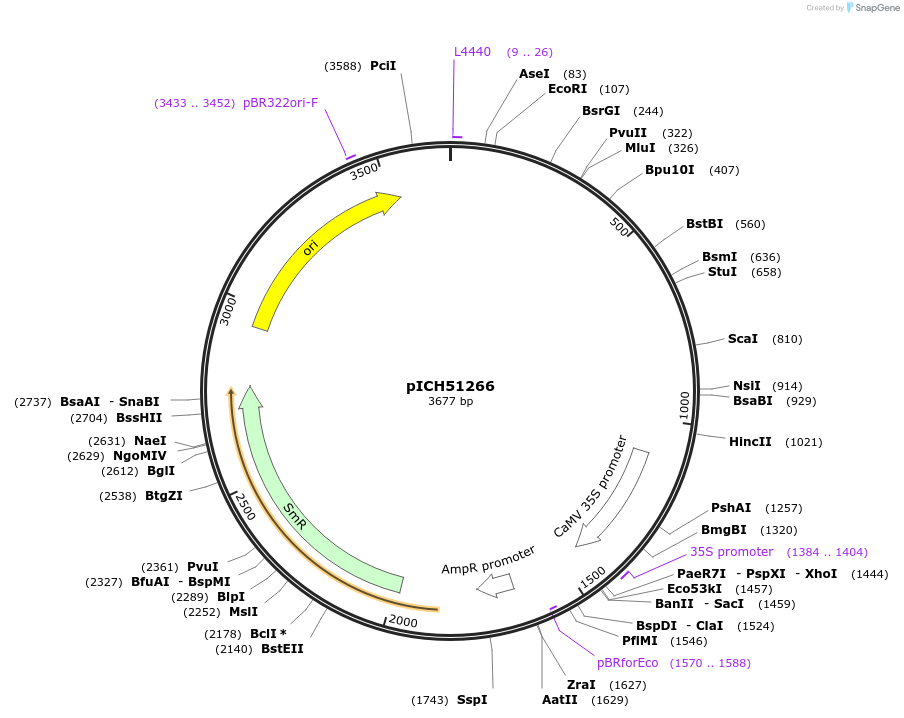
pICH51266
Plasmid#50267DepositorInsertpromoter (1.3 kb ), 35s (Cauliflower Mosaic Virus) + 5'UTR omega (Tobacco Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
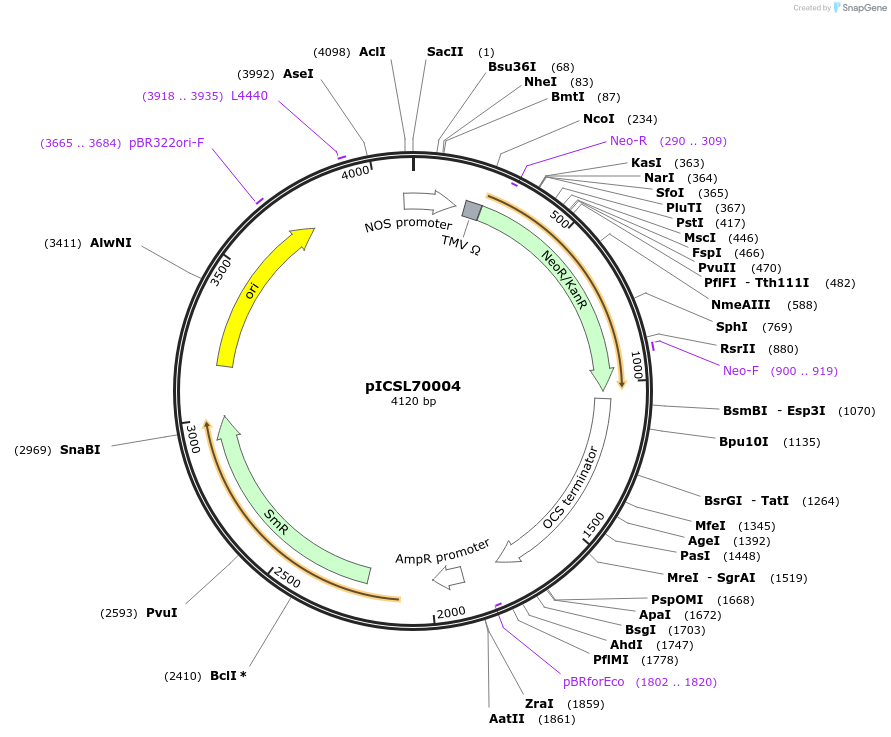
pICSL70004
Plasmid#50334DepositorInsertkanamycin/neomycin/paromomycin resistance cassette (Nos promoter (A. tumefaciens) + NptII (E.coli) + Ocs terminator (A. tumefaci
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
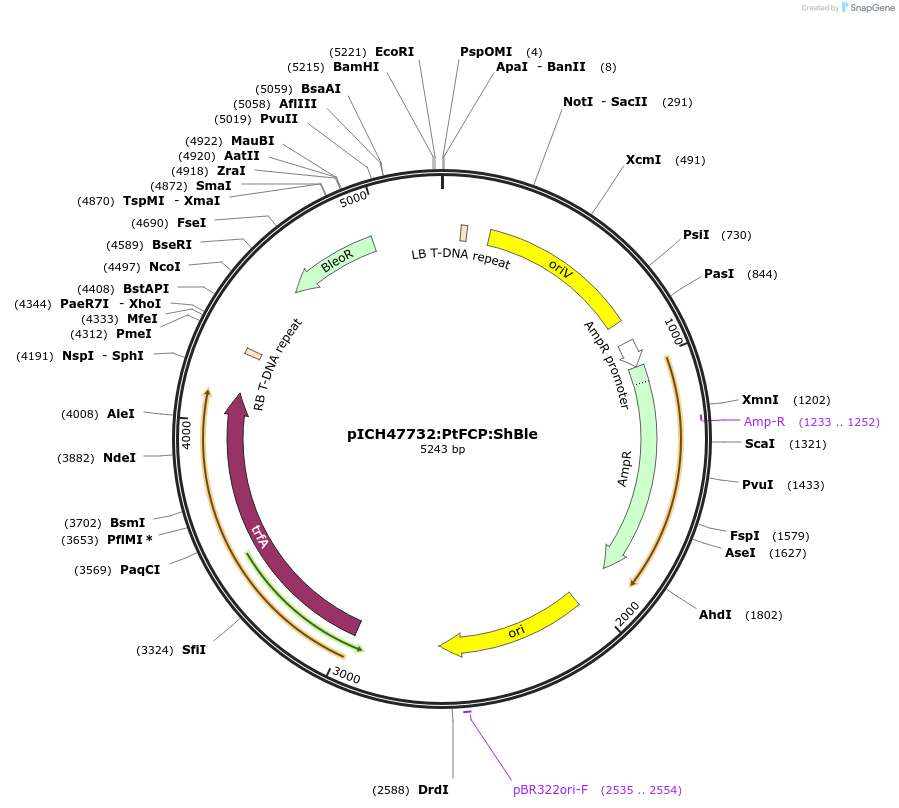
pICH47732:PtFCP:ShBle
Plasmid#104893PurposeExpresses ShBle with FCP promoter/terminator, Golden Gate Assembly, for Phaeodactylum transformation.DepositorInsertShBle
UsePhaeodactylum; golden gate assemblyTagsPtFCP promoter and terminatorAvailable SinceDec. 28, 2017AvailabilityAcademic Institutions and Nonprofits only