We narrowed to 2,377 results for: pET28
-
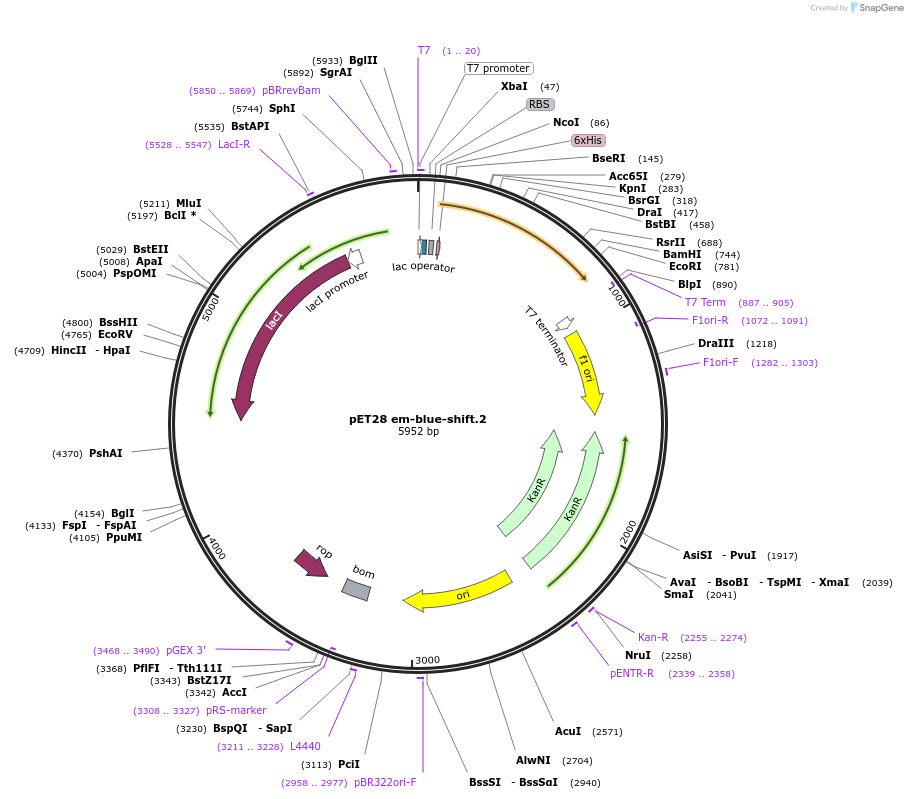
Plasmid#191901PurposeEncodes a GFP design, em-blue-shift.2DepositorInsertem-blue-shift.2
TagsHis TagExpressionBacterialMutationT65S S72A T108I Y145F V150IPromoterT7Available SinceDec. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
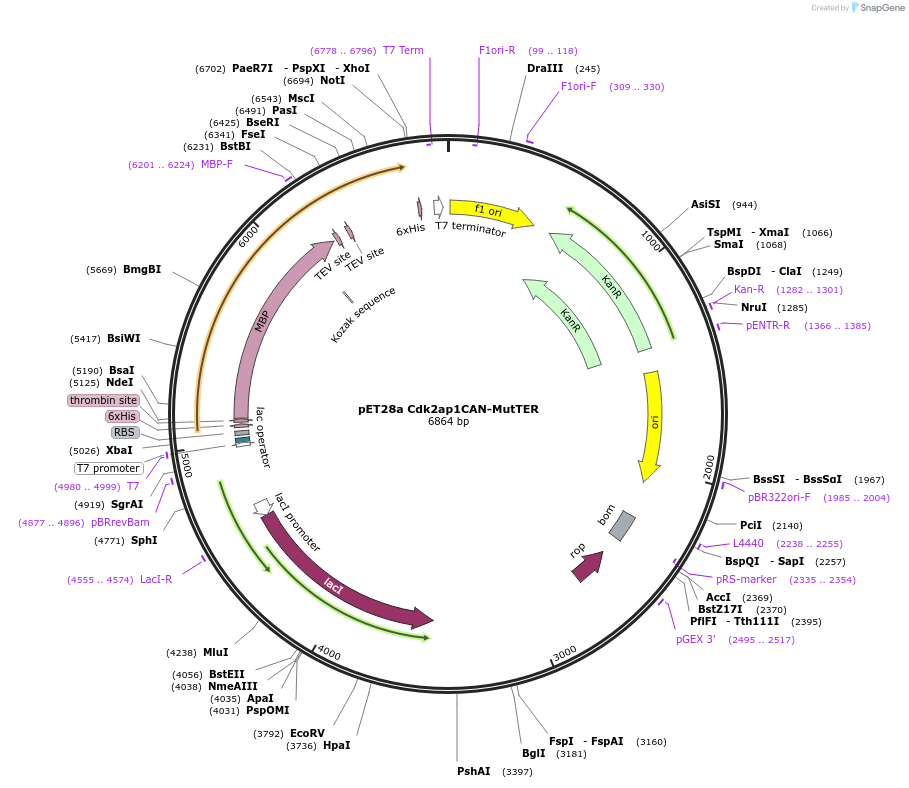
pET28a Cdk2ap1CAN-MutTER
Plasmid#178033PurposeExpression vector for the purpose of protein purification.DepositorInsertCdk2ap1 (Cdk2ap1 Mouse)
UseNonviralTags6xHIS - Thrombin - MBP- TEV-TRSExpressionBacterialMutationChanged T(108)A, E(109)A, R(110)APromoterT7LacIAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
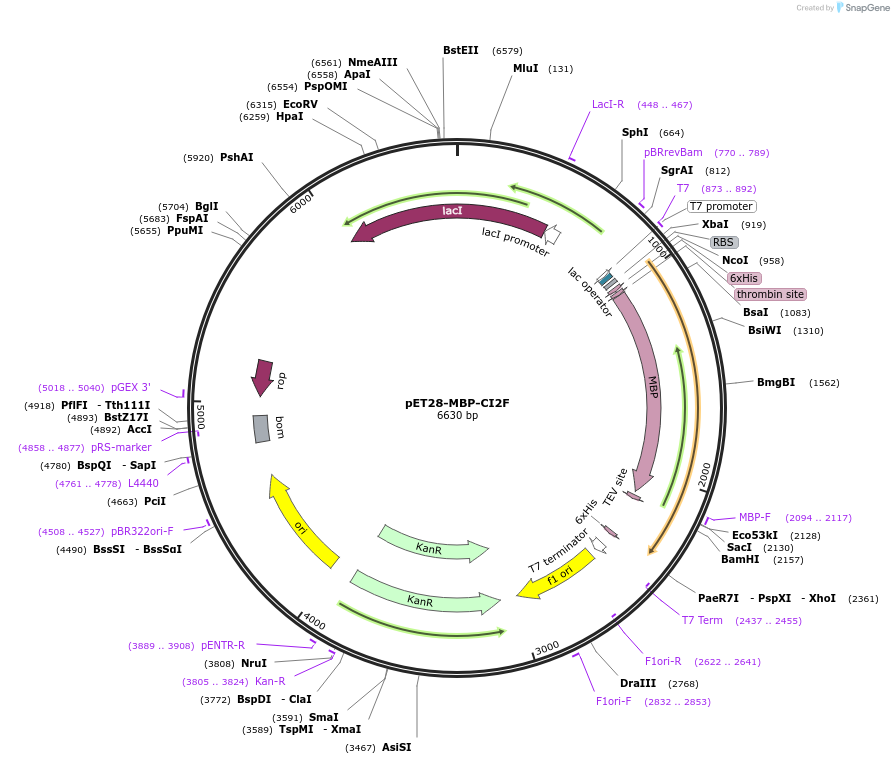
pET28-MBP-CI2F
Plasmid#191254PurposeExpresses chymotrypsin inhibitor with an N-terminal MBP fusion proteinDepositorInsertChymotrypsin Inhibitor 2
TagsMaltose Binding ProteinExpressionBacterialAvailable SinceOct. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
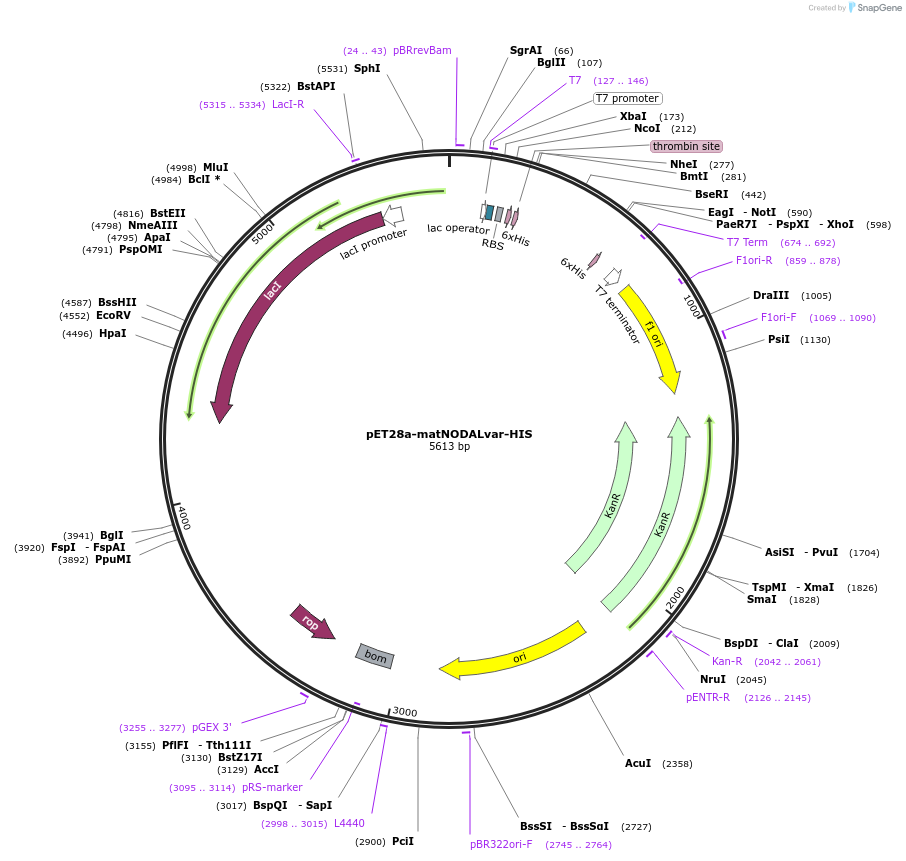
pET28a-matNODALvar-HIS
Plasmid#115265PurposeFor bacterial recombinant protein production of the mature peptide of a human NODAL splice variantDepositorAvailable SinceSept. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
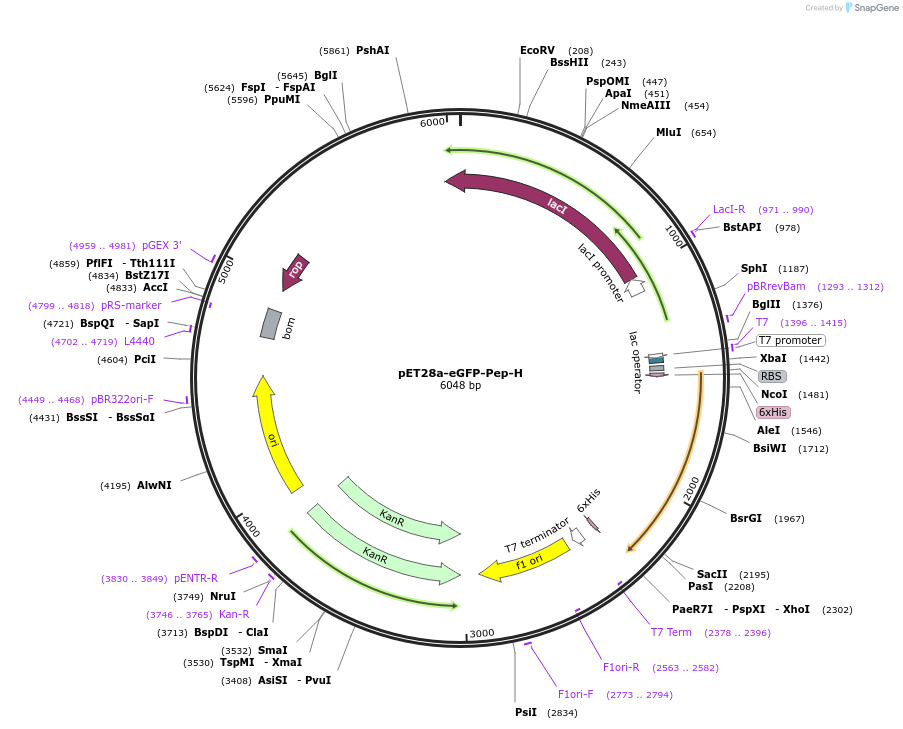
pET28a-eGFP-Pep-H
Plasmid#185788PurposeEncodes enhanced green flourescent protein linked to Pep-H self-assembling peptide via a GS-linker sequenceDepositorInserteGFP-Pep4
TagsHis-tagExpressionBacterialPromoterT7Available SinceAug. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
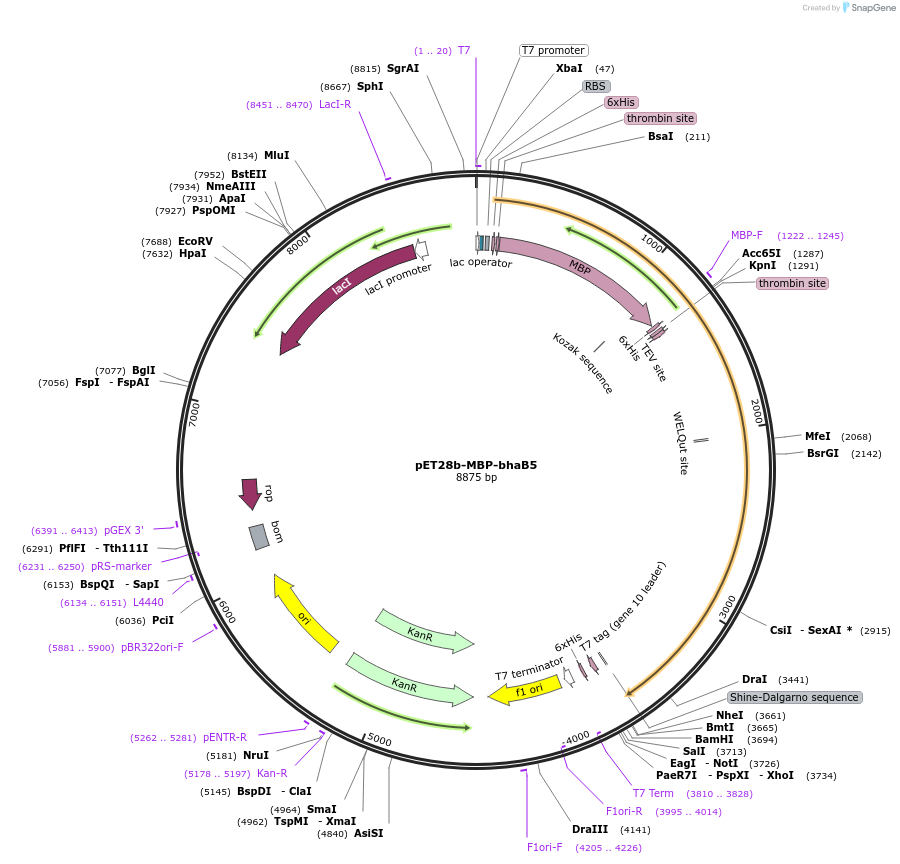
pET28b-MBP-bhaB5
Plasmid#167812PurposeExpresses His6- and MBP-tagged BhaB5 in E. coliDepositorInsertBhaB5
TagsHis6, MBPExpressionBacterialPromoterT7Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
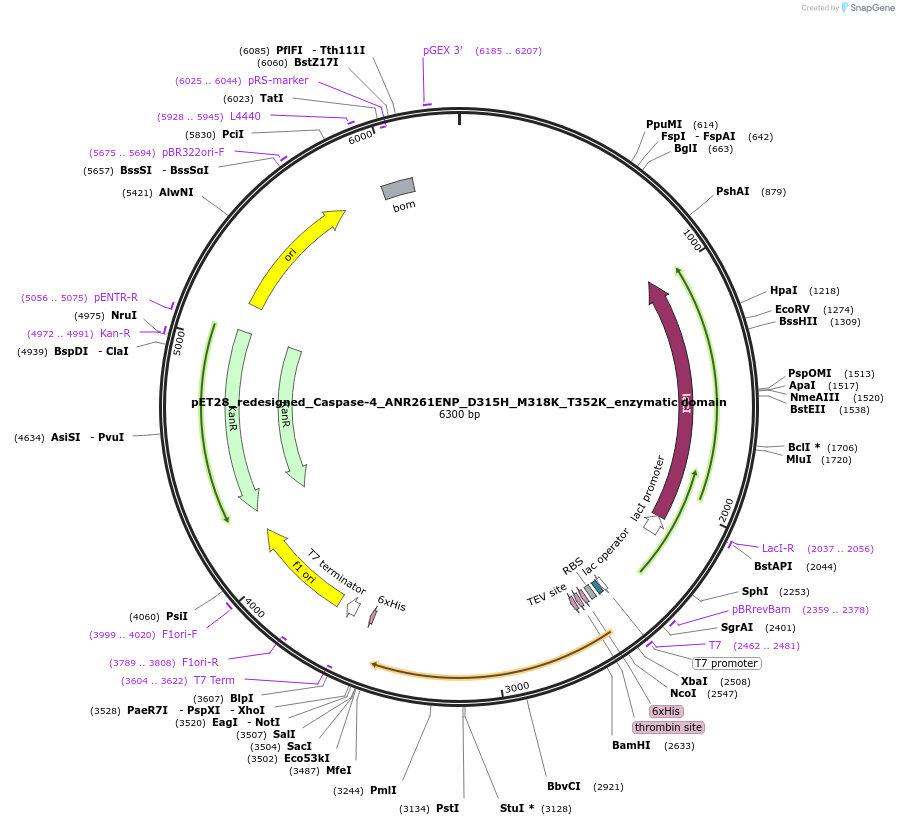
pET28_redesigned_Caspase-4_ANR261ENP_D315H_M318K_T352K_enzymatic domain
Plasmid#183387PurposeOverexpression of catalytic domain of inflammatory caspase in bacteriaDepositorInsertredesigned Caspase-4 (AA94-377) (CASP4 Human)
TagsHis-TEVExpressionBacterialMutationANR261ENP, D315H, M318K, T352KPromoterT7Available SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
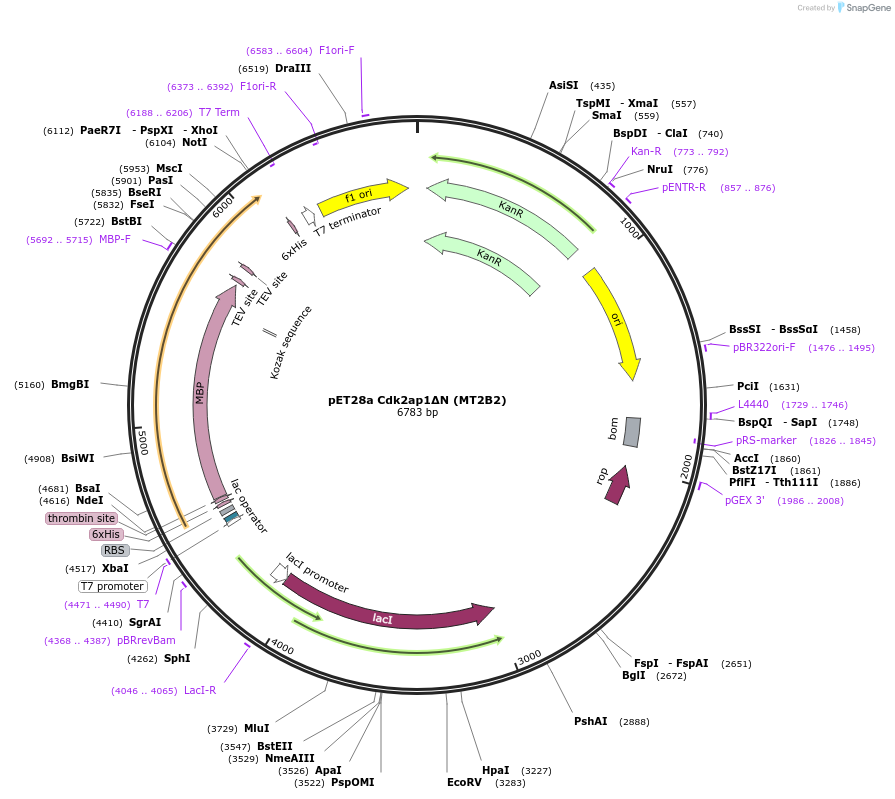
pET28a Cdk2ap1ΔN (MT2B2)
Plasmid#178034PurposeExpression vector for the purpose of protein purification.DepositorInsertCdk2ap1 (Cdk2ap1 Mouse)
UseNonviralTags6xHIS - Thrombin - MBP- TEV-TRSExpressionBacterialMutationFirst N-Terminal 27aa are deletedPromoterT7LacIAvailable SinceFeb. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
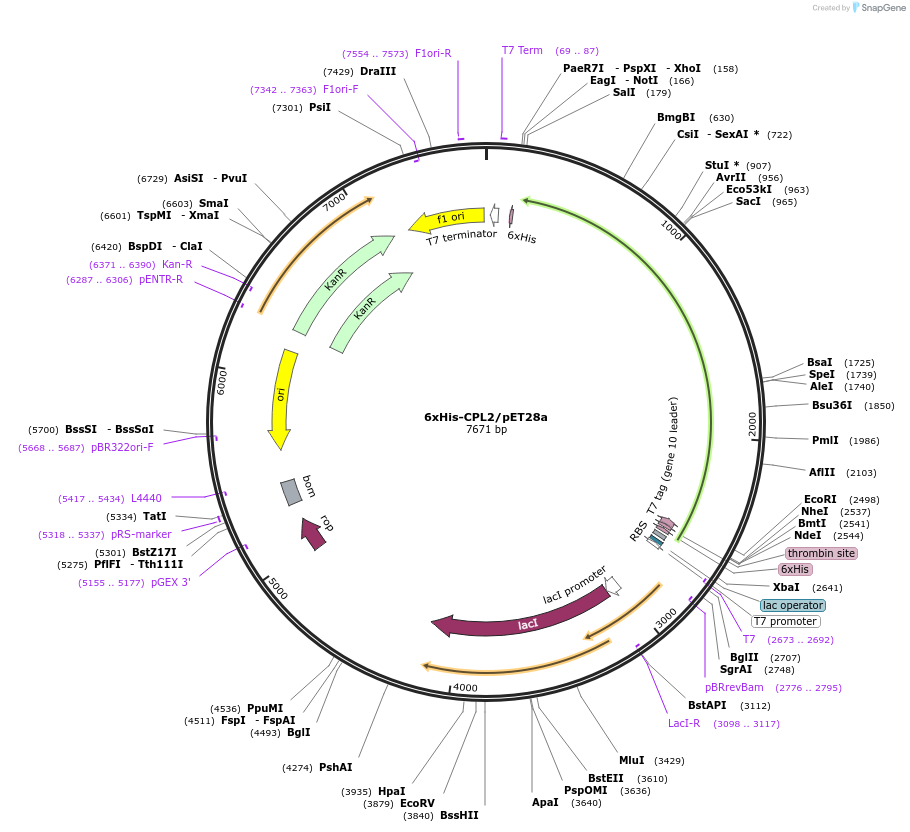
6xHis-CPL2/pET28a
Plasmid#172095PurposecDNA of CER11/CPL2 cloned into pET28a for protein expressionDepositorAvailable SinceOct. 20, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
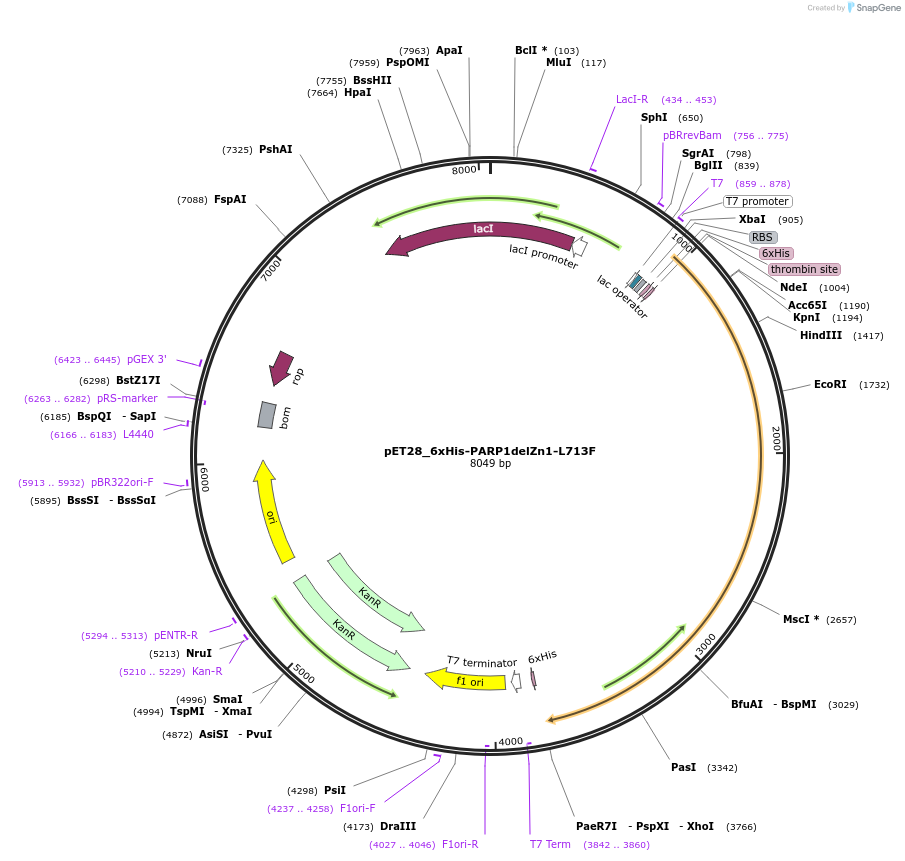
pET28_6xHis-PARP1delZn1-L713F
Plasmid#173937PurposeBacterial expression of PARP1 lacking Zn1 domain but containing L713F gain-of-function mutationDepositorInsertPARP1delZn1-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 1-96; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
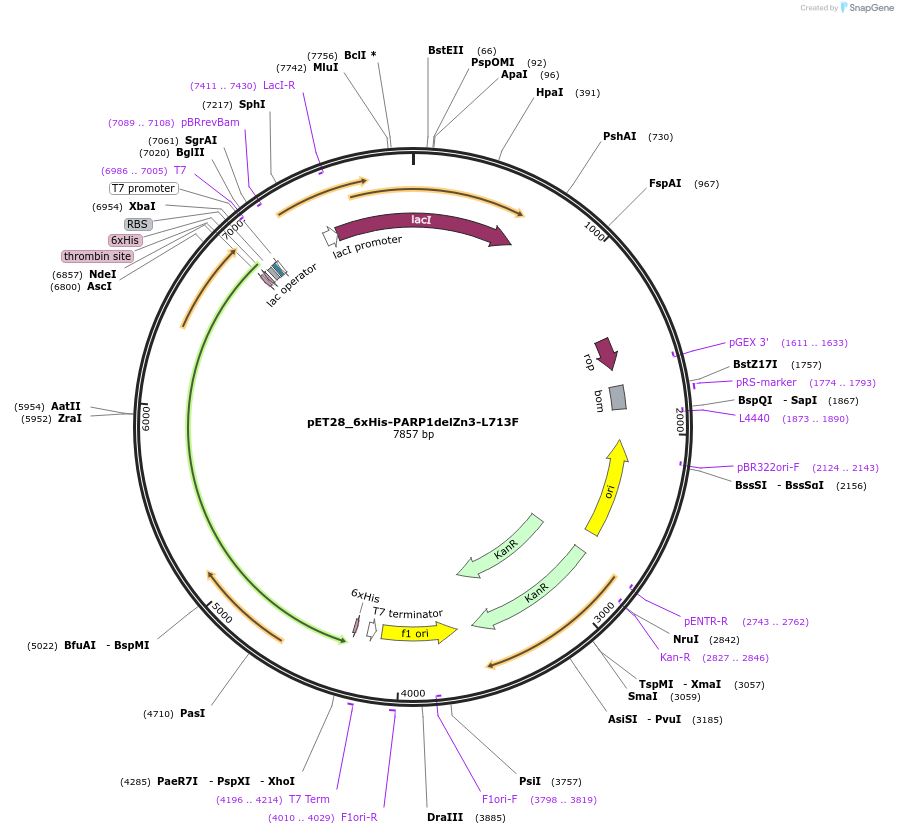
pET28_6xHis-PARP1delZn3-L713F
Plasmid#173934PurposeBacterial expression of PARP1 lacking Zn3 domain but containing L713F gain-of-function mutationDepositorInsertPARP1delZn3-L713F (PARP1 Human)
Tags6xHisExpressionBacterialMutationDeletion of residues 214-373; L713F, V762AAvailable SinceSept. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
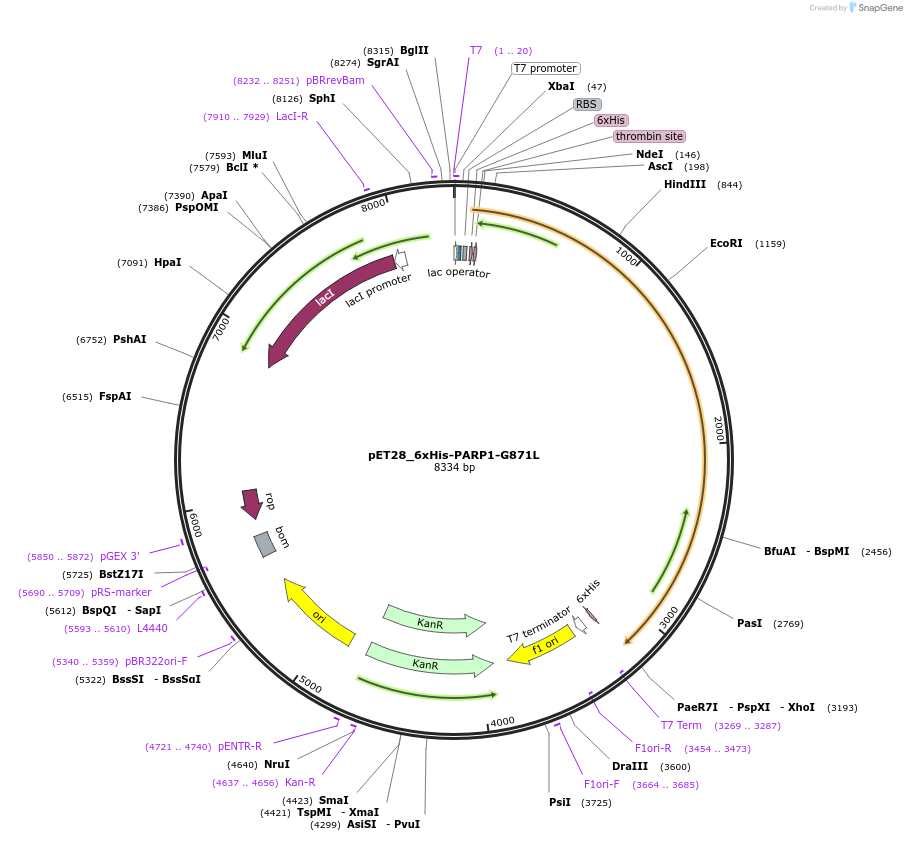
pET28_6xHis-PARP1-G871L
Plasmid#174798PurposeBacterial expression of a hyperactive PARP1 mutant (G871L destabilizes the autoinhibitory HD subdomain)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
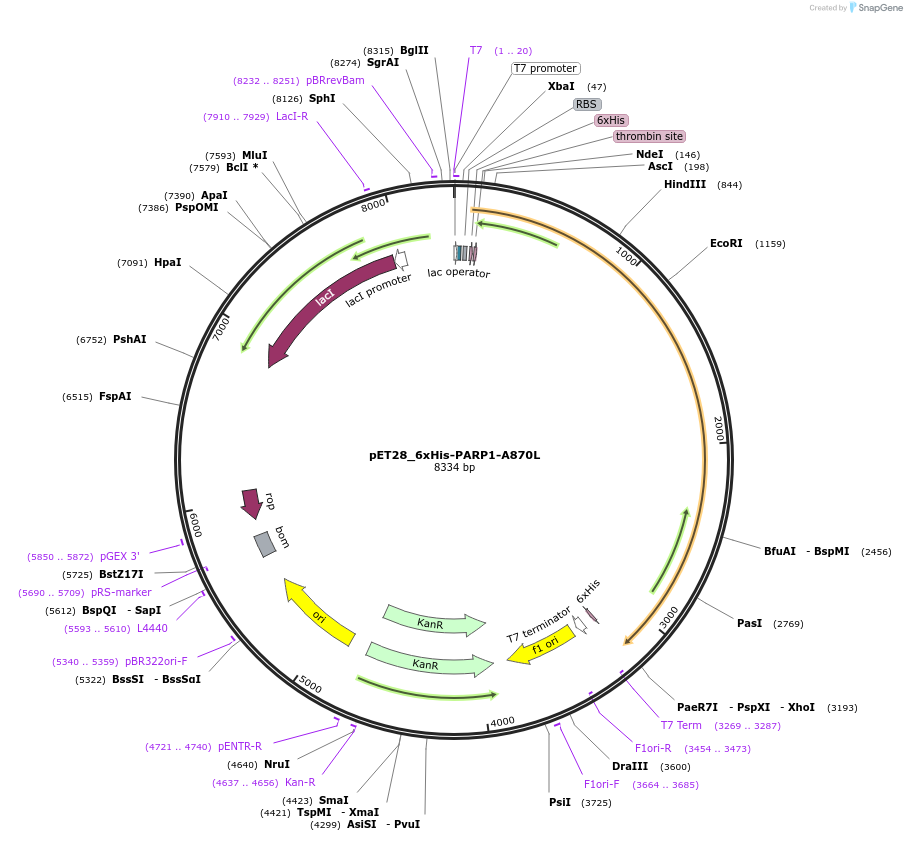
pET28_6xHis-PARP1-A870L
Plasmid#174797PurposeBacterial expression of a hyperactive PARP1 mutant (A870L destabilizes the autoinhibitory HD subdomain)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
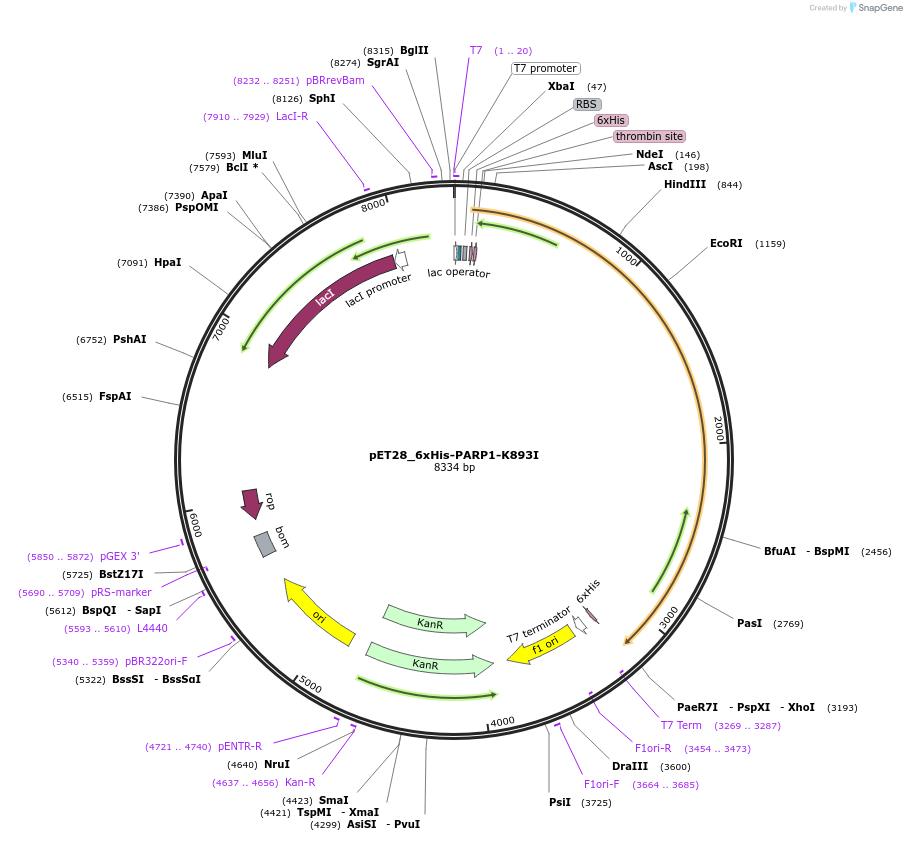
pET28_6xHis-PARP1-K893I
Plasmid#174799PurposeBacterial expression of an inactive mutant (K893I may disrupt NAD+ binding)DepositorAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
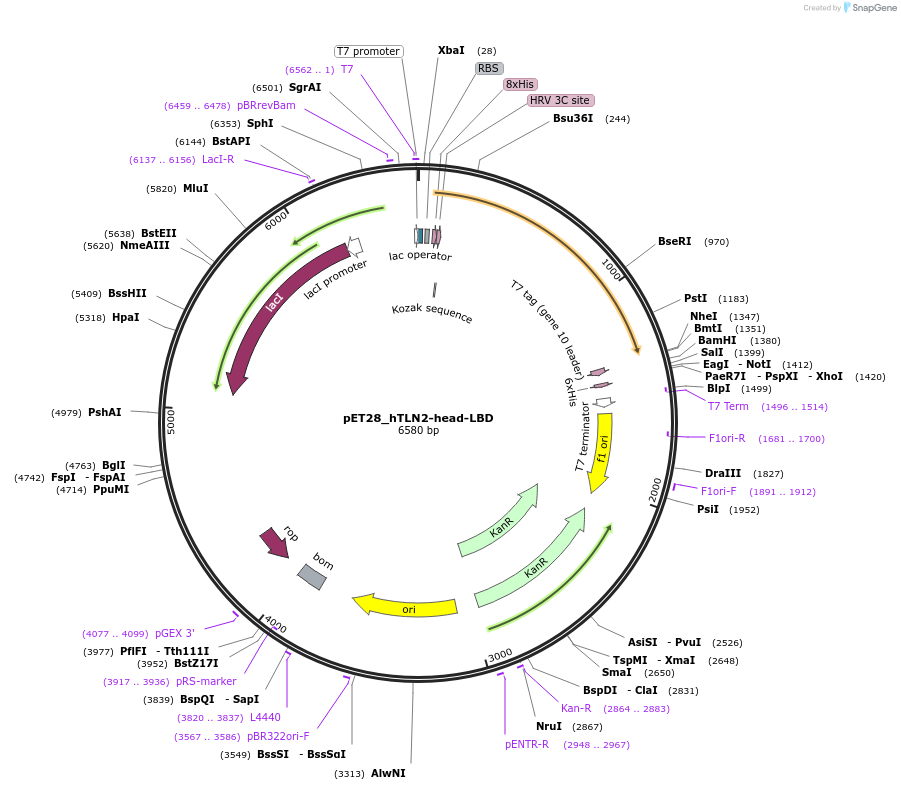
pET28_hTLN2-head-LBD
Plasmid#164837PurposeExpression of human talin2 head mutantDepositorAvailable SinceMarch 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
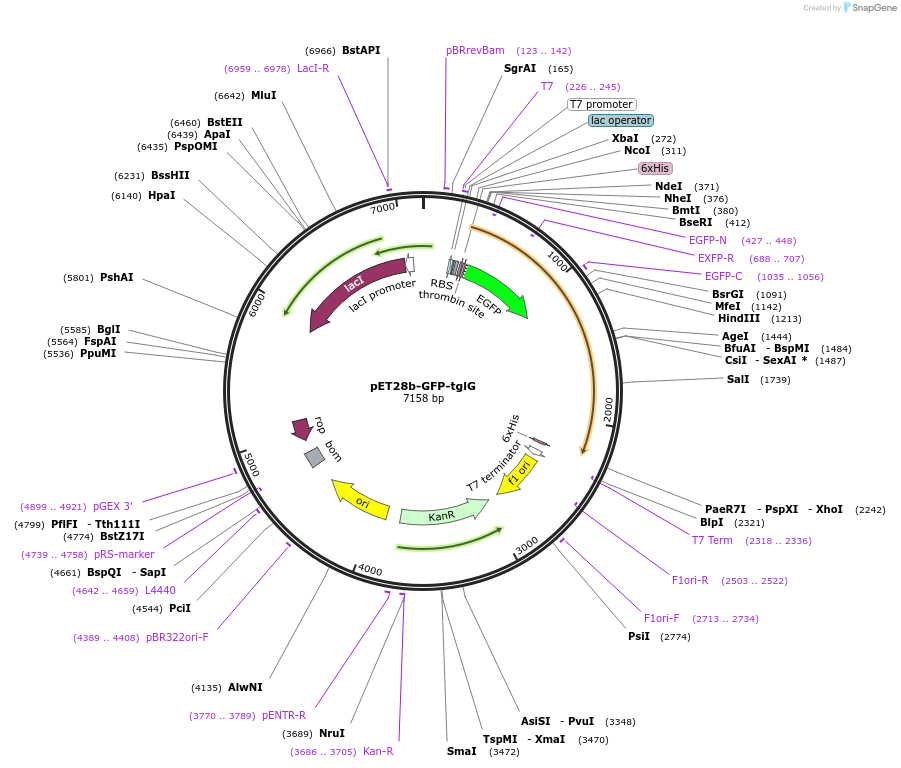
pET28b-GFP-tglG
Plasmid#134679PurposeExpresses His6-GFP-TglG fusion in E. ColiDepositorInsertTglG
TagsHis6-GFPExpressionBacterialPromoterT7Available SinceJuly 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
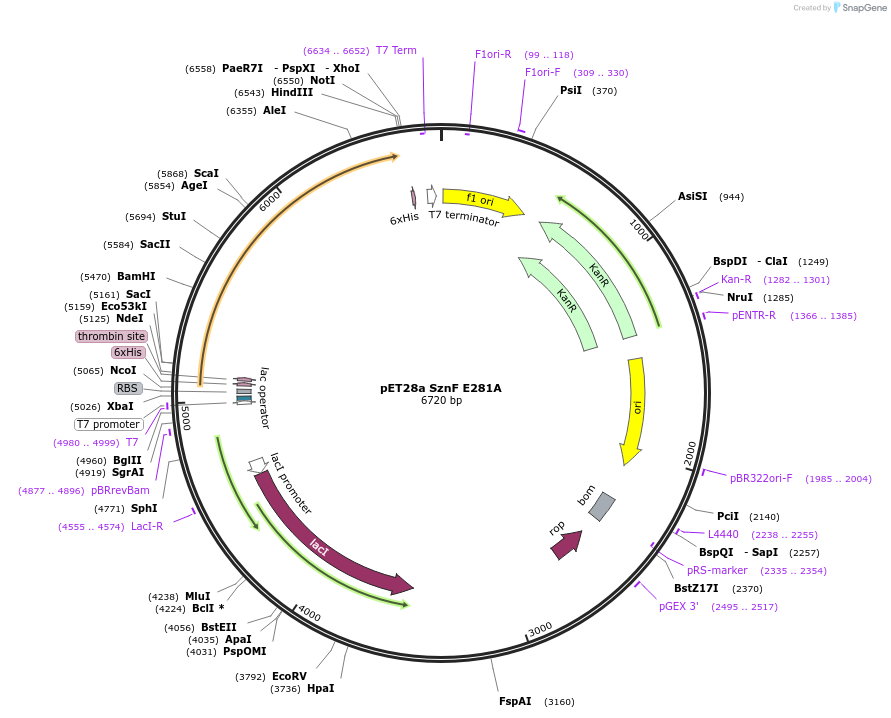
pET28a SznF E281A
Plasmid#139591Purposeexpresses SznF E281ADepositorInsertSznF E281A
TagshexahistidineExpressionBacterialMutationchanged glutamate 281 to alaninePromoterT7Available SinceMarch 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
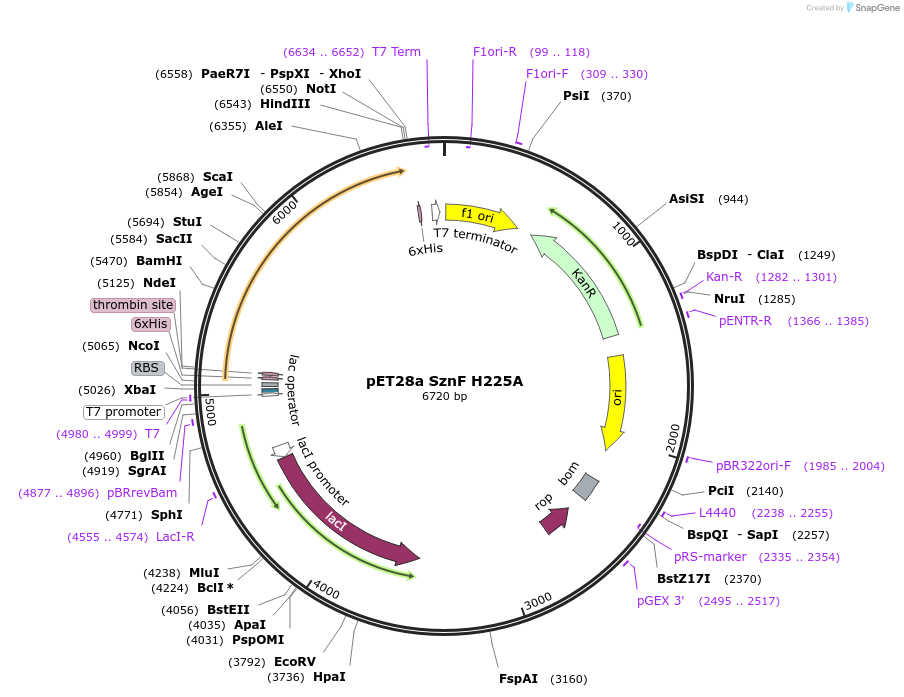
pET28a SznF H225A
Plasmid#139590Purposeexpresses SznF H225ADepositorInsertSznF H225A
TagshexahistidineExpressionBacterialMutationchanged histidine 225 to alaninePromoterT7Available SinceMarch 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
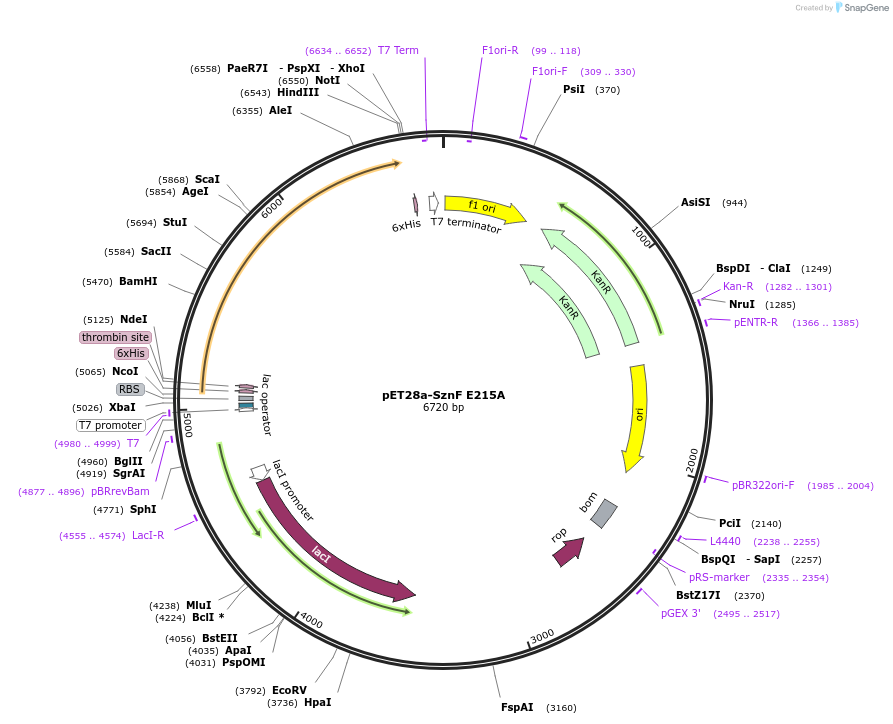
pET28a-SznF E215A
Plasmid#139565Purposeexpresses SznF E215A in E. coliDepositorInsertSznF E215A
TagshexahistidineExpressionBacterialMutationchanged glutamate 215 to alaninePromoterT7Available SinceMarch 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
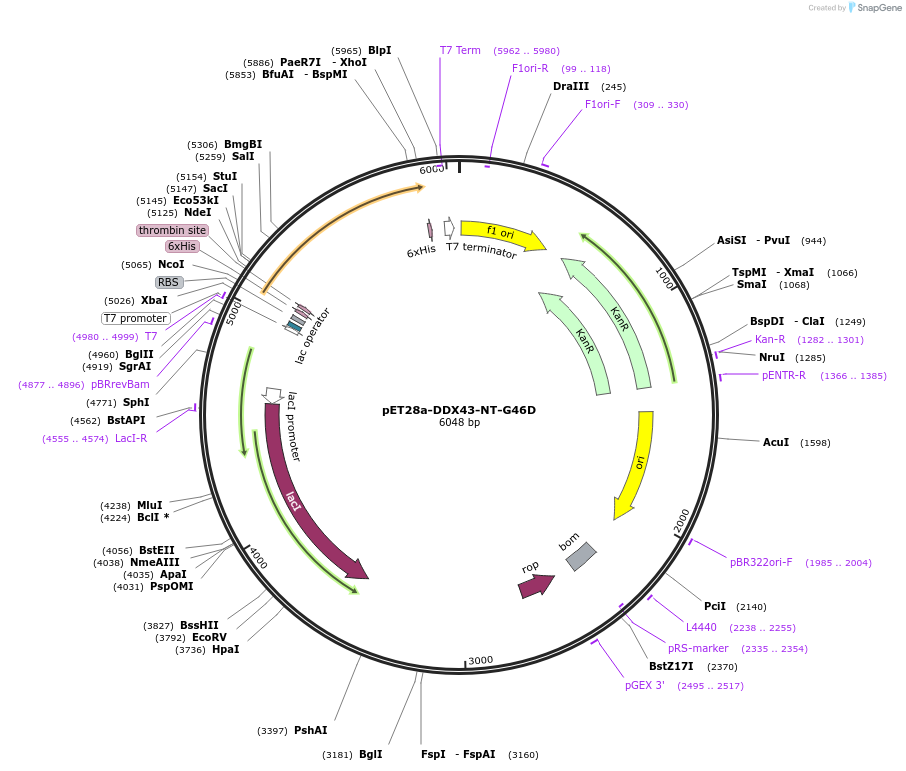
pET28a-DDX43-NT-G46D
Plasmid#134588PurposeExpress human DDX43 N-terminal region (1-253 aa, with G46D mutation) in E. coliDepositorInsertDDX43-N-terminal (1-253 aa) with G46D mutation (DDX43 Human)
TagsHisExpressionBacterialMutationfull-length (G154D)PromoterT7Available SinceJan. 6, 2020AvailabilityAcademic Institutions and Nonprofits only