We narrowed to 1,791 results for: plasmid for e coli
-
Plasmid#69659PurposeUntargetted Bacterial MutagenesisDepositorInsertpolB
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
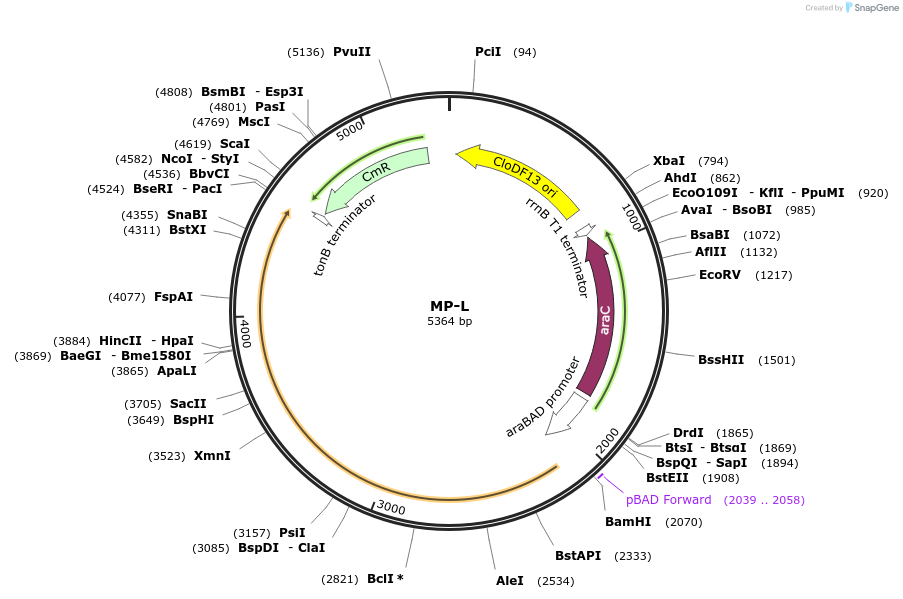
MP-J3
Plasmid#69649PurposeUntargetted Bacterial MutagenesisDepositorInsertrpsD16
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
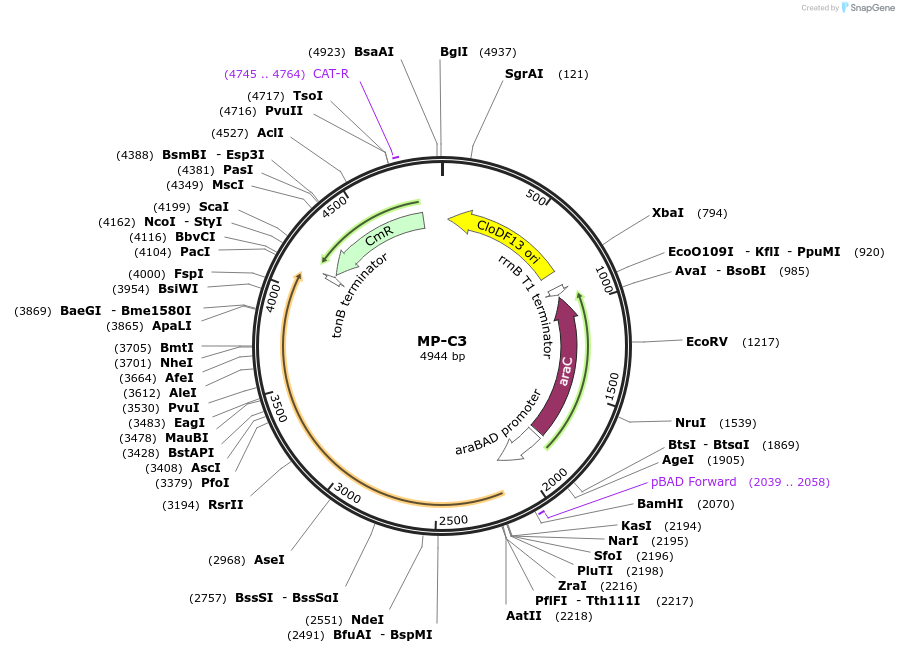
MP-C3
Plasmid#69633PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaX2016
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
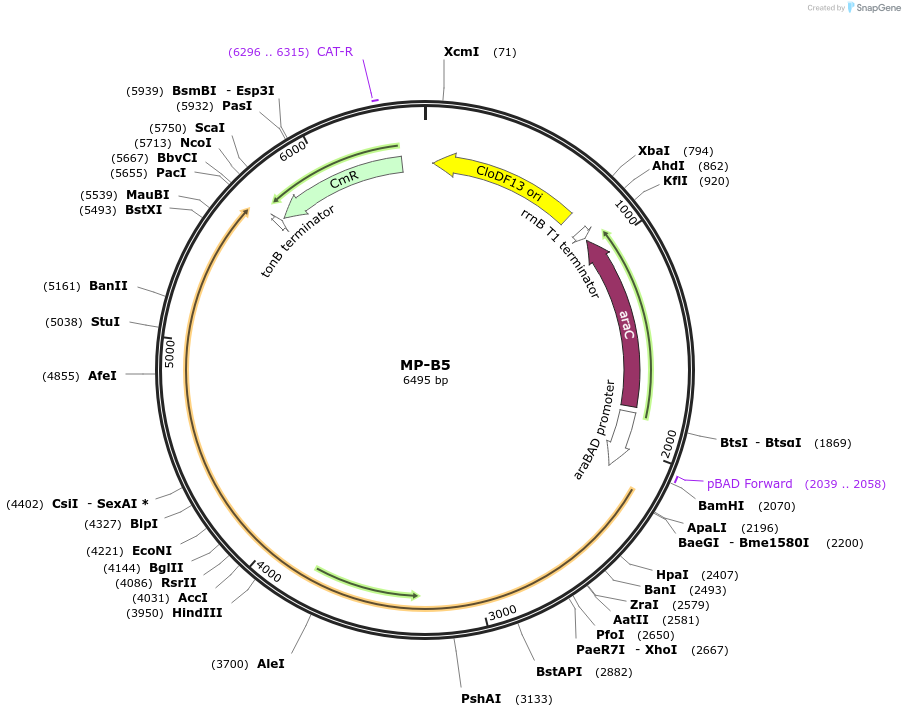
MP-B5
Plasmid#69631PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaE1026
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
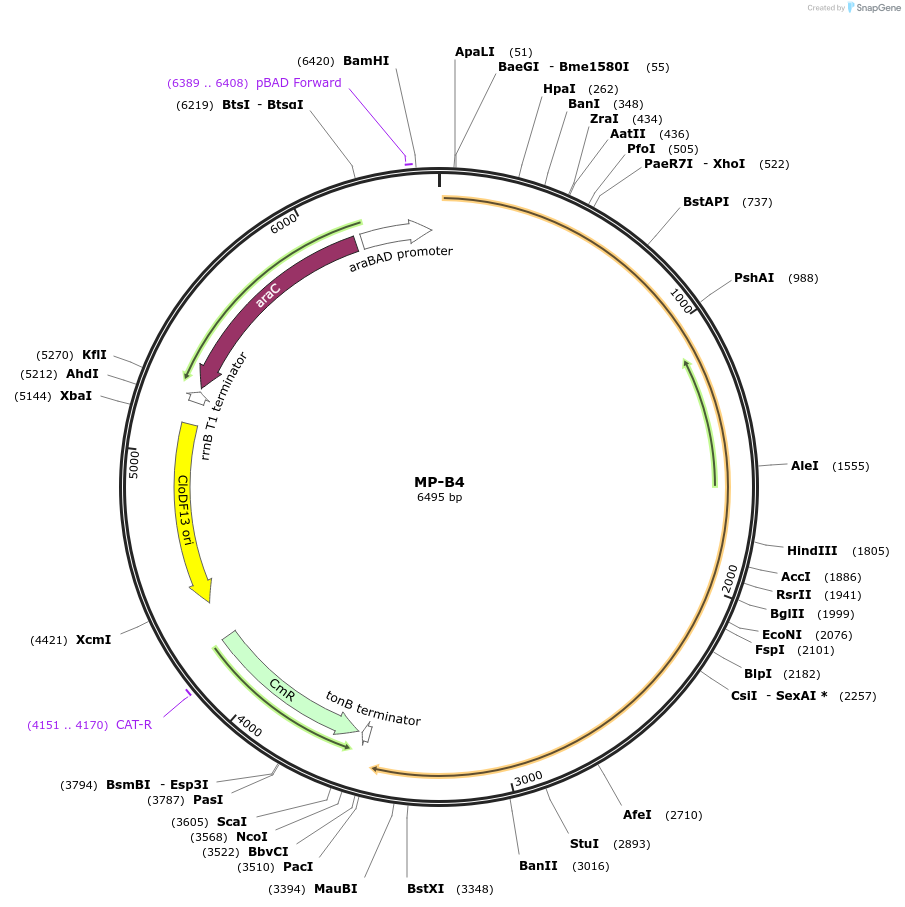
MP-B4
Plasmid#69630PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaE486
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
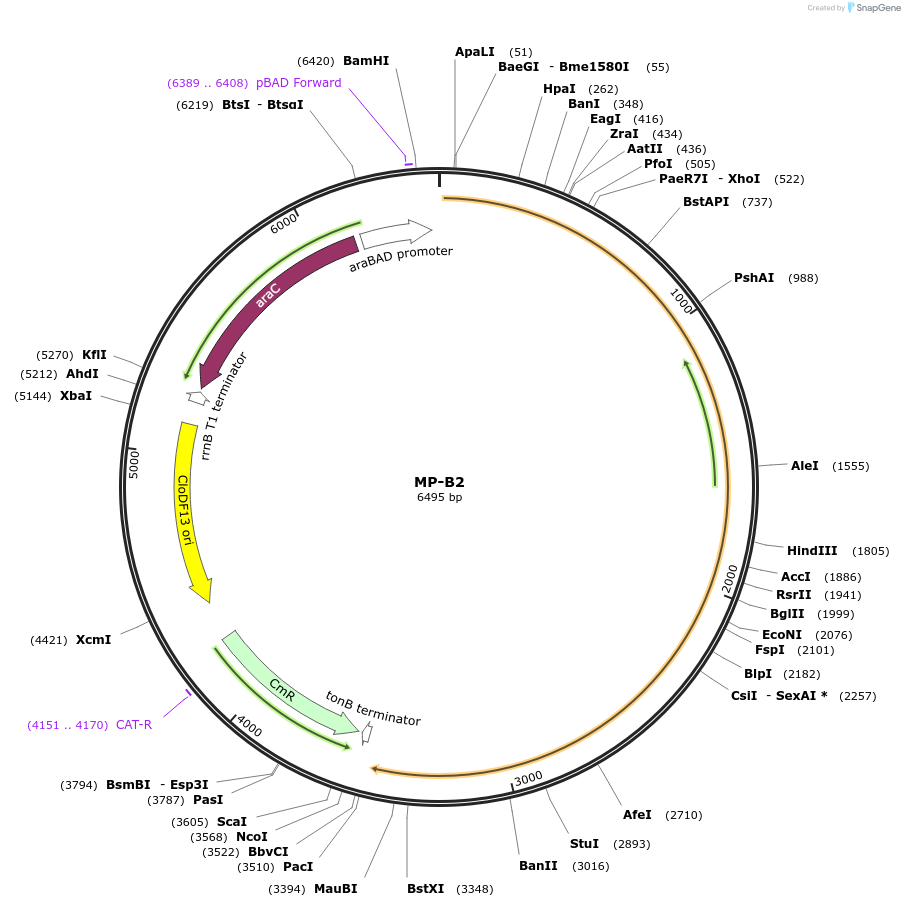
MP-B2
Plasmid#69629PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaE74
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
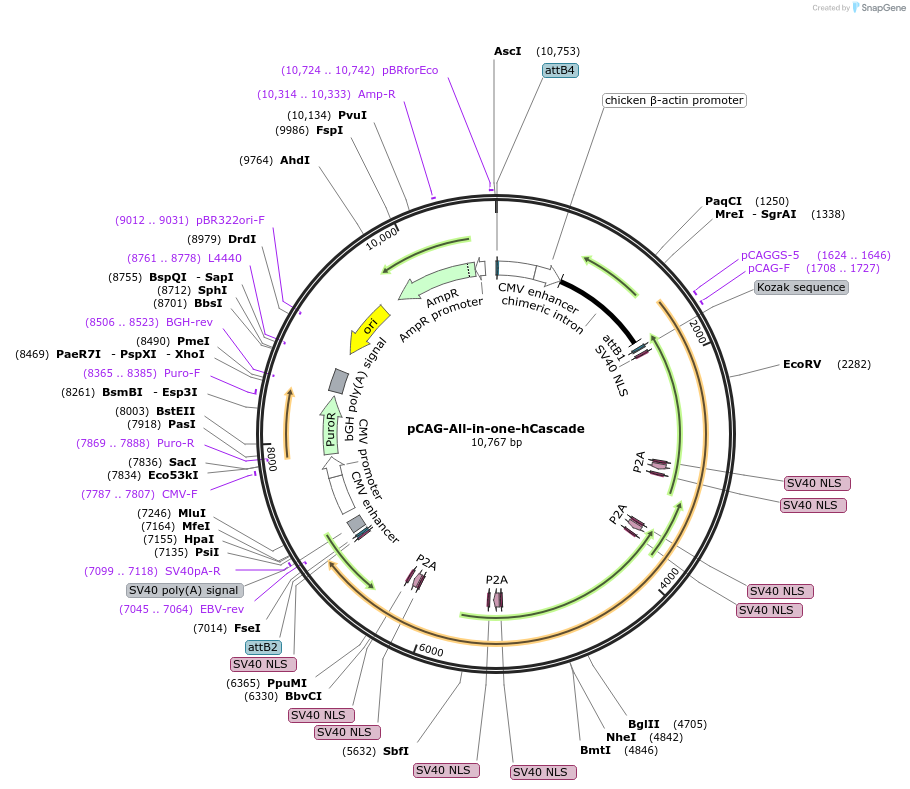
pCAG-All-in-one-hCascade
Plasmid#134919PurposeComponents for genome editing in mammalian cells with pPB-CAG-hCas3 and pBS-U6-crRNA-targeted. This plasmid coexpresses PuroR protein.DepositorInsertCascade factors (Cse1(Cas8), Cse2(Cas11), Cas5, Cas6, Cas7) with bpNLS
ExpressionMammalianPromoterCAGAvailable SinceFeb. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
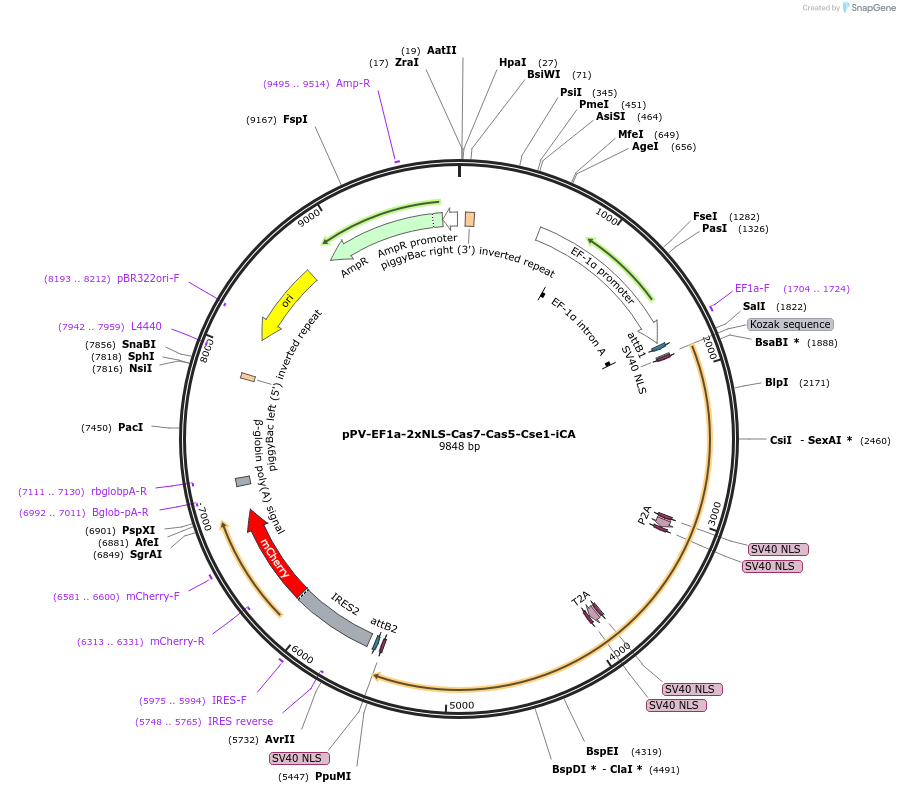
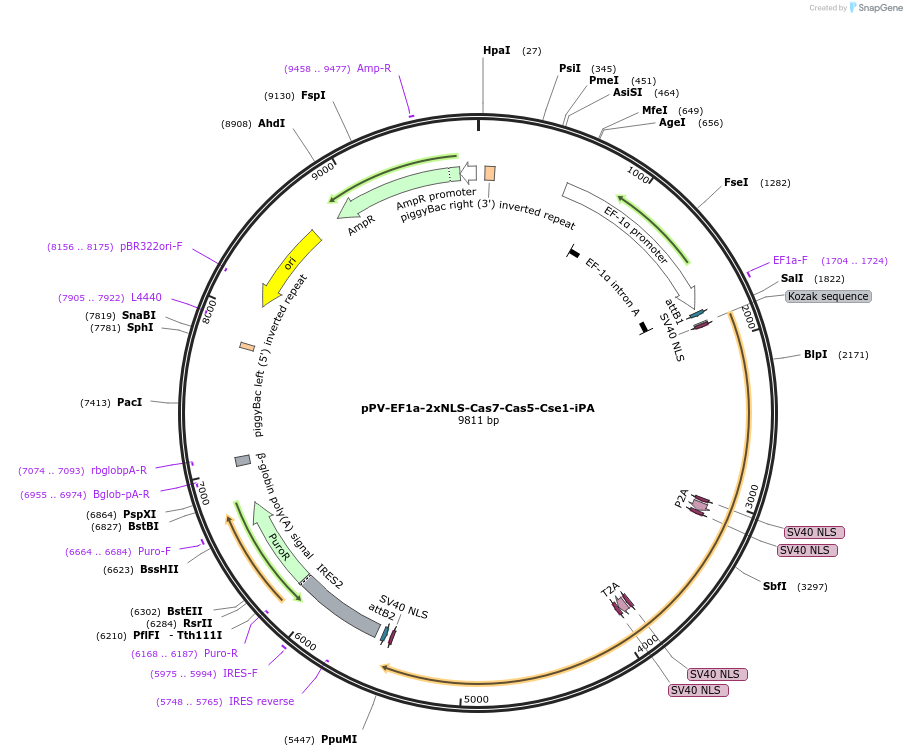
pPV-EF1a-2xNLS-Cas7-Cas5-Cse1-iCA
Plasmid#134922PurposeComponents for genome editing in mammalian cells with pPV-EF1a-2xNLS-Cse2-Cas6-Cas3-iCA and pBS-U6-crRNA-targeted. This plasmid was optimized for iPS cells and coexpresses mCherry protein.DepositorInsertCascade factors (Cse1(Cas8), Cas5,Cas7) with bpNLS
ExpressionMammalianPromoterEF1aAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
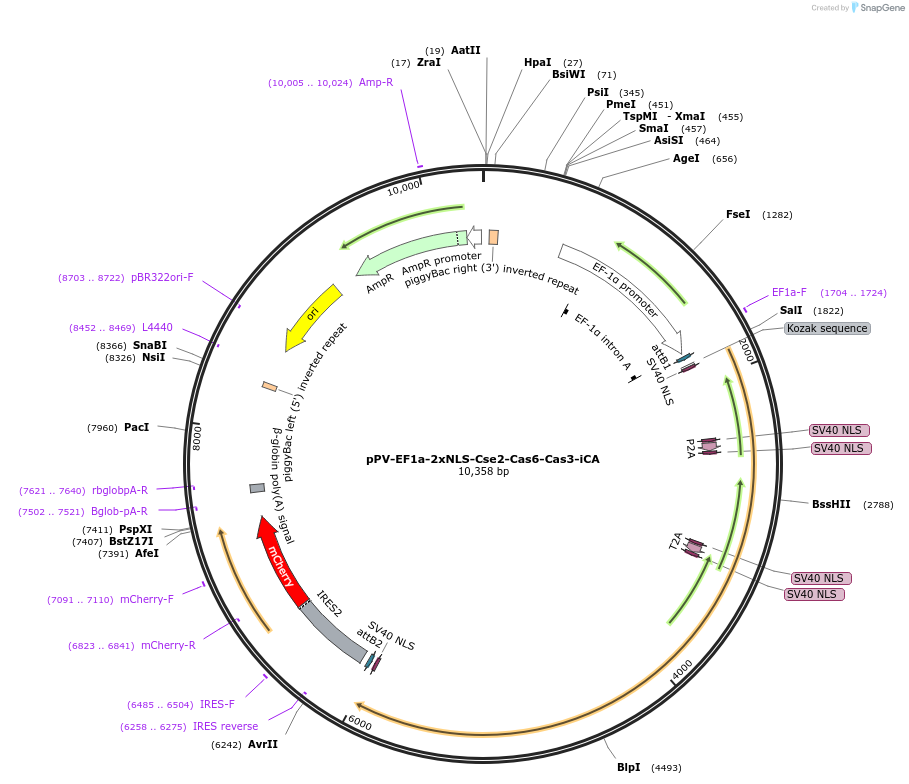
pPV-EF1a-2xNLS-Cse2-Cas6-Cas3-iCA
Plasmid#134923PurposeComponents for genome editing in mammalian cells with pPV-EF1a-2xNLS-Cas7-Cas5-Cse1-iCA and pBS-U6-crRNA-targeted. This plasmid was optimized for iPS cells and coexpresses mCherry protein.DepositorInsertCascade factors (Cse2(Cas11), Cas3, Cas6) with bpNLS
ExpressionMammalianPromoterEF1aAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
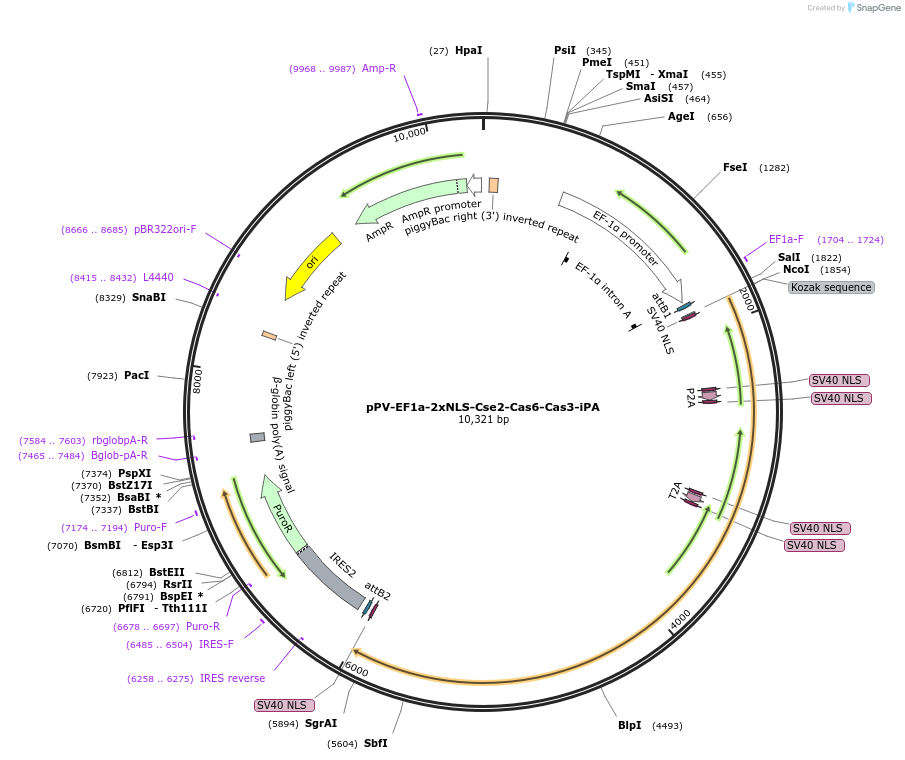
pPV-EF1a-2xNLS-Cse2-Cas6-Cas3-iPA
Plasmid#134925PurposeComponents for genome editing in mammalian cells with pPV-EF1a-2xNLS-Cas7-Cas5-Cse1-iPA and pBS-U6-crRNA-targeted. This plasmid was optimized for iPS cells and coexpresses PuroR protein.DepositorInsertCascade factors (Cse2(Cas11), Cas3, Cas6) with bpNLS
ExpressionMammalianPromoterEF1aAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPV-EF1a-2xNLS-Cas7-Cas5-Cse1-iPA
Plasmid#134924PurposeComponents for genome editing in mammalian cells with pPV-EF1a-2xNLS-Cse2-Cas6-Cas3-iPA and pBS-U6-crRNA-targeted. This plasmid was optimized for iPS cells and coexpresses PuroR protein.DepositorInsertCascade factors (Cse1(Cas8), Cas5,Cas7) with bpNLS
ExpressionMammalianPromoterEF1aAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
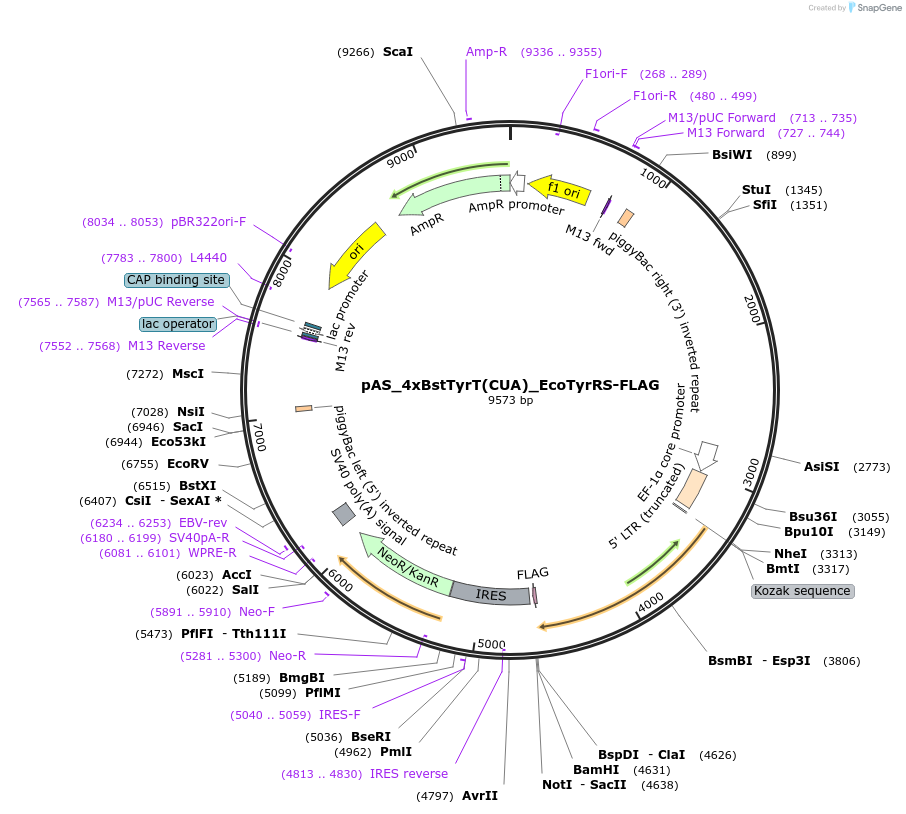
pAS_4xBstTyrT(CUA)_EcoTyrRS-FLAG
Plasmid#140018PurposePlasmid with 4xBstTyrT(CUA) cassette and E. coli TyrRS for amber suppression; for transient or stable piggyBac-mediated integrationDepositorInsertEcoTyrRS
TagsFLAGExpressionMammalianMutationY37L, D182S, F183M, L186APromoterEF1Available SinceMay 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
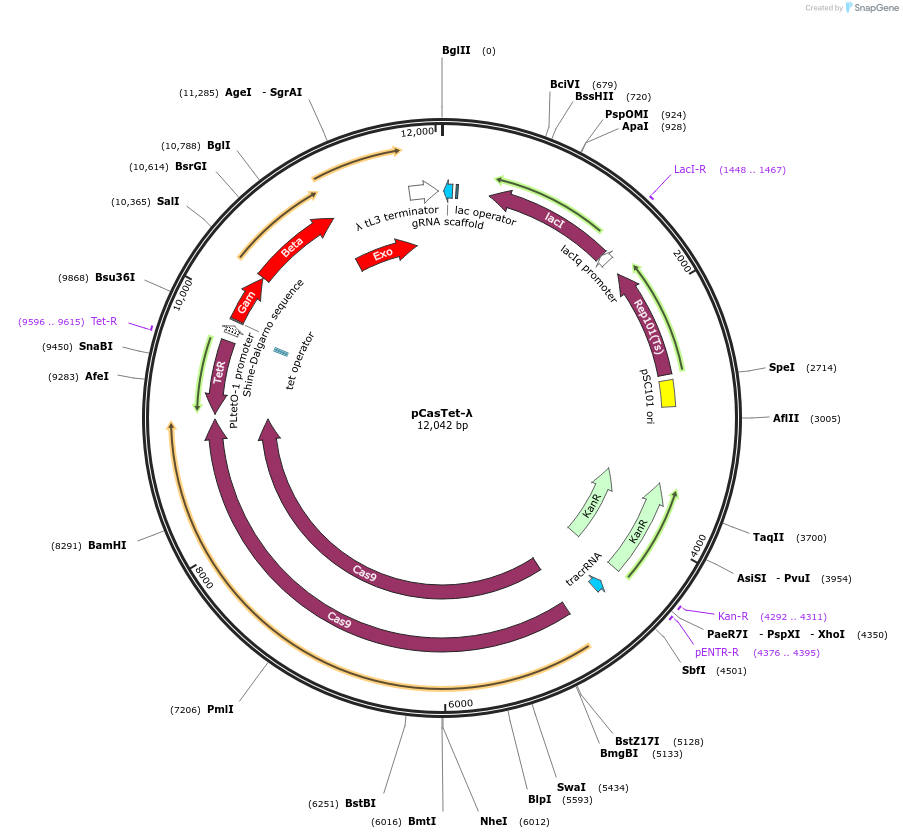
pCasTet-λ
Plasmid#128318PurposeConstitutive expression of cas9 and anhydrotetracycline/tetracycline inducible expression of lamda RED. Useful variant of Plasmid #62225 when arabinose induction is not possible.DepositorInsertTetR and pTetR/TetO
UseCRISPR and Synthetic BiologyExpressionBacterialMutationConstitutive expression of cas9 and anhydro-tetra…Promotertet promoterAvailable SinceAug. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
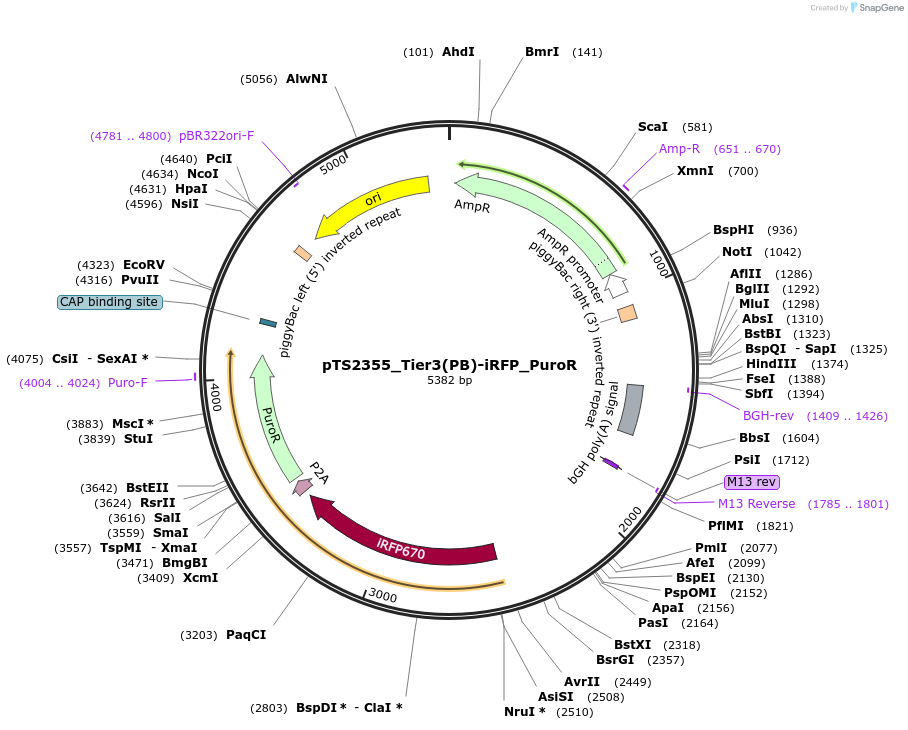
pTS2355_Tier3(PB)-iRFP_PuroR
Plasmid#169654PurposeTier-3 vector for stable stable integration of up to three cassettes via Piggy Bac transposase including a iRFP670 marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-iRFP670-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven iRFP670 and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
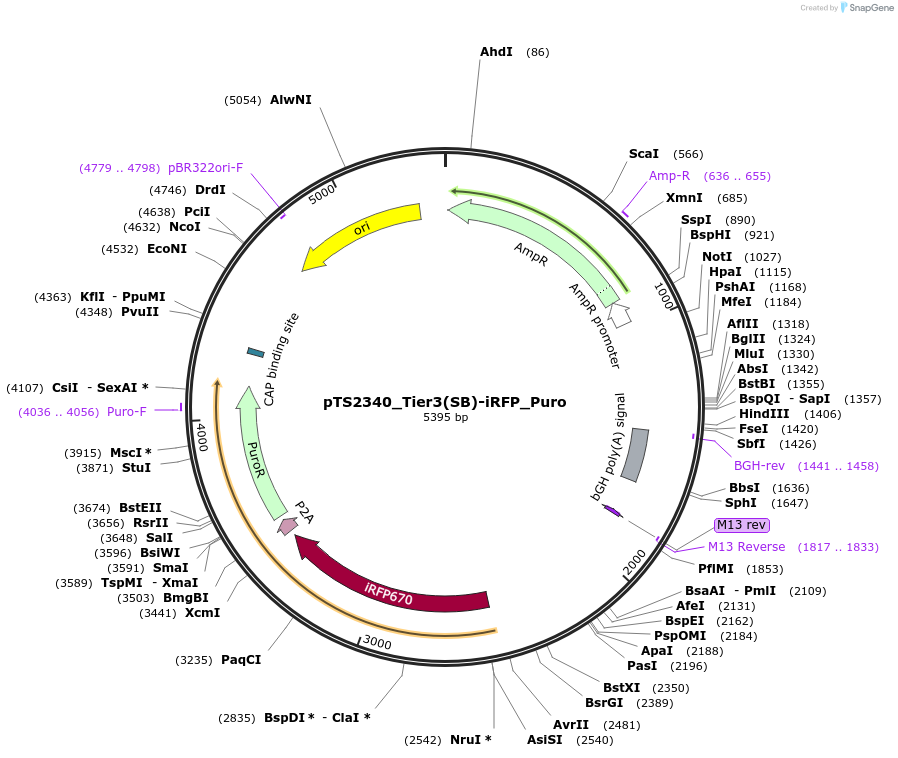
pTS2340_Tier3(SB)-iRFP_Puro
Plasmid#169638PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a iRFP670 marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-iRFP670-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven iRFP670 and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJune 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
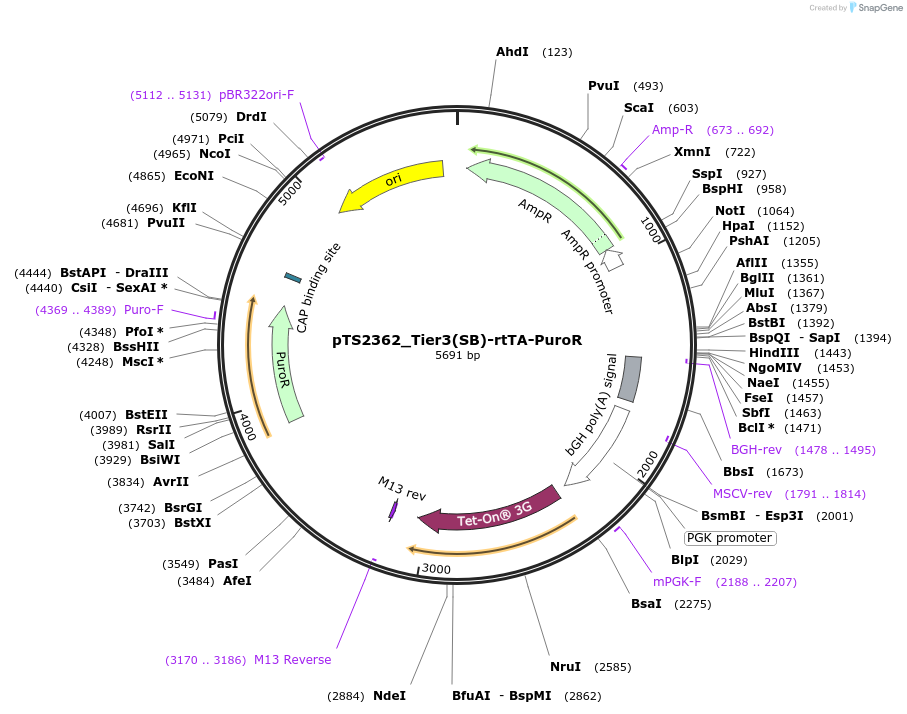
pTS2362_Tier3(SB)-rtTA-PuroR
Plasmid#169658PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a PuroR resistance gene and a PPGK-driven rtTA transactivator (5'ITR-A1-pA::PPGK-rtTA-pA::PRPBSA-PuroR-pA-3'ITR)DepositorInsertPPGK-driven rtTA expression and PRPBSA-driven PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
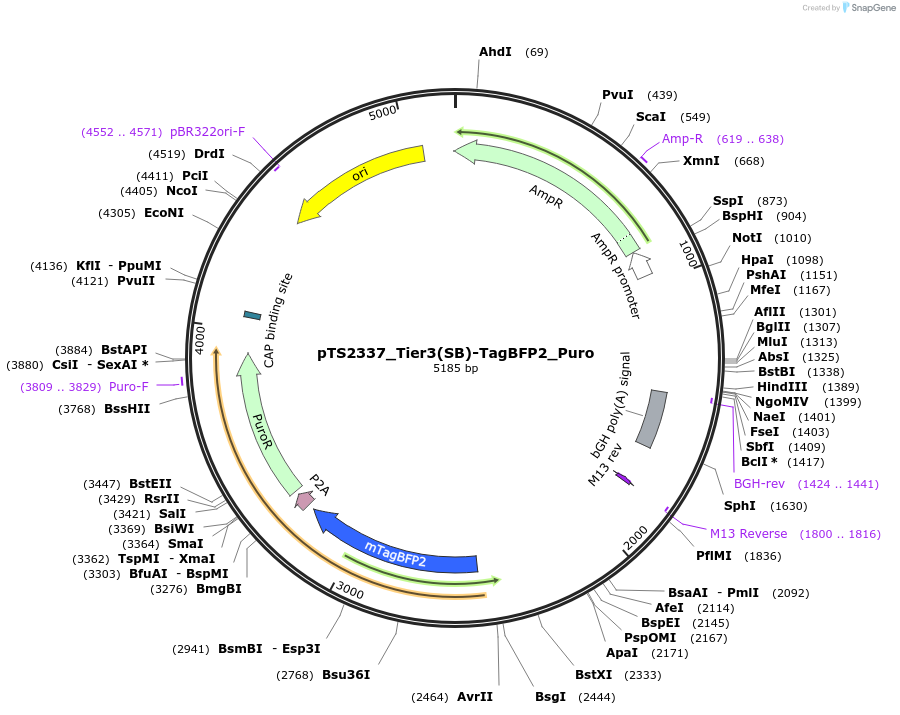
pTS2337_Tier3(SB)-TagBFP2_Puro
Plasmid#169635PurposeTier-3 vector for stable integration of up to three cassettes via sleeping beauty transposase including a TagBFP marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-mTagBFP2-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven mTagBFP2 and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
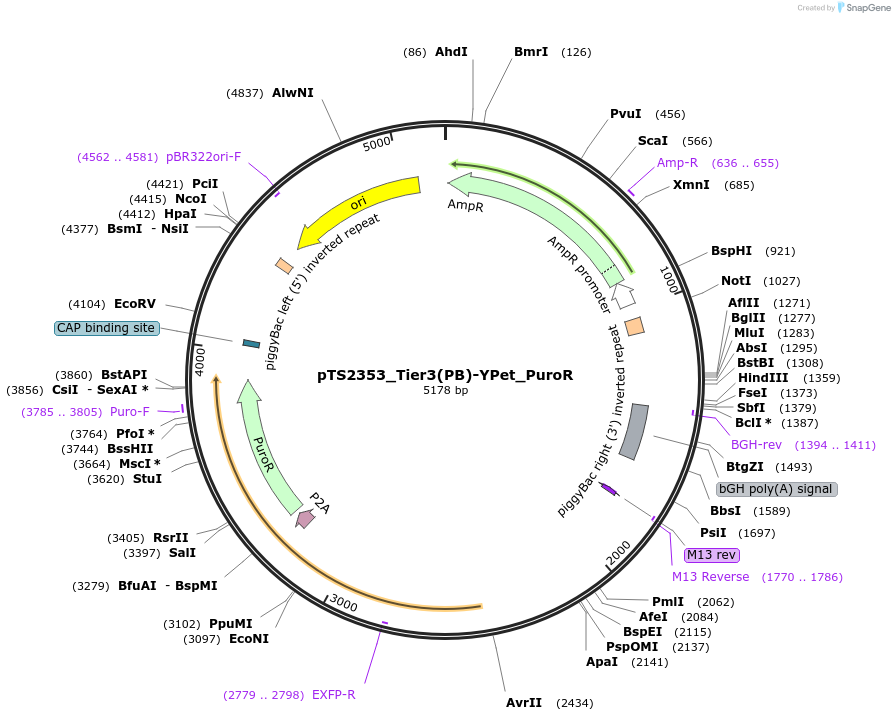
pTS2353_Tier3(PB)-YPet_PuroR
Plasmid#169652PurposeTier-3 vector for stable stable integration of up to three cassettes via Piggy Bac transposase including a YPet marker and PuroR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-YPet-p2A-PuroR-pA-3'ITR)DepositorInsertPRPBSA-driven YPet and PuroR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pTS2343_Tier3(SB)-YPet_Blast
Plasmid#169641PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a YPet marker and BlastR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-YPet-p2A-BlastR-pA-3'ITR)DepositorInsertPRPBSA-driven YPet and BlastR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
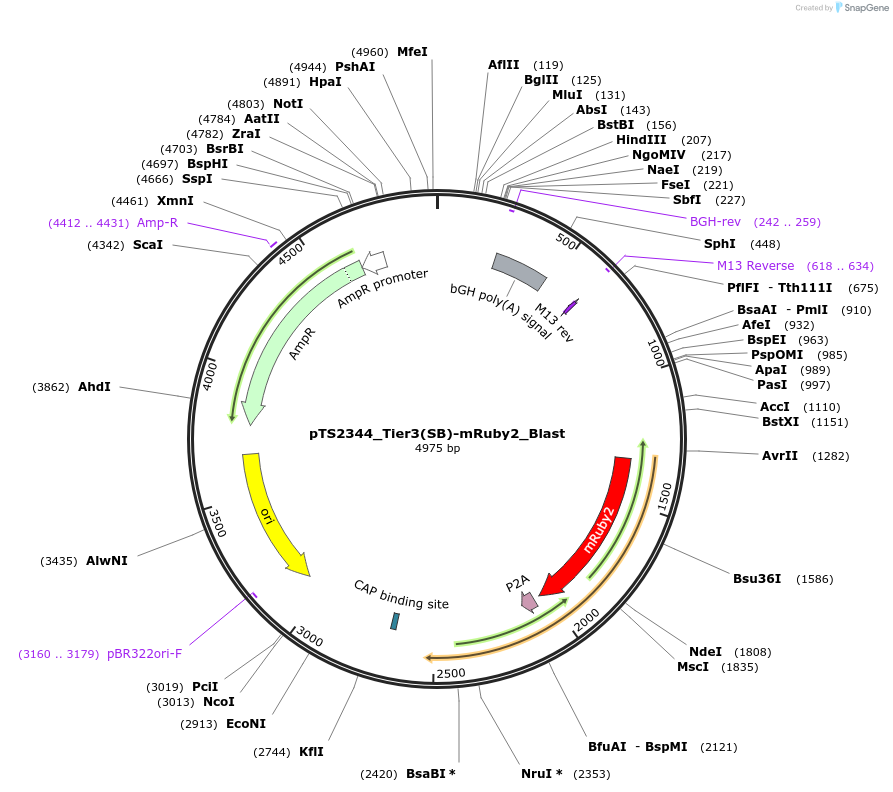
pTS2344_Tier3(SB)-mRuby2_Blast
Plasmid#169642PurposeTier-3 vector for stable stable integration of up to three cassettes via sleeping beauty transposase including a mRuby marker and BlastR resistance gene (5'ITR-A1-pA::A2-pA::PRPBSA-mRuby2-p2A-BlastR-pA-3'ITR)DepositorInsertPRPBSA-driven mRuby2 and BlastR expression
ExpressionMammalianPromoterPRPBSAAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only