We narrowed to 1,791 results for: plasmid for e coli
-
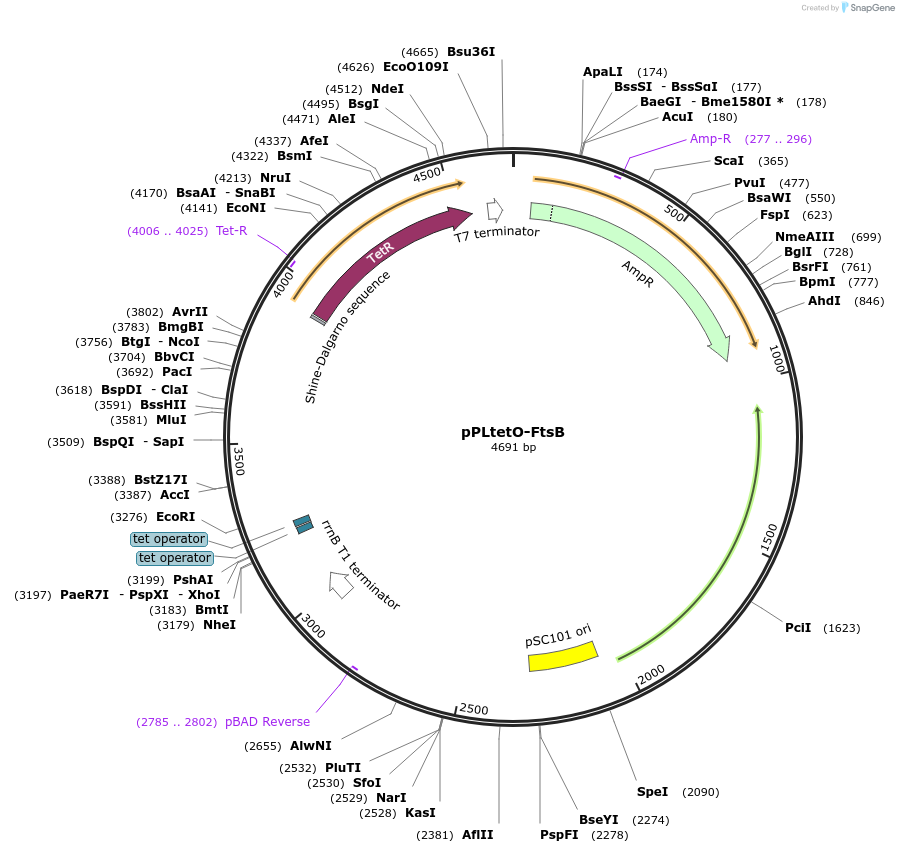
Plasmid#107559PurposePlasmid for titrating synthesis of FtsB with doxycyclineDepositorInsertftsB
UseSynthetic BiologyPromoterpLtetOAvailable SinceMay 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
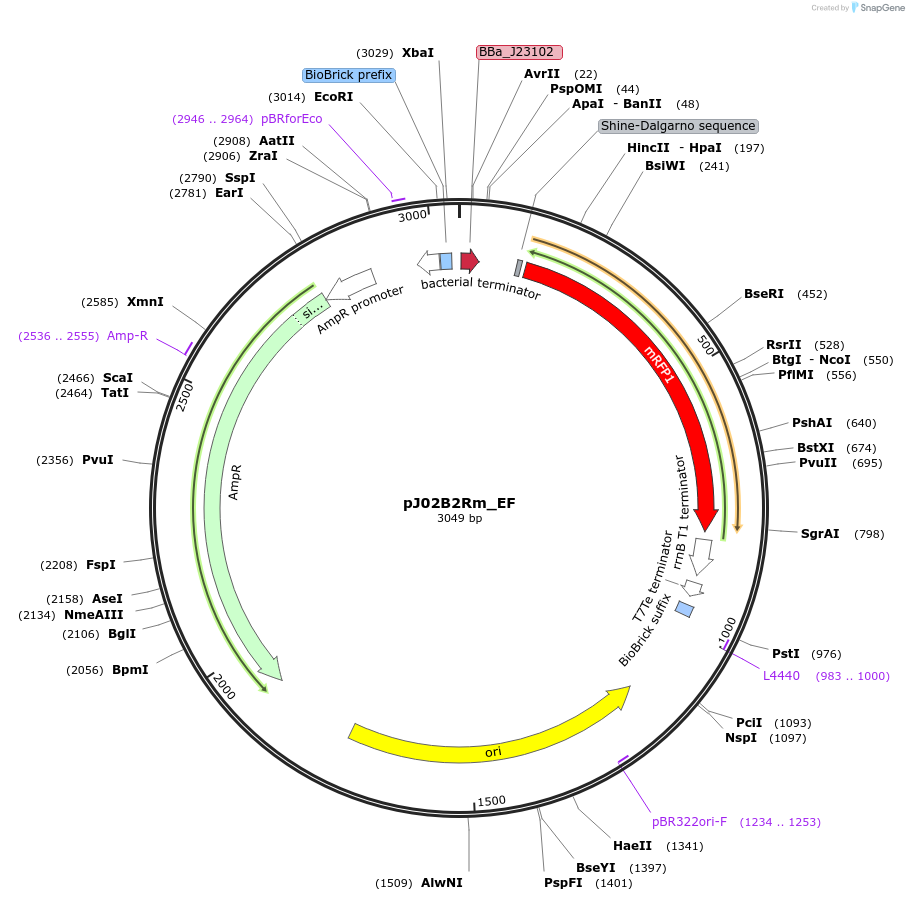
pJ02B2Rm_EF
Plasmid#66060PurposeMoClo Transcriptional Unit: FACS Standard Color Controls - High RFP expression cassette - Ampicillin plasmid [E:J23102:B:BCD2:C:E1010m:D:B0015:F]DepositorInsertFACS Controls
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
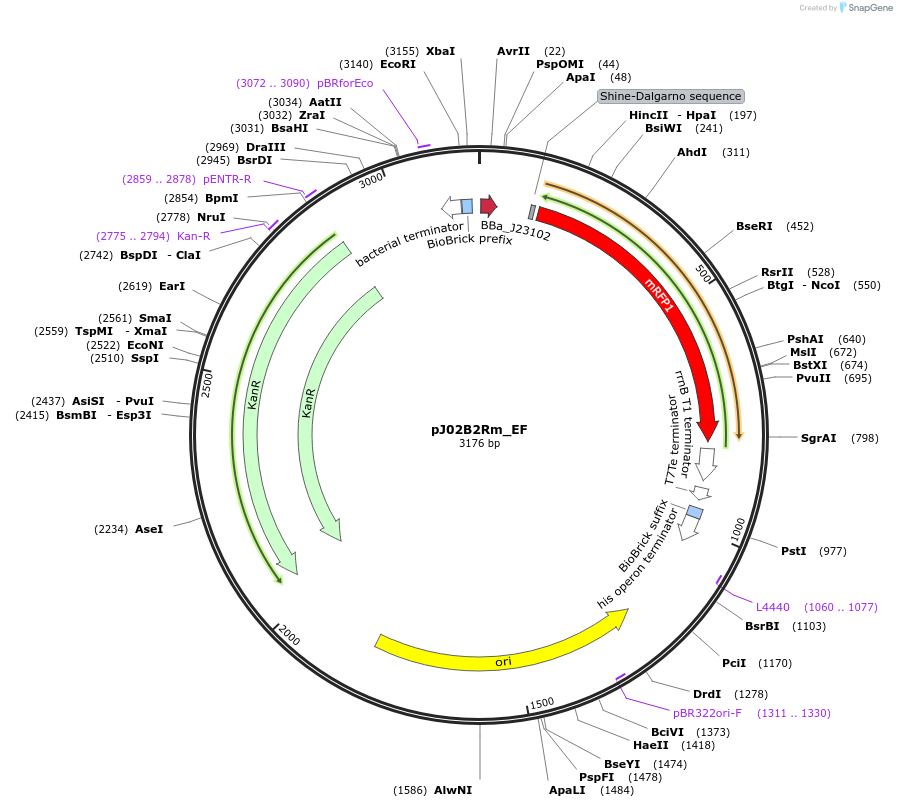
pJ02B2Rm_EF
Plasmid#66066PurposeMoClo Transcriptional Unit: FACS Standard Color Controls - High RFP expression cassette - Kanamycin plasmid [E:J23102:B:BCD2:C:E1010m:D:B0015:F]DepositorInsertFACS Controls
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
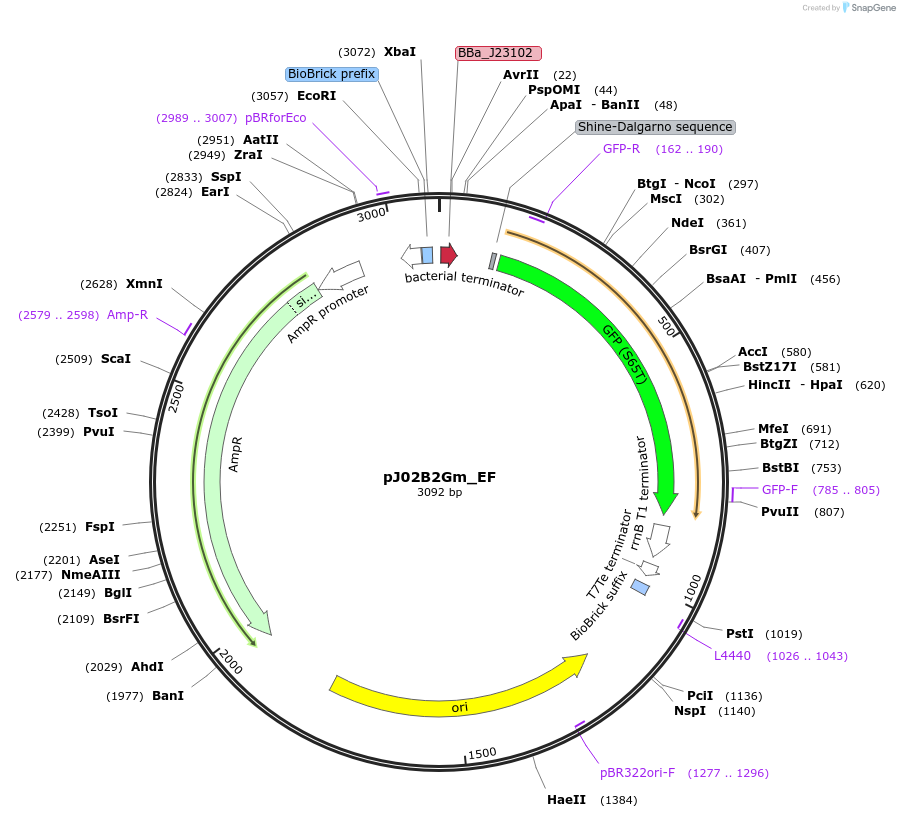
pJ02B2Gm_EF
Plasmid#66058PurposeMoClo Transcriptional Unit: FACS Standard Color Controls - High GFP expression cassette - Ampicillin plasmid [E:J23102:B:BCD2:C:E0040m:D:B0015:F]DepositorInsertFACS Controls
UseSynthetic BiologyAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
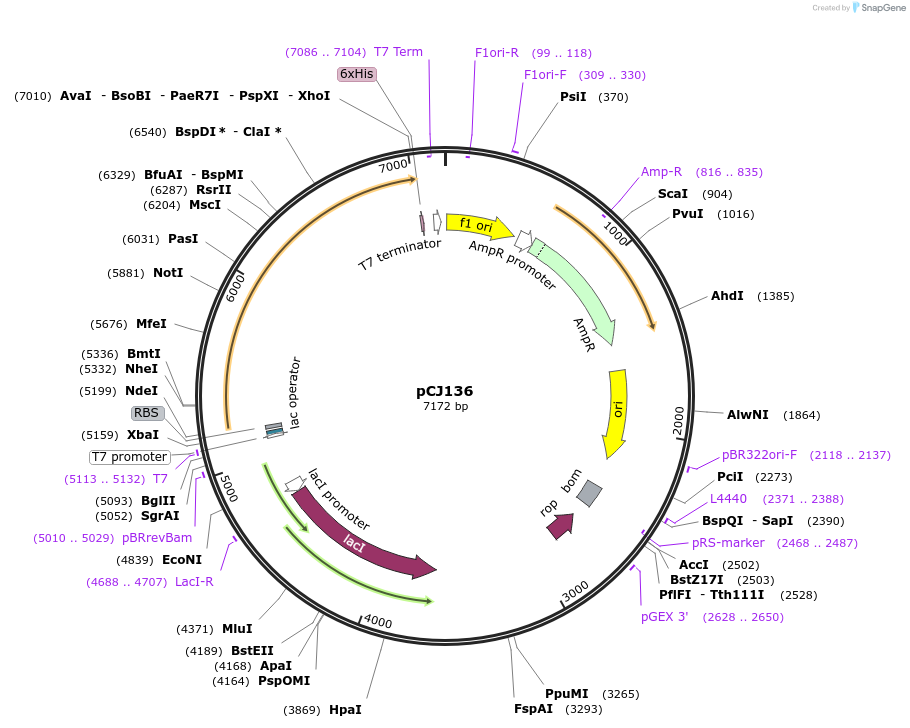
pCJ136
Plasmid#162665PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
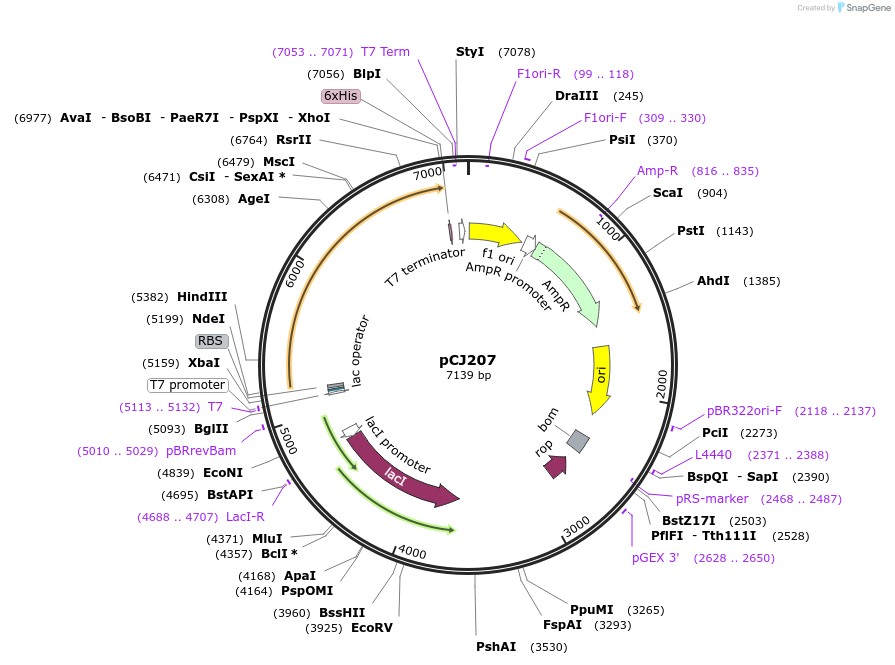
pCJ207
Plasmid#162680PurposepET-21b(+) based plasmid for expression of the putative MHETase from Hydrogenophaga sp. PML113 (Genbank WP_083293388.1) with C-terminal His tag, codon optimized for expression in E. coli K12.DepositorInsertPutative MHETase from Hydrogenophaga sp. PML113 (Genbank WP_083293388.1) with signal peptide
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
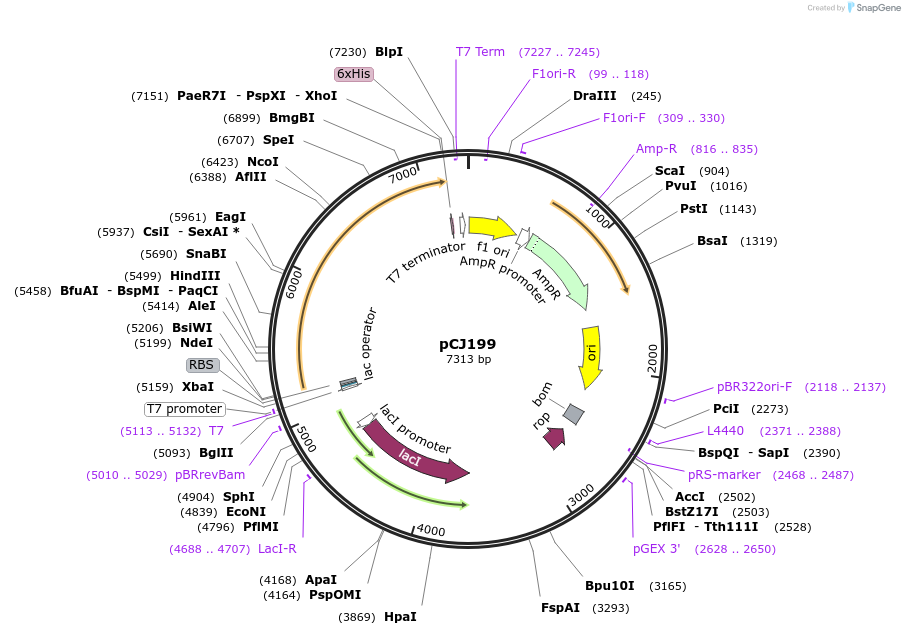
pCJ199
Plasmid#162672PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with C-terminal His tag, codon optimized for expression in E. coli K12.DepositorInsertPutative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with signal peptide
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
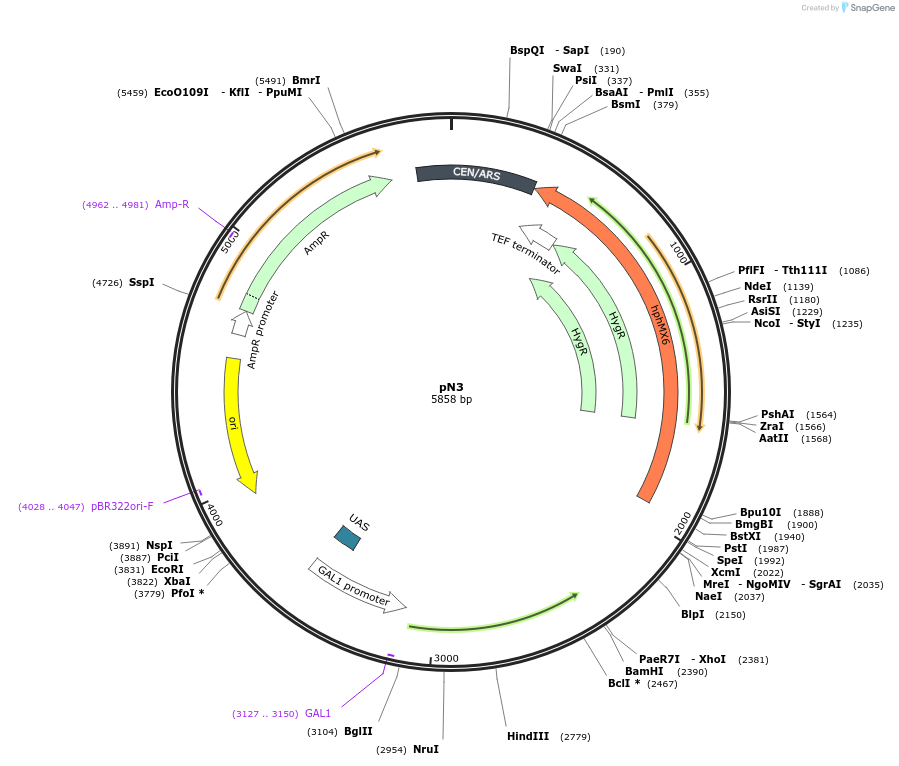
pN3
Plasmid#233520PurposeNb Vector Plasmid (under galactose induction), AmpR (E. coli) and HygroR (yeast), Cen6/ARSDepositorTypeEmpty backboneExpressionBacterial and YeastPromoterpGAL1Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
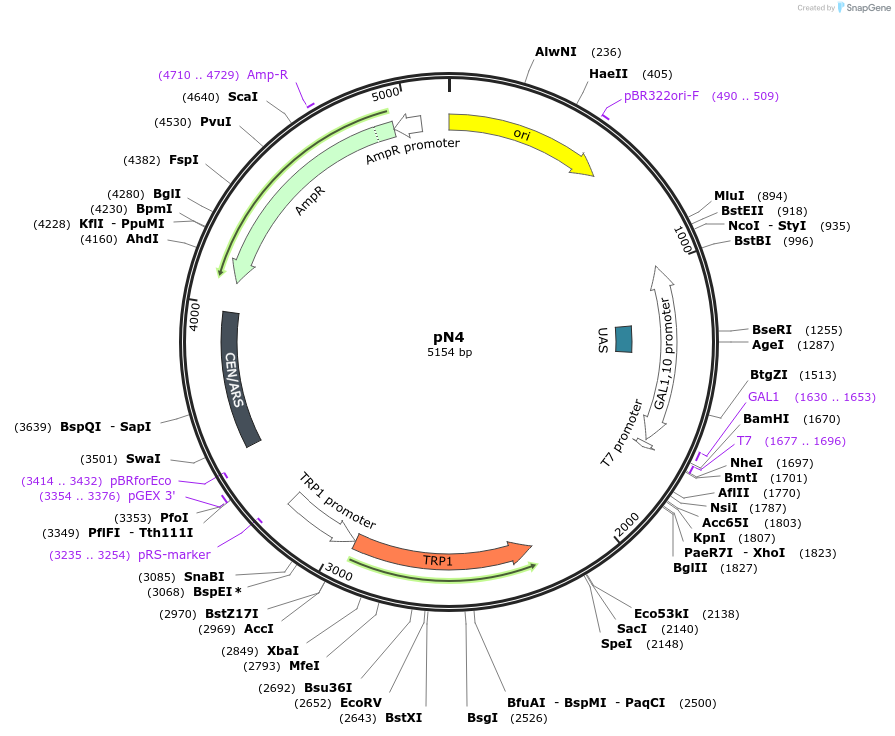
pN4
Plasmid#233521PurposeNb Vector Plasmid (under galactose induction), AmpR (E. coli) and TRP1 (yeast), Cen6/ARSDepositorTypeEmpty backboneExpressionBacterial and YeastPromoterpGAL1-10Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
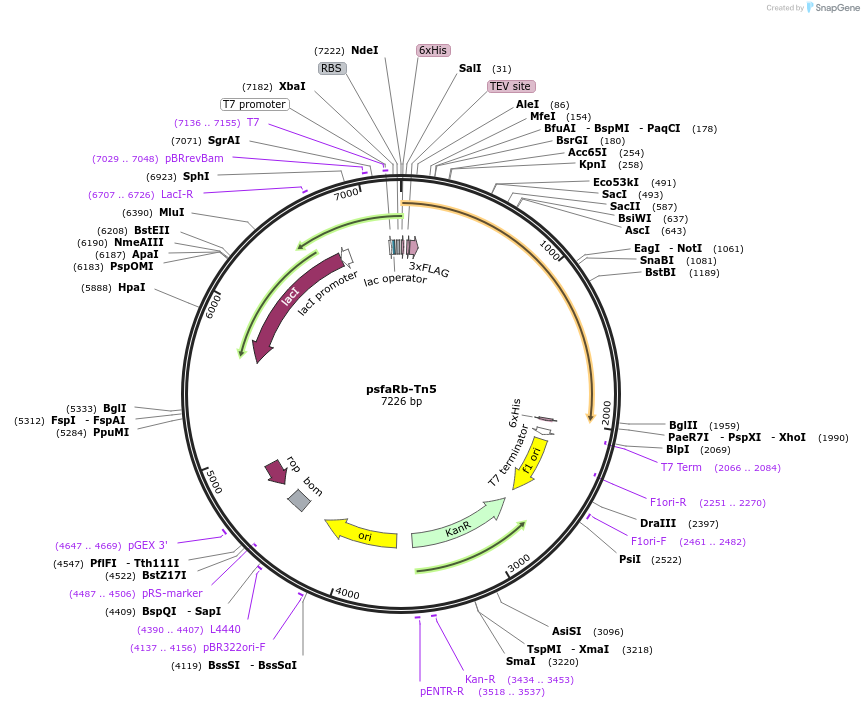
psfaRb-Tn5
Plasmid#183638PurposeExpresses anti-rabbit nanobody fused with Tn5DepositorInsertanti rabbit secondary nanobody fused to Tn5
ExpressionBacterialAvailable SinceJune 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
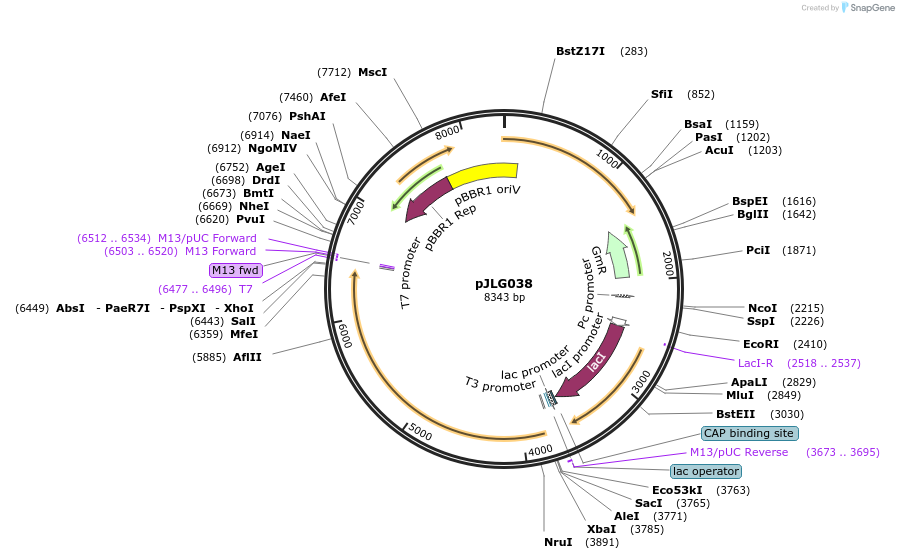
pJLG038
Plasmid#192981PurposeBHR plasmid (pBBR1 ori) expressing T7 polymerase under control of the lac promoterDepositorInsertT7 polymerase
ExpressionBacterialPromoterlacAvailable SinceMay 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
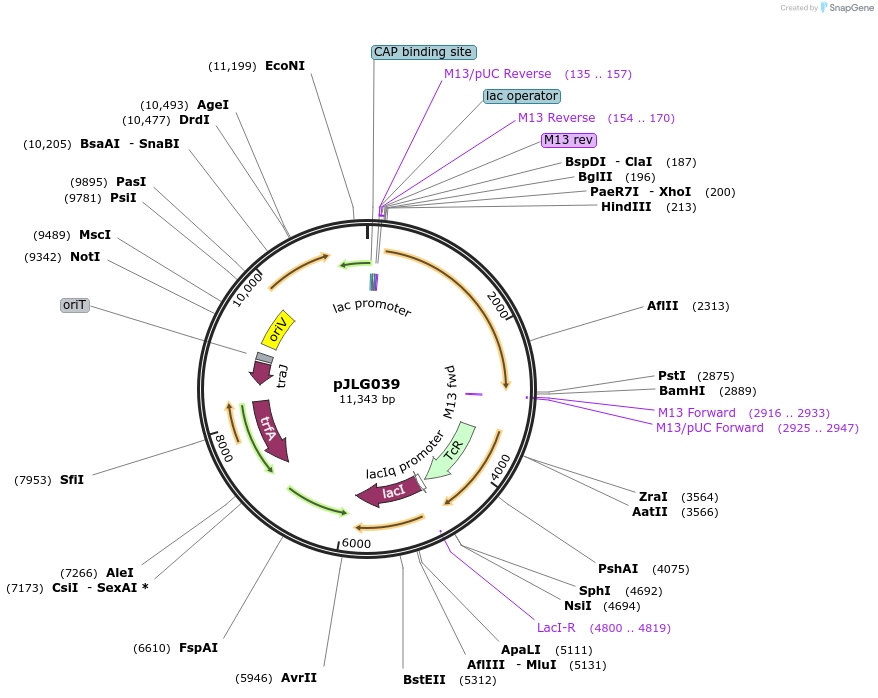
pJLG039
Plasmid#192982PurposeBHR plasmid (IncP ori) expressing T7 polymerase under control of the lac promoterDepositorInsertT7 polymerase
ExpressionBacterialPromoterlacAvailable SinceMay 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
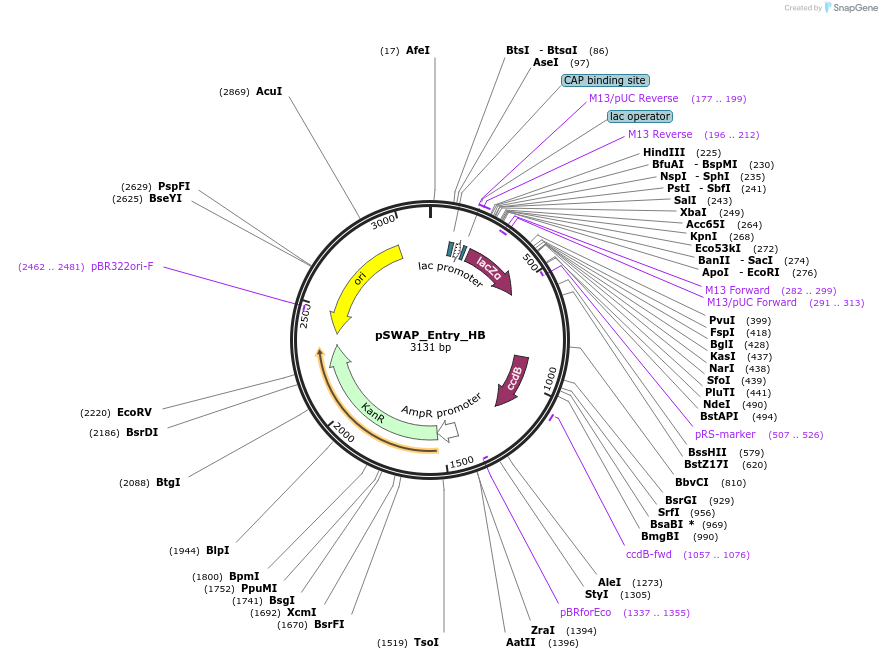
pSWAP_Entry_HB
Plasmid#184868PurposepSwap plasmid part with HB homology armsDepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
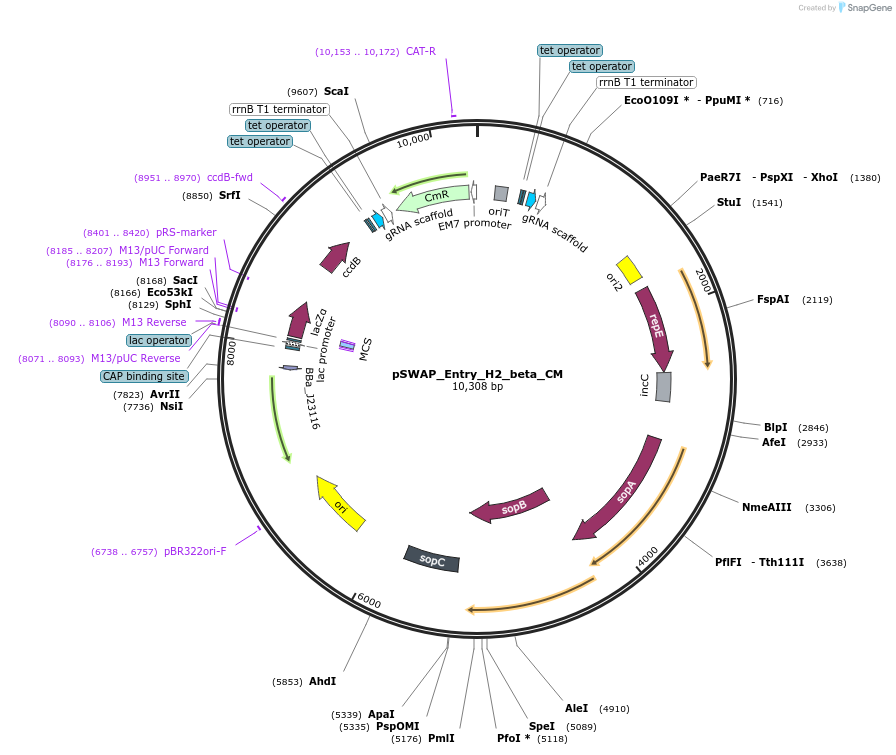
pSWAP_Entry_H2_beta_CM
Plasmid#184870PurposepSwap plasmid part with H2_beta homology arms and chloramphenicol cassetteDepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceMarch 2, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
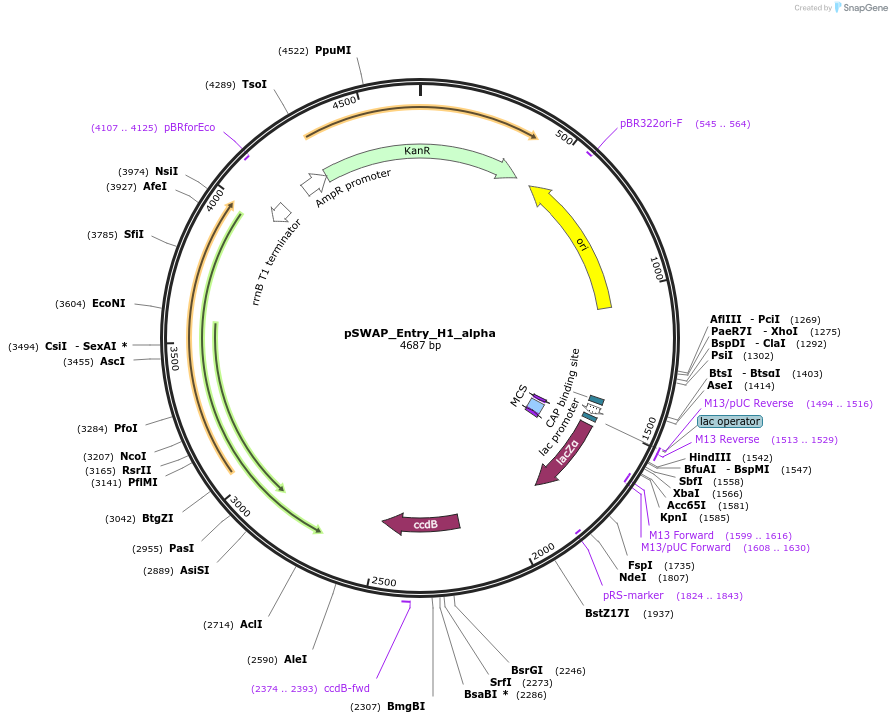
pSWAP_Entry_H1_alpha
Plasmid#184865PurposepSwap plasmid part with H1_alpha homology armsDepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
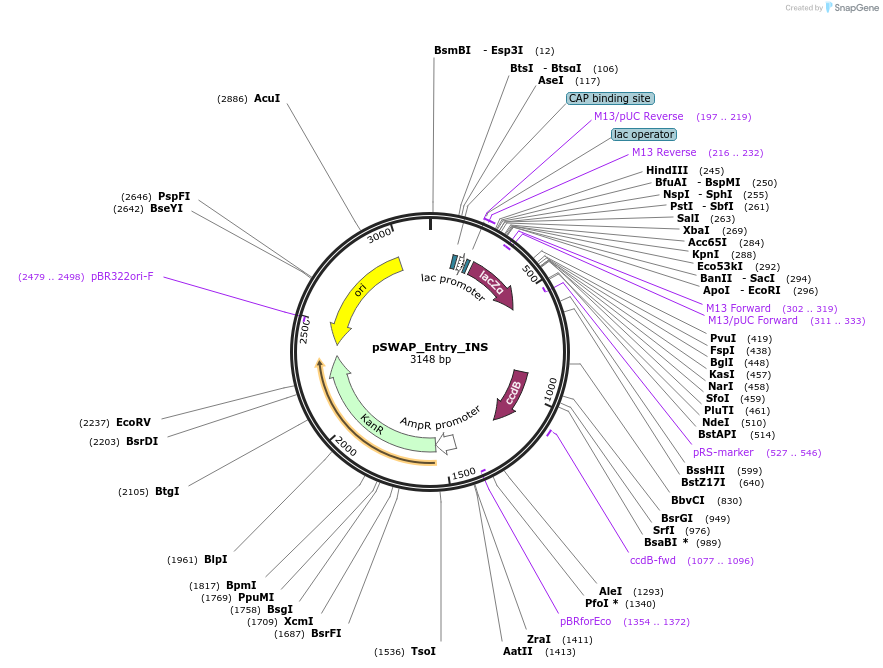
pSWAP_Entry_INS
Plasmid#184867PurposepSwap plasmid part with insert fragment moduleDepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
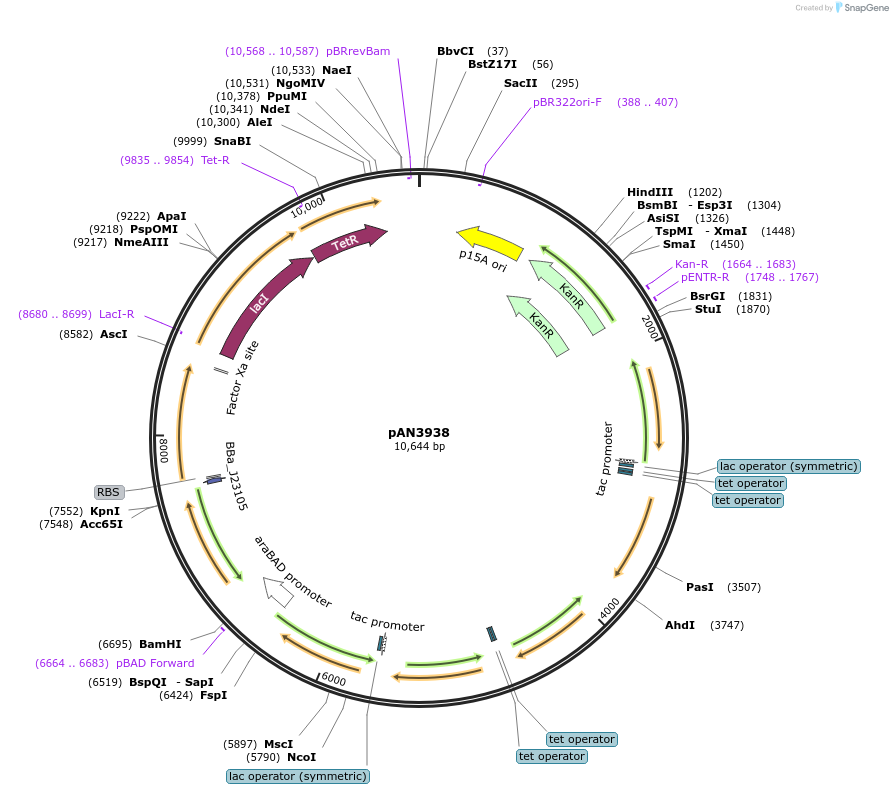
pAN3938
Plasmid#74697PurposeCircuit plasmid: 0xF6DepositorInsertCircuit 0xF6
ExpressionBacterialAvailable SinceMay 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
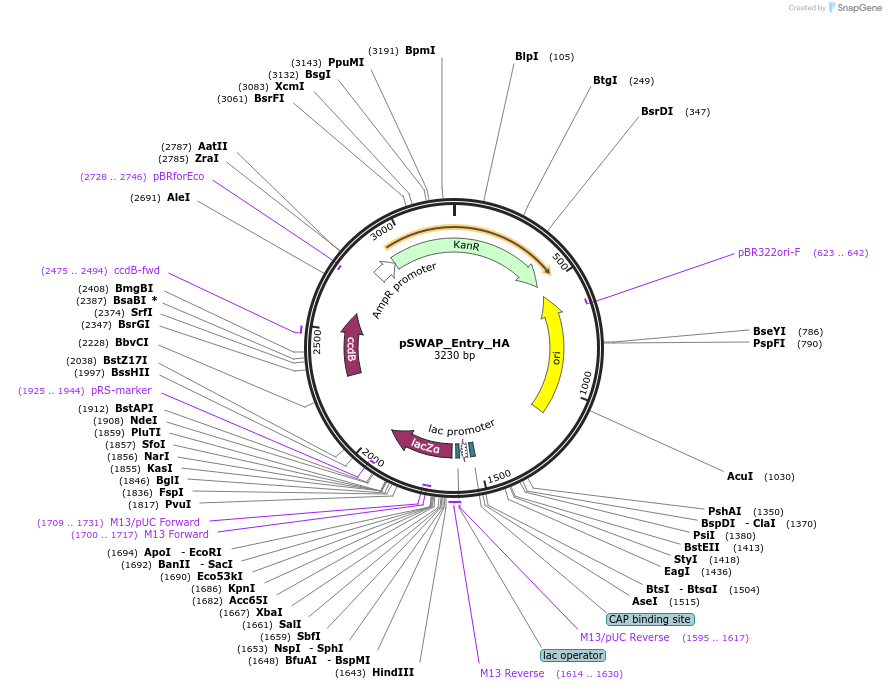
pSWAP_Entry_HA
Plasmid#184866PurposepSwap plasmid part with HA homology armsDepositorInsertlacZalpha,ccdB
ExpressionBacterialAvailable SinceJan. 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
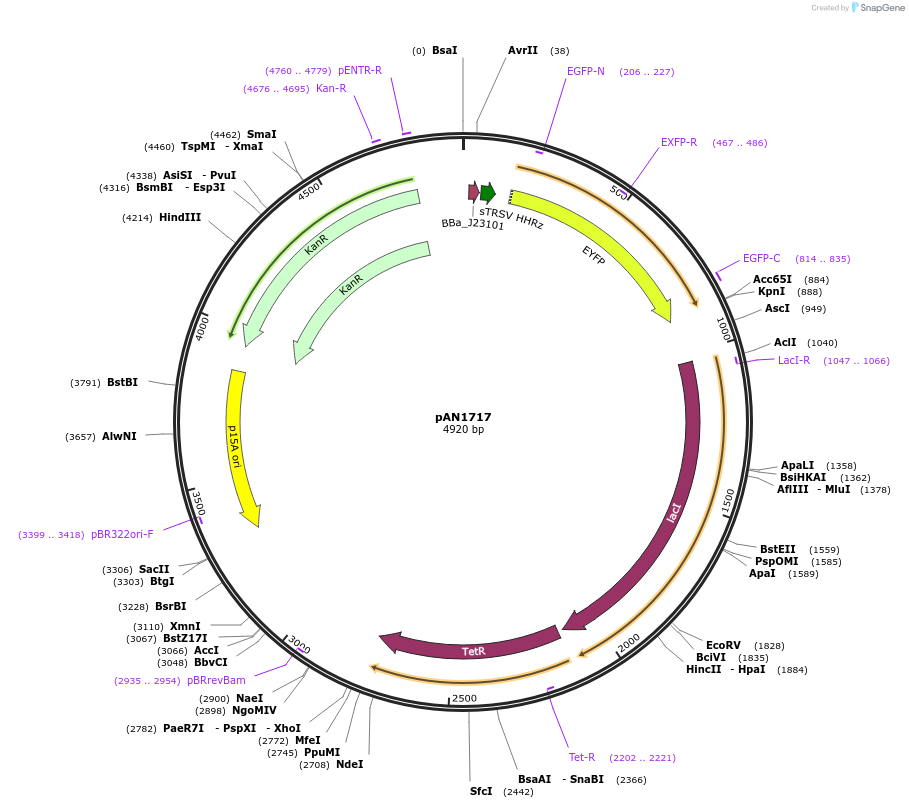
pAN1717
Plasmid#74696PurposeRPU standard plasmidDepositorInsertInsulated J23101-YFP
ExpressionBacterialAvailable SinceMay 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
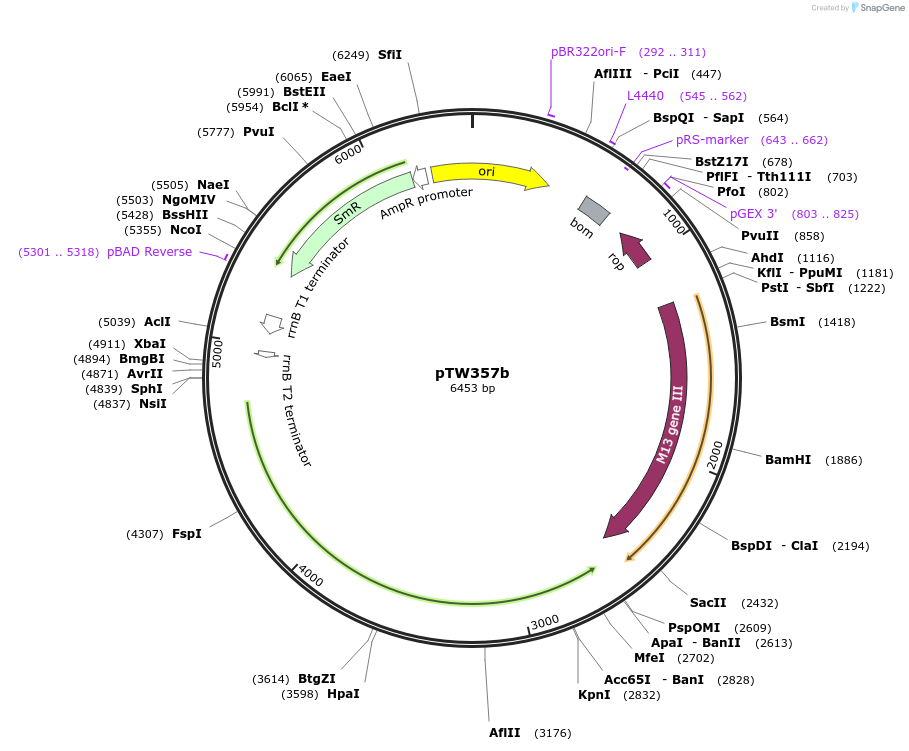
pTW357b
Plasmid#227930PurposeLow-stringency accessory plasmid for selectionDepositorInsertsgIII (recoded)
T7c
ExpressionBacterialMutationR632SAvailable SinceNov. 26, 2024AvailabilityAcademic Institutions and Nonprofits only