We narrowed to 40,709 results for: kan
-
Pooled Library#224585PurposePyrrolysine tRNA synthetase library to generate new genetic code expansion (GCE) systems for site-specific incorporation of non-canonical amino acids (ncAAs) into proteins.DepositorExpressionBacterialSpeciesMethanomethylophilus alvusAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only
-
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 1
Pooled Library#168776PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 2
Pooled Library#168777PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
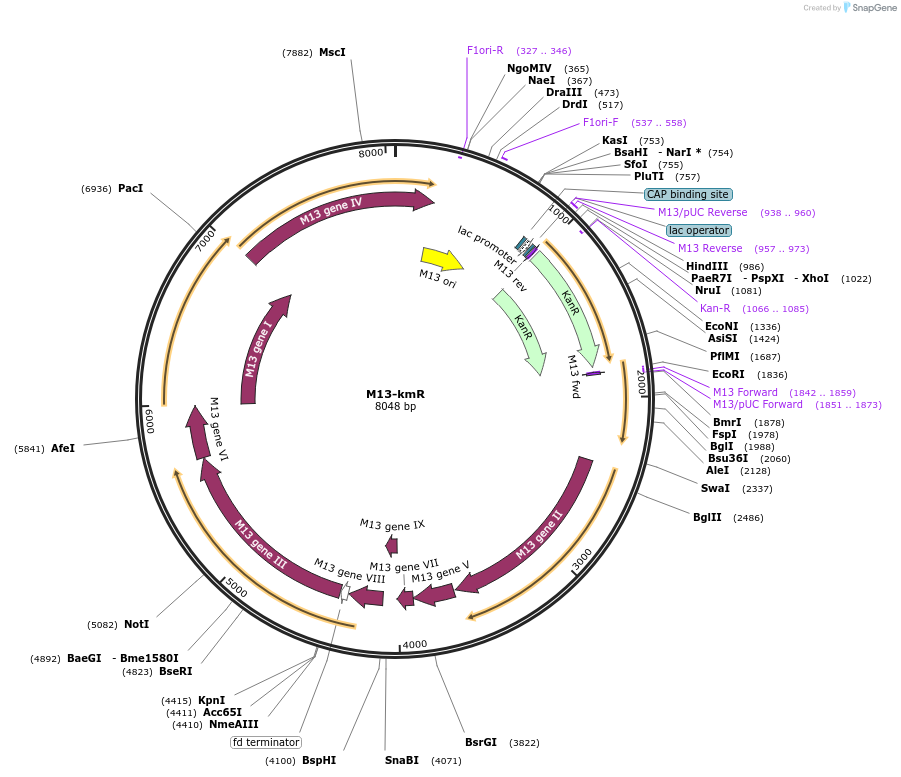
M13-kmR
Plasmid#206861PurposeM13KE from NEB modified with kanamycin resistance gene and NotI restriction site on g3p ( M13KE-NotI-Kan)DepositorInsertKanamycin resistance gene
ExpressionBacterialPromoterlacAvailable SinceNov. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
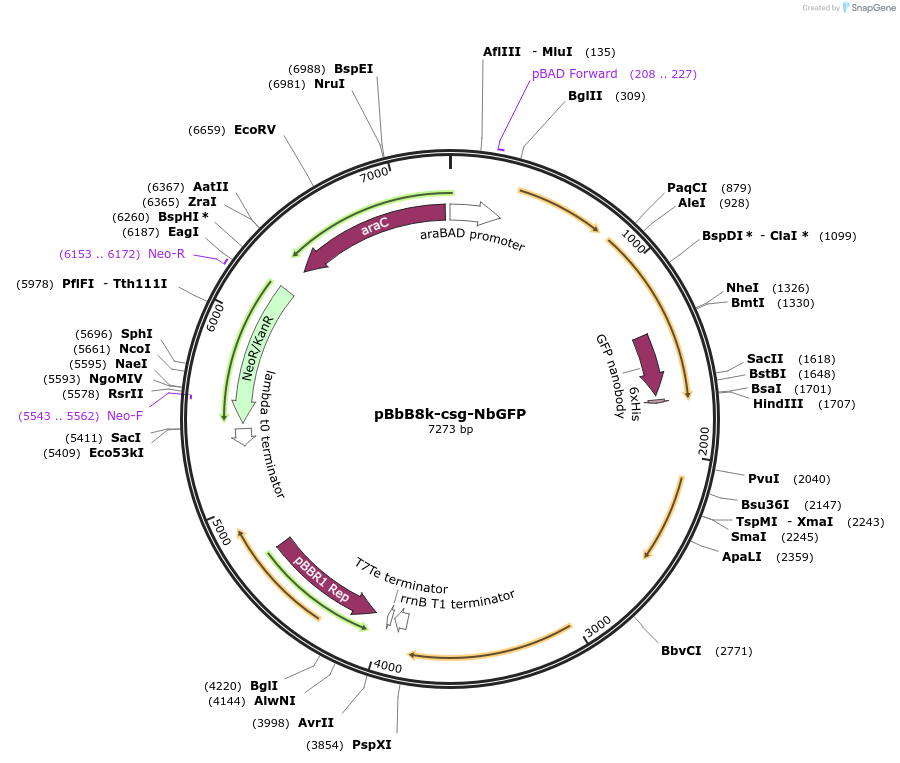
pBbB8k-csg-NbGFP
Plasmid#166858PurposeArabinose-inducible csgBACEFG operon for curli fiber synthesis in which curli subunit CsgA has been fused to a GFP-specific nanobody via a flexible linker. Derived from https://www.addgene.org/35363/.DepositorInsertcsgBACEFG, with GFP-specific nanobody fused to CsgA via flexible linker
UseSynthetic BiologyExpressionBacterialAvailable SinceMarch 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
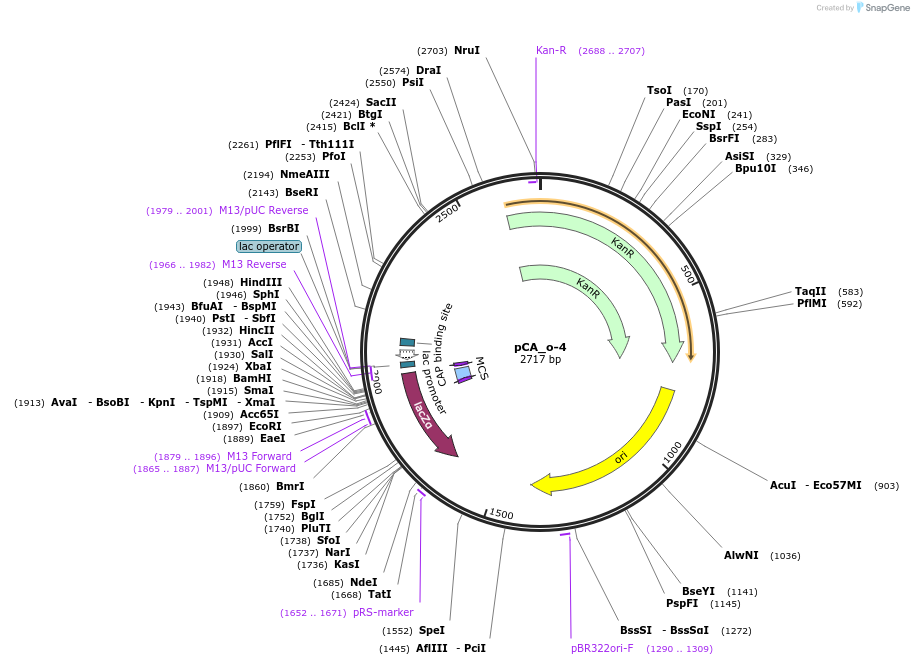
pCA_o-4
Plasmid#202094PurposeReceiver cloning vector for odd-4 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
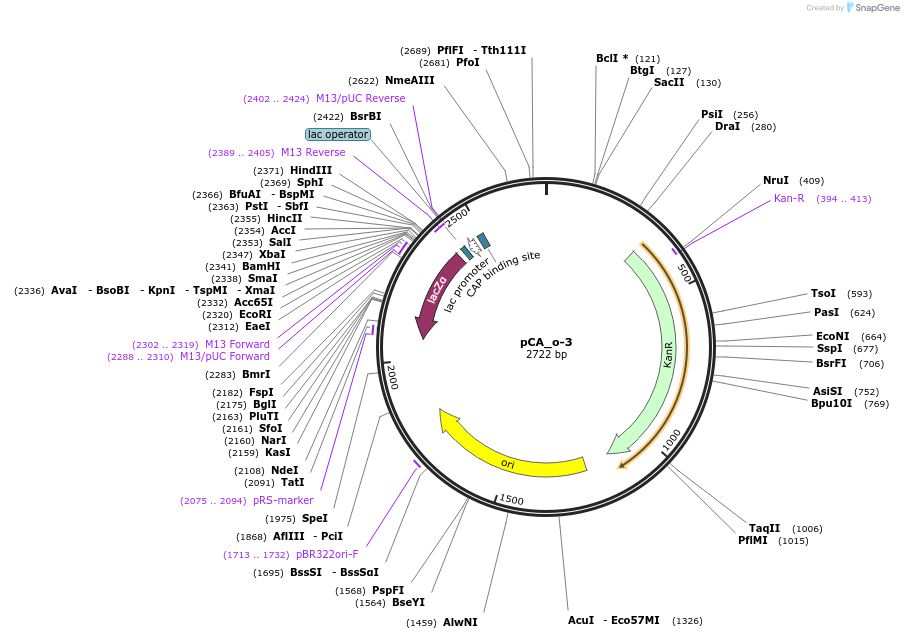
pCA_o-3
Plasmid#202093PurposeReceiver cloning vector for odd-3 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
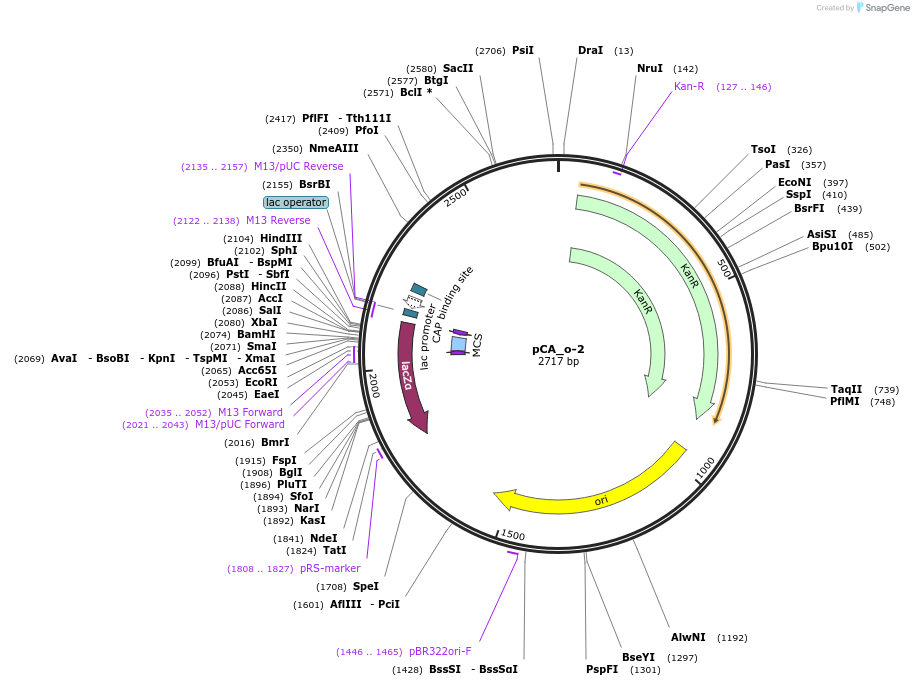
pCA_o-2
Plasmid#202092PurposeReceiver cloning vector for odd-2 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
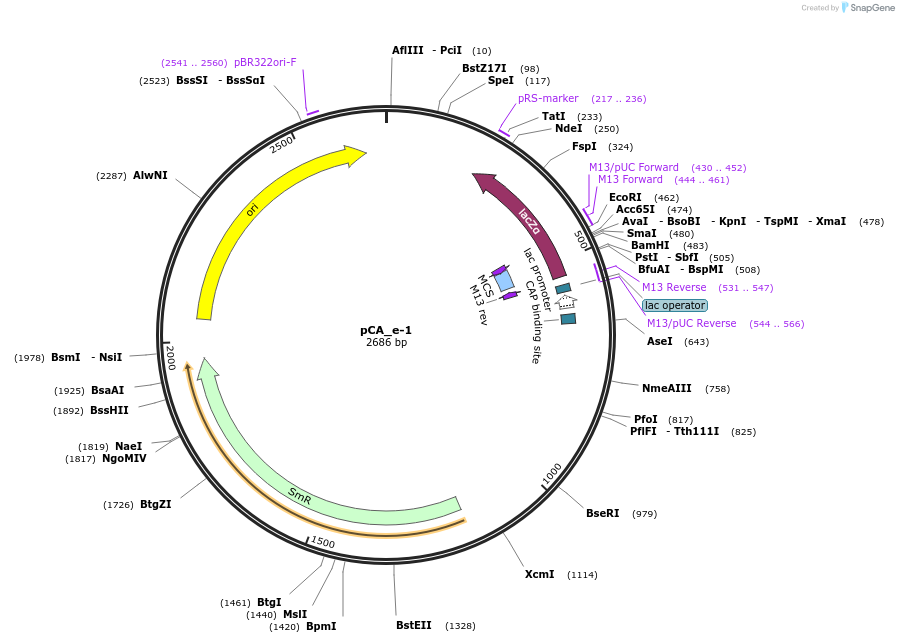
pCA_e-1
Plasmid#202096PurposeReceiver cloning vector for even-1 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
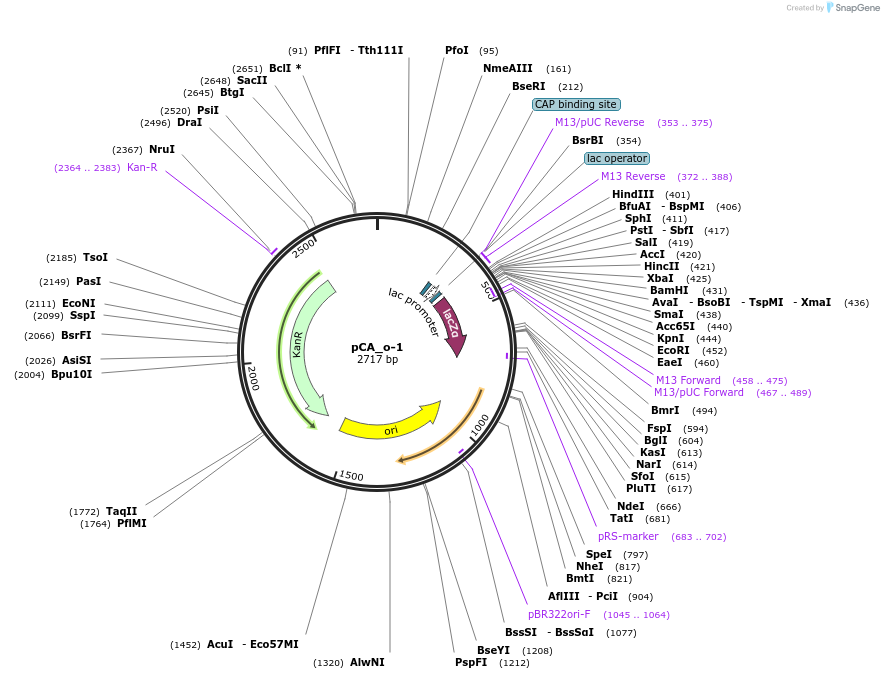
pCA_o-1
Plasmid#202091PurposeReceiver cloning vector for odd-1 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
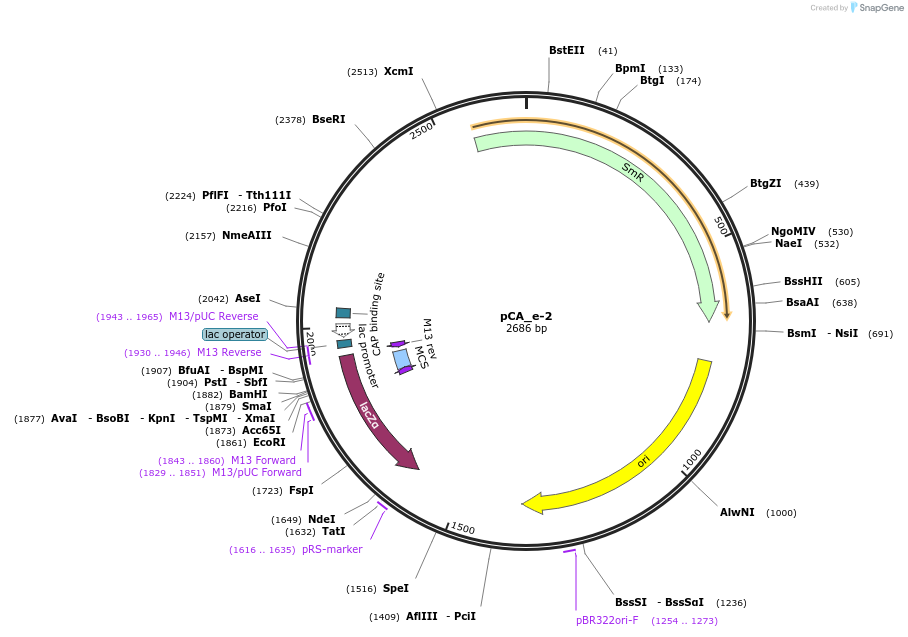
pCA_e-2
Plasmid#202097PurposeReceiver cloning vector for even-2 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
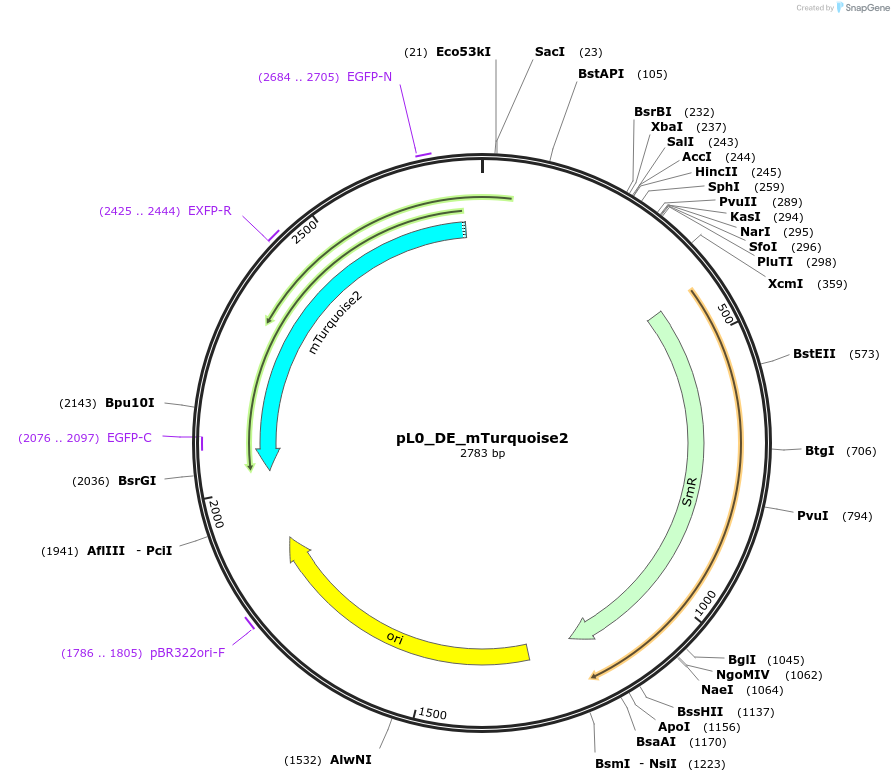
pL0_DE_mTurquoise2
Plasmid#202155PurposeLevel 0 plasmid containing a mTurquoise2 fluorescent protein with DE overhangs used to build a level 1 construct.DepositorInsertmTurquoise2 gene
UseSynthetic BiologyMutationNoneAvailable SinceAug. 25, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
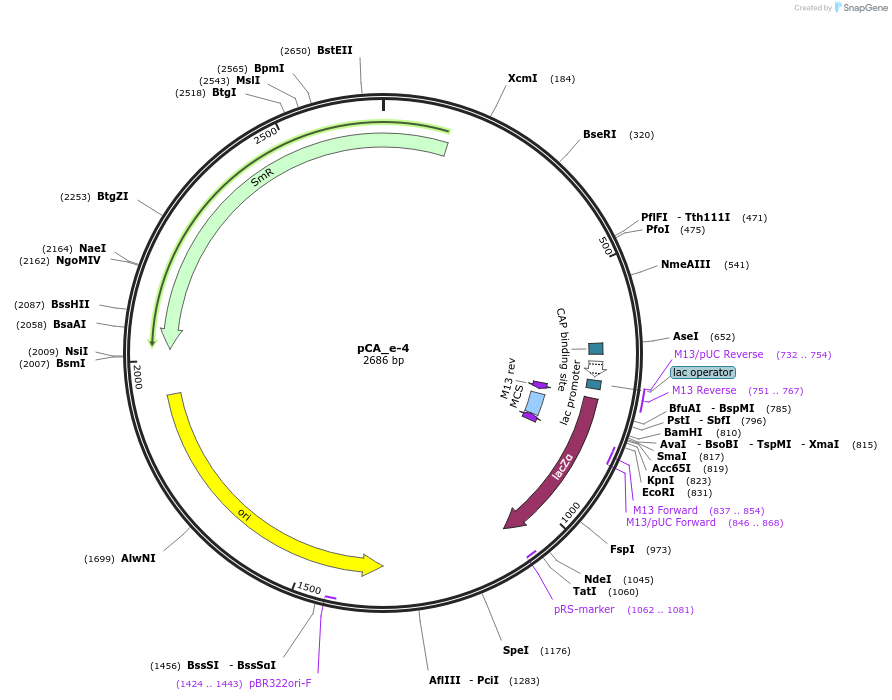
pCA_e-4
Plasmid#202099PurposeReceiver cloning vector for even-4 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
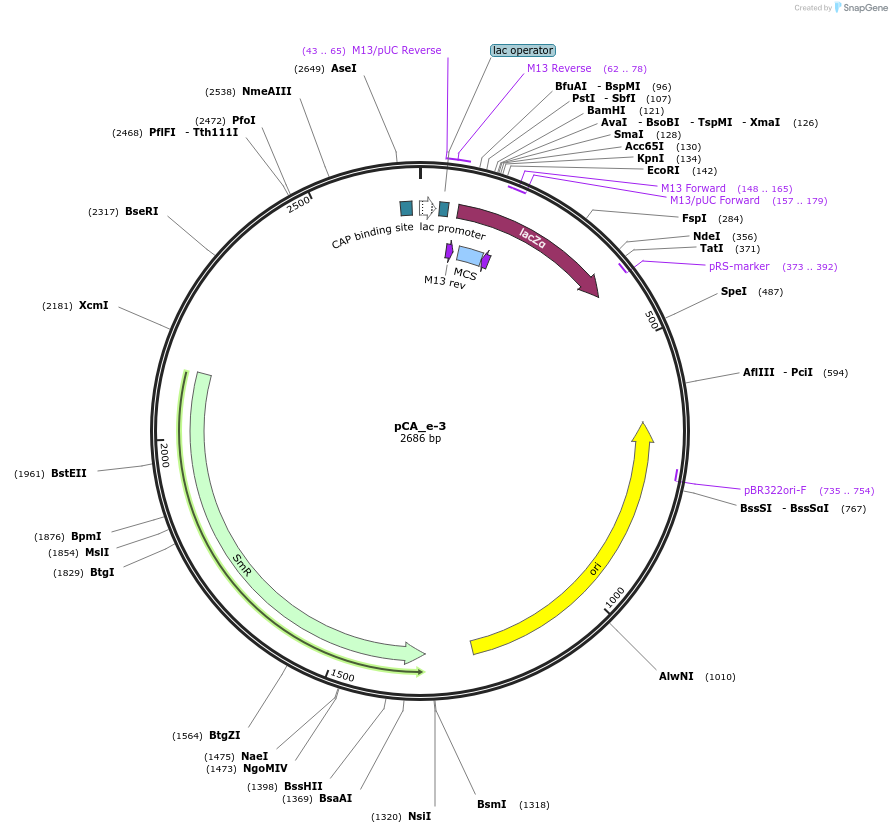
pCA_e-3
Plasmid#202098PurposeReceiver cloning vector for even-3 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon5mut
Pooled Library#229137PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 5 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon6mut
Pooled Library#229138PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 6 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon7mut
Pooled Library#229139PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 7 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon8mut
Pooled Library#229140PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 8 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only