We narrowed to 2,507 results for: mia
-
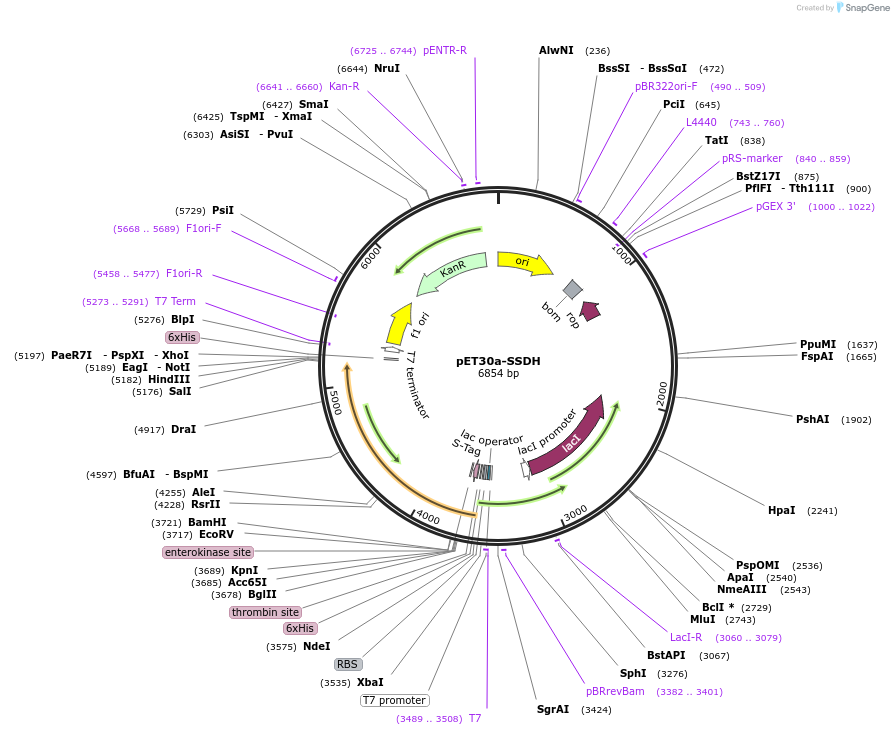
Plasmid#239573PurposeExpresses E. coli succinate semialdehyde dehydrogenase (SSDH/gabD) for recombinant protein purification. Contains a His6-tag on its C-terminus for Ni2+ affinity chromatography.DepositorAvailable SinceJune 27, 2025AvailabilityAcademic Institutions and Nonprofits only
-
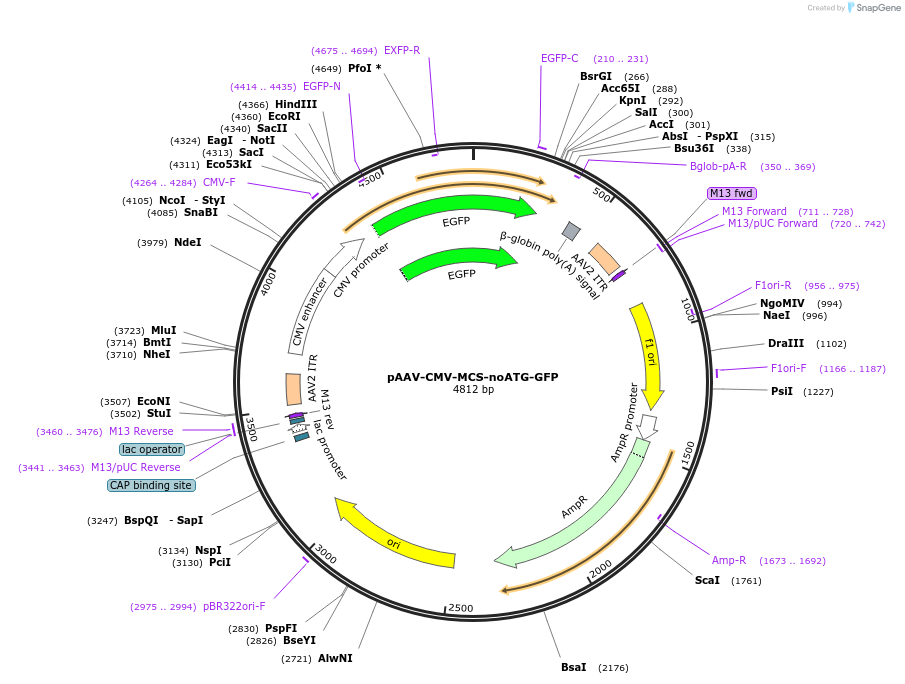
pAAV-CMV-MCS-noATG-GFP
Plasmid#223181PurposeA multicloning site (MCS) was synthesized and inserted into AAV-CMV-GFP to construct the AAV-CMV-MCS-(no ATG)-GFP negative control plasmid.DepositorTypeEmpty backboneUseAAVAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
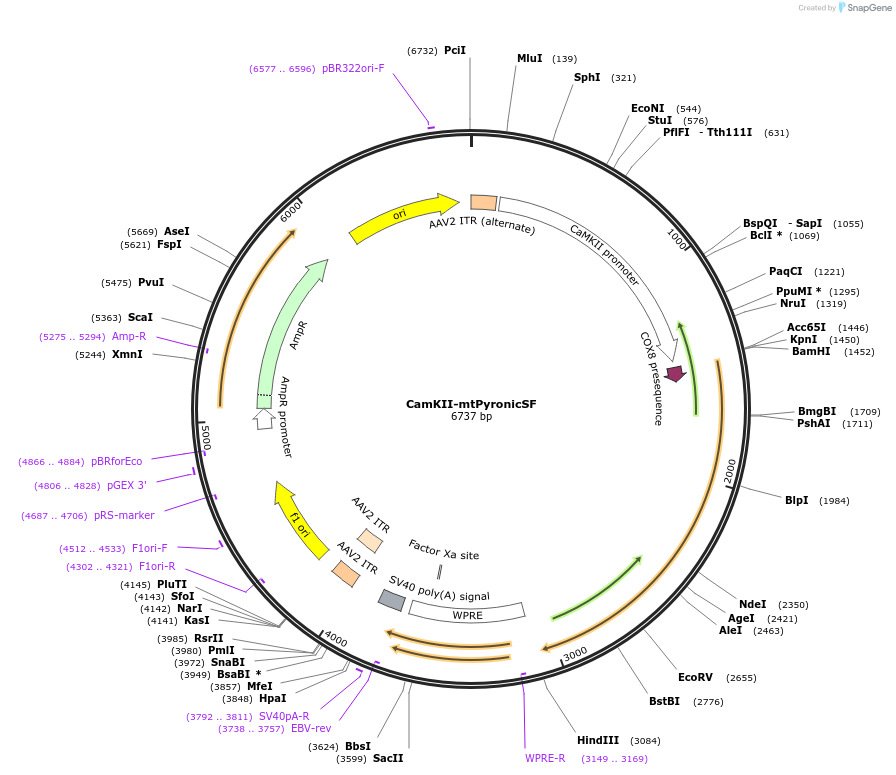
CamKII-mtPyronicSF
Plasmid#228906PurposeExpression of mtPyronicSF in neurons targeted to the mitochondria serving as an optical sensor for mitochondrial pyruvate uptake. Backbone from Addgene plasmid 100834DepositorInsertpyronic SF
UseAAVTagsCox8 targettingExpressionMammalianPromoterCamKIIAvailable SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
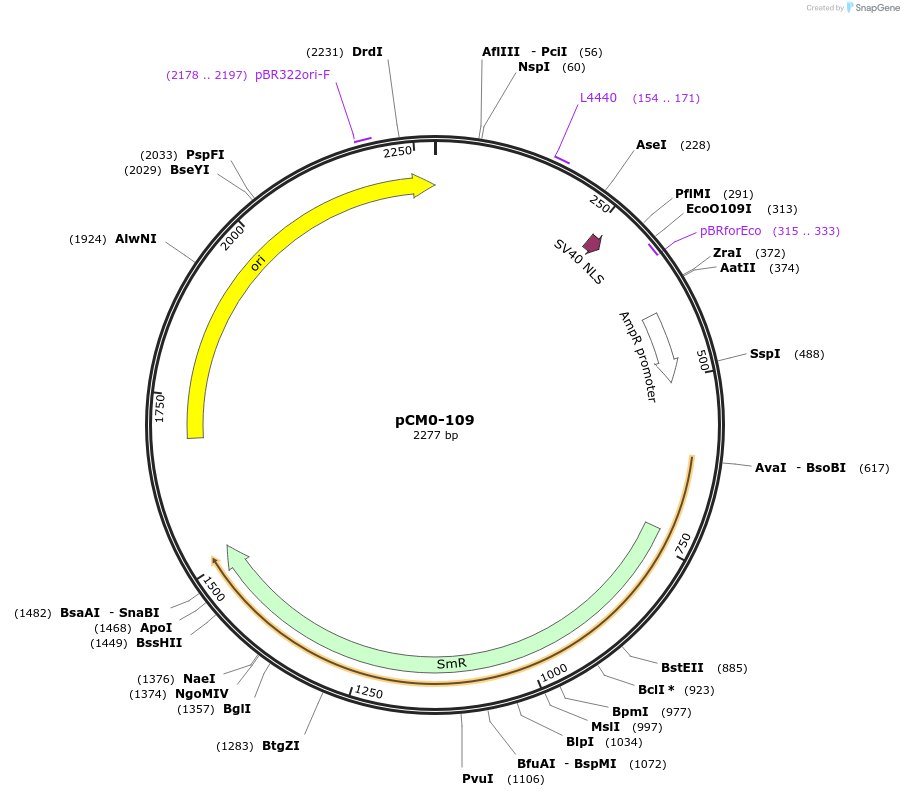
pCM0-109
Plasmid#218200PurposeSV40 NLS for protein targeting to nucleusDepositorInsertSV40 NLS
UseSynthetic BiologyAvailable SinceSept. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
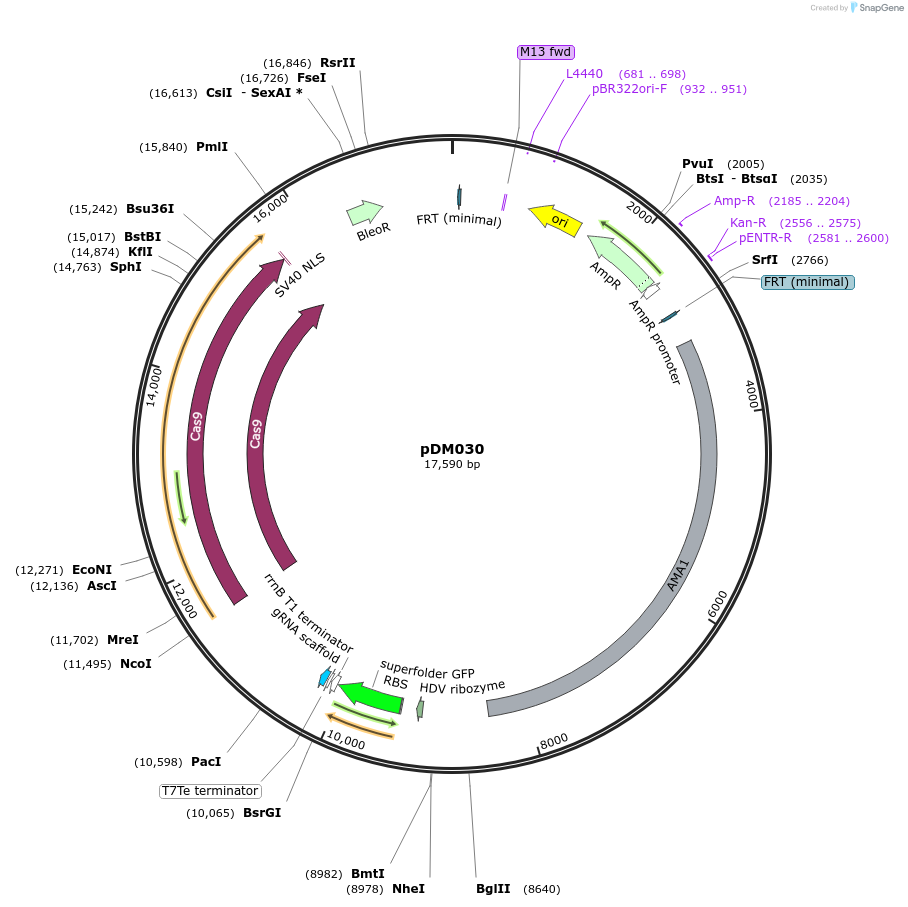
pDM030
Plasmid#216810PurposeVector for Aspergillus CRISPR-Cas9 genetic engineering with Golden Gate cloning drop-out cassette for protospacer insertion and ble selectable marker.DepositorTypeEmpty backboneUseCRISPR; Fungal expressionExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
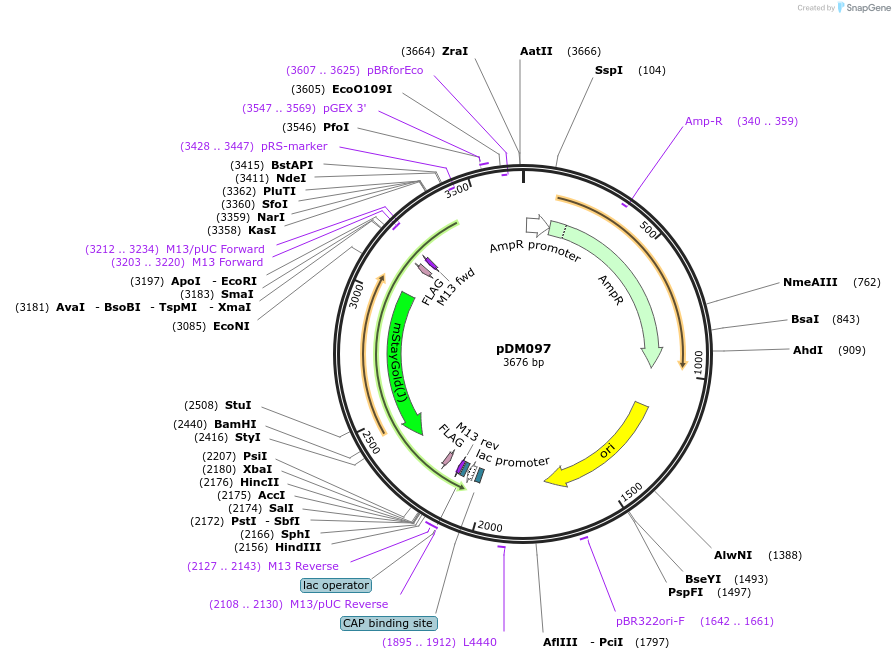
pDM097
Plasmid#216819PurposeAspergillus nidulans codon-adjusted mStayGold fluorescent protein, includes linker for N-terminal or internal tagging.DepositorInsertmStayGold
TagsFLAG-(SGGS)x2-XTEN16-(GGGGS)x3 and c4-(GGGGS)x2-X…ExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
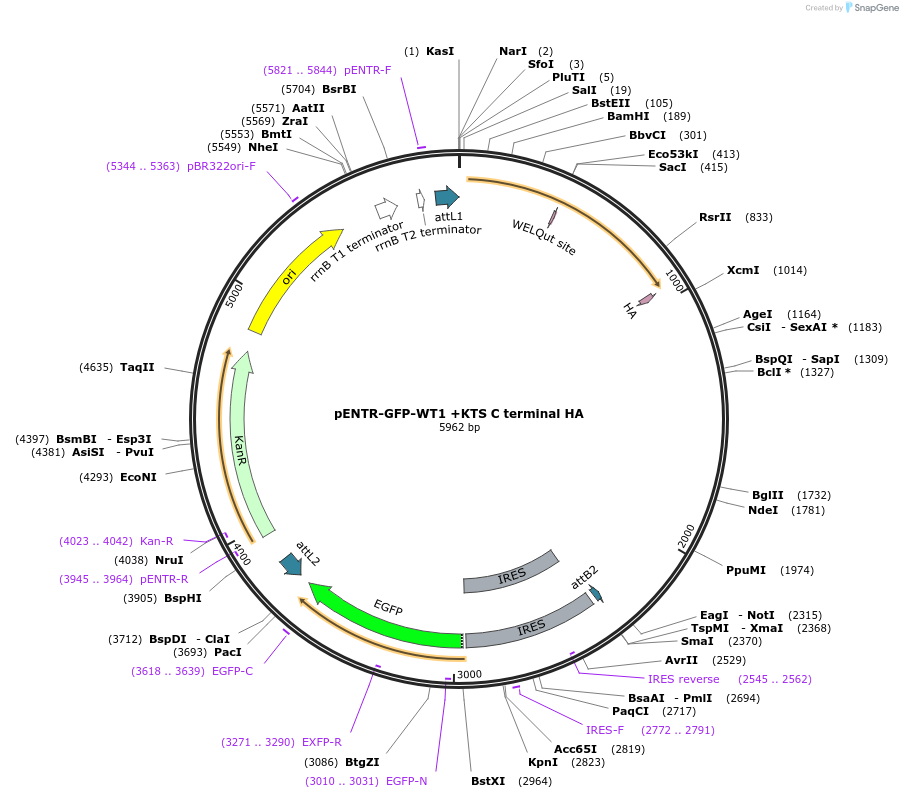
pENTR-GFP-WT1 +KTS C terminal HA
Plasmid#217640PurposeEntry vector with WT1 +KTS mutant with C terminal HA for gateway cloningDepositorAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
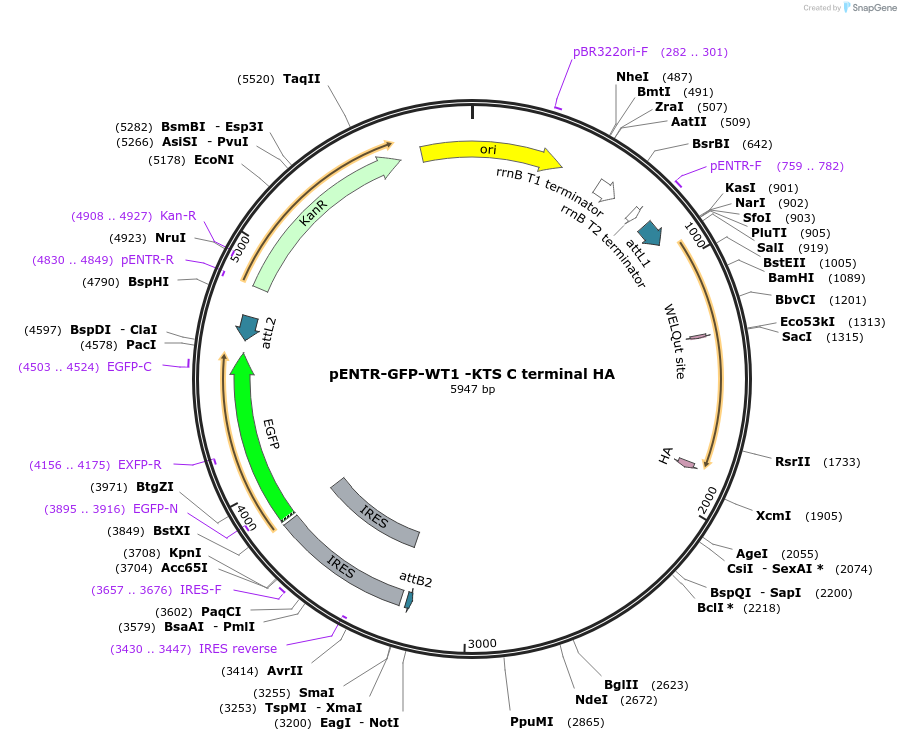
pENTR-GFP-WT1 -KTS C terminal HA
Plasmid#217642PurposeEntry vector with WT1 -KTS mutant with C terminal HA for gateway cloningDepositorAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
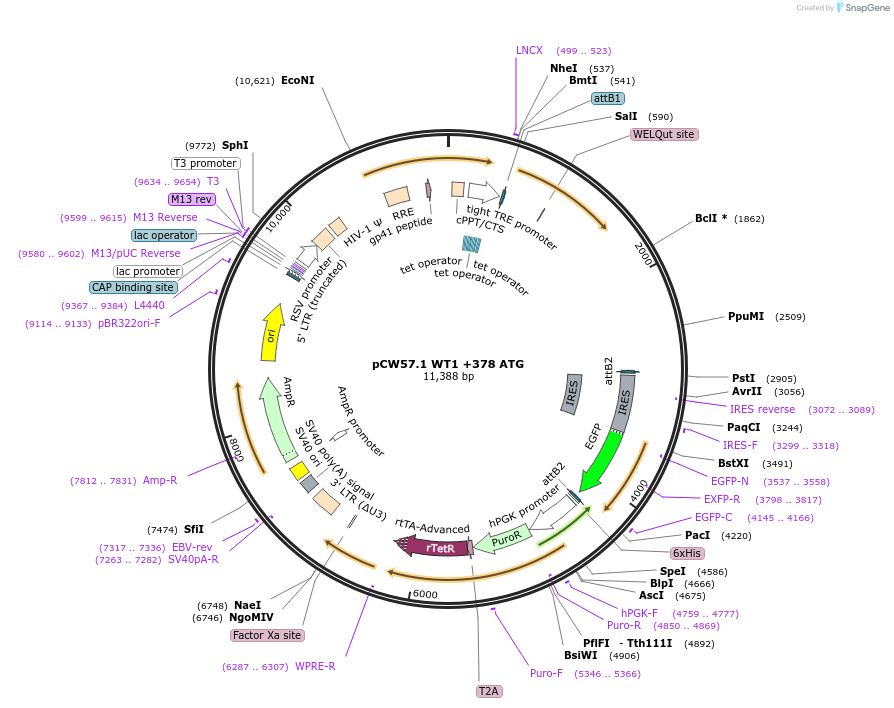
pCW57.1 WT1 +378 ATG
Plasmid#217644PurposeDox inducible WT1 cDNA - as transcribed from +378ATG (short isoform)DepositorInsertWT1 short isoform (WT1 Human)
ExpressionMammalianAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
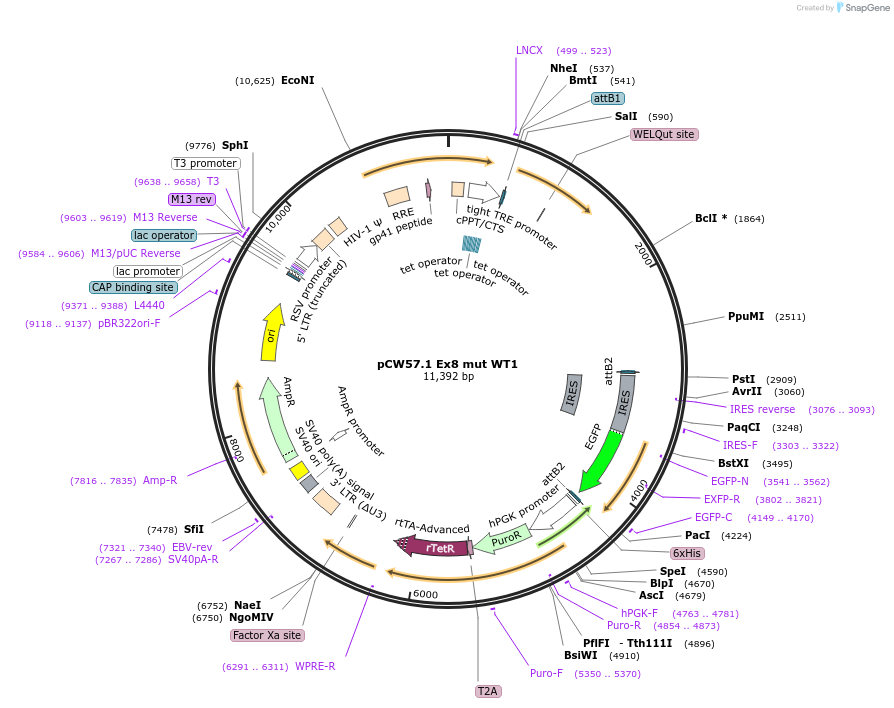
pCW57.1 Ex8 mut WT1
Plasmid#217645PurposeDox inducible WT1 cDNA with exon 8 mutationDepositorAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
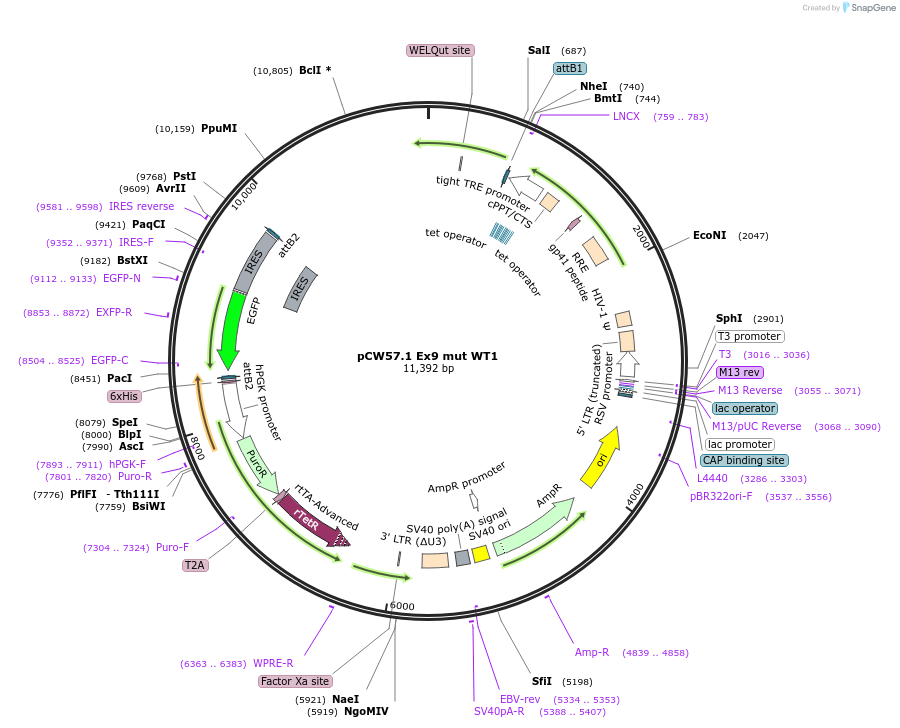
pCW57.1 Ex9 mut WT1
Plasmid#217646PurposeDox inducible WT1 cDNA with exon 9 mutationDepositorAvailable SinceMay 22, 2024AvailabilityAcademic Institutions and Nonprofits only -
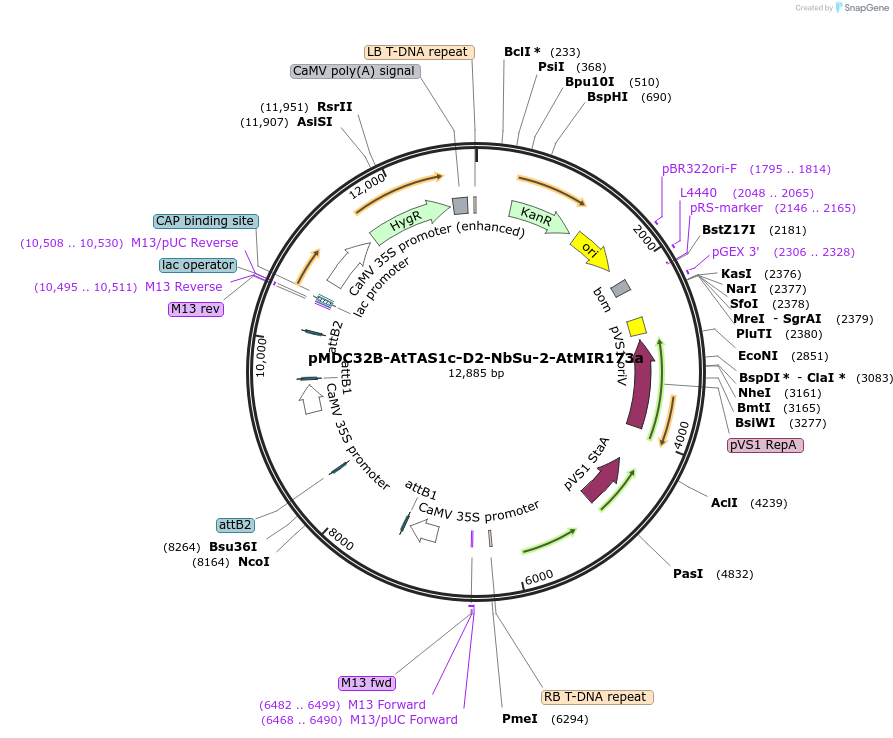
pMDC32B-AtTAS1c-D2-NbSu-2-AtMIR173a
Plasmid#213401PurposePlant expression vector (2x35S) for expressing a syn-tasiRNA against Nicotiana benthamiana SULFUR gene from AtTAS1c precursorDepositorInsertArabidopsis TAS1c with a syn-tasiRNA sequence at D2 for silencing N. benthamiana SULFUR gene. MIR173 cassette.
ExpressionPlantPromoter2x35SAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
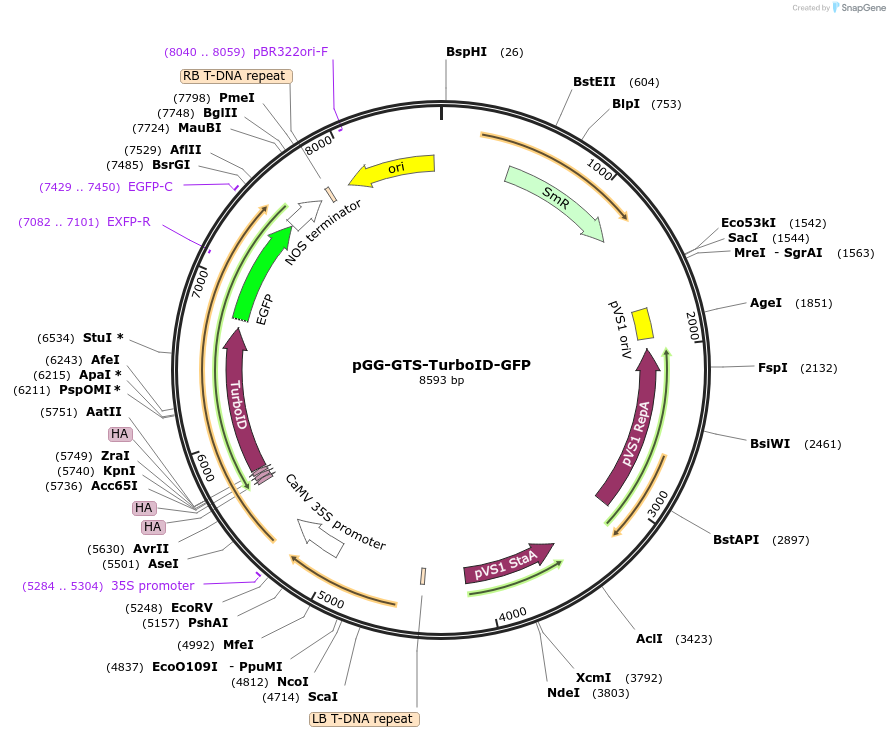
pGG-GTS-TurboID-GFP
Plasmid#209405PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Golgi targeting sequence (GTS) in plantaDepositorInsertGTS-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
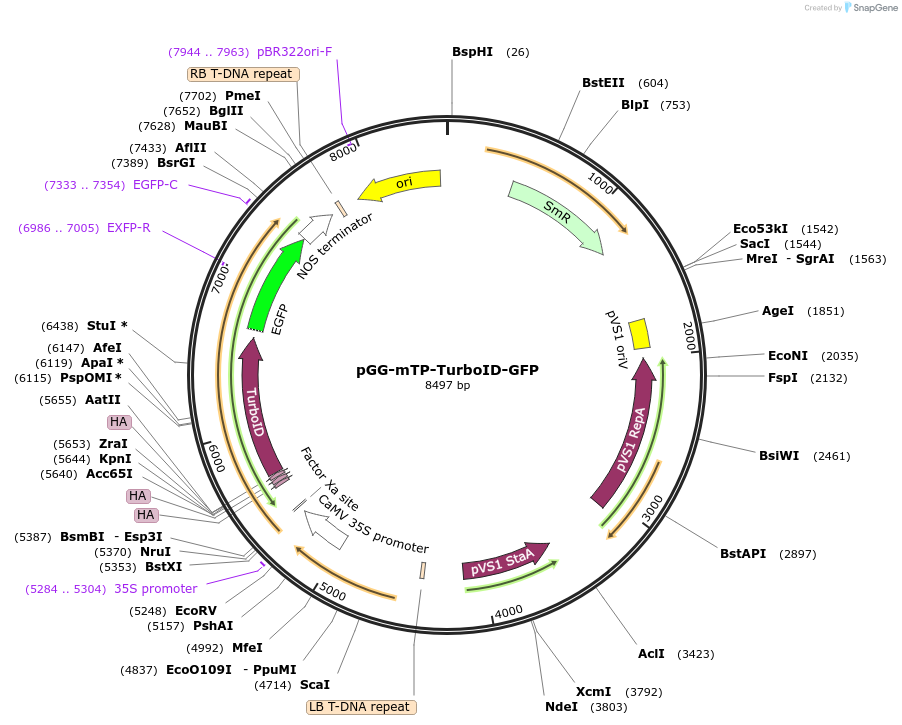
pGG-mTP-TurboID-GFP
Plasmid#209404PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Mitochondrial transit peptide (mTP) in plantaDepositorInsertmTP-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
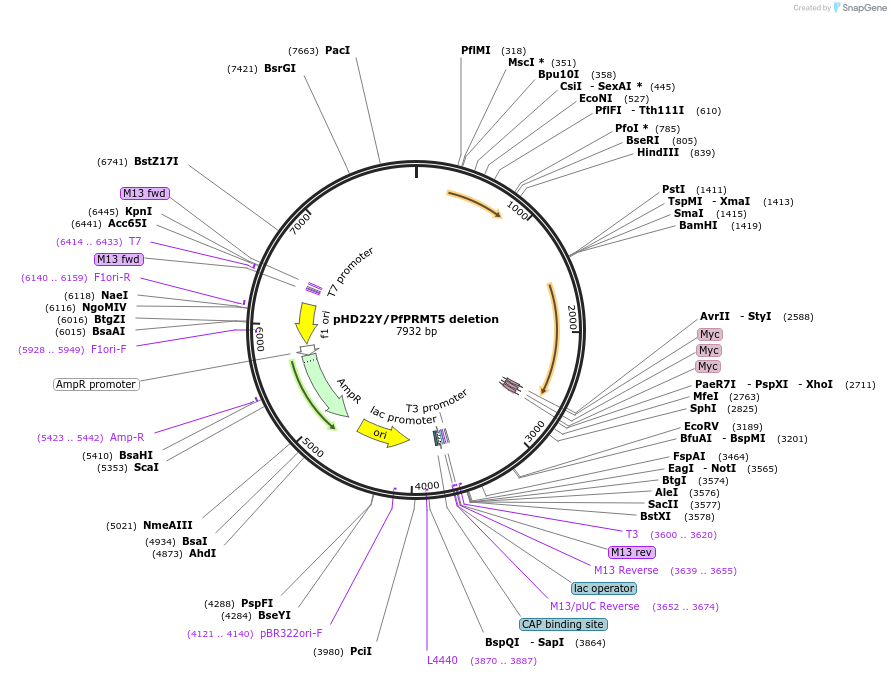
pHD22Y/PfPRMT5 deletion
Plasmid#203161Purposefor disruption of PfPRMT5 geneDepositorInsertPfPRMT5 fragment
Tags3 myc tags and 3' UTR of P. berghei dihydrof…ExpressionMammalianPromoterNoAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
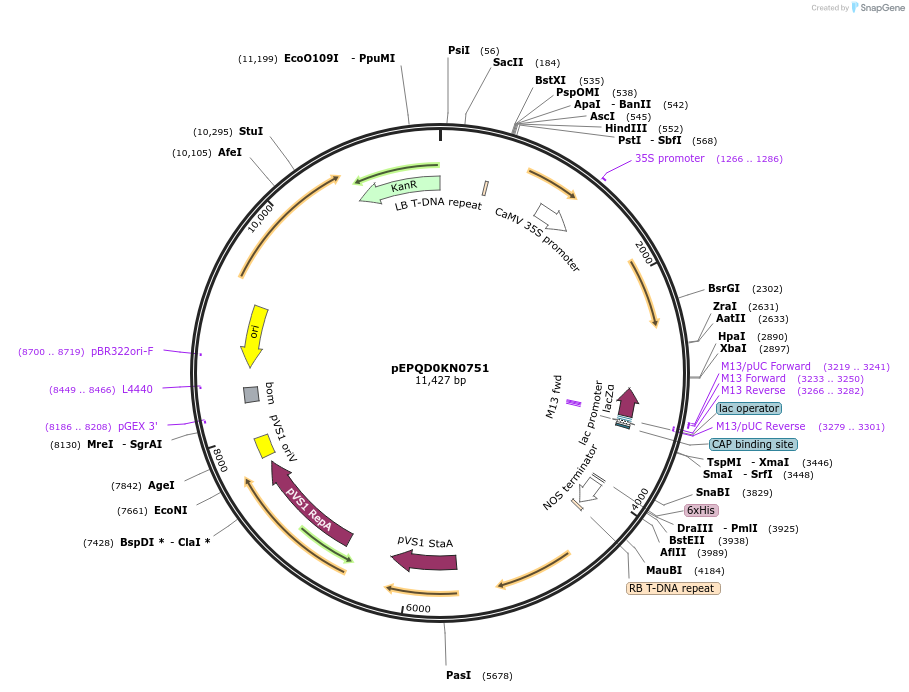
pEPQD0KN0751
Plasmid#185628PurposeModified tobacco rattle virus RNA2 acceptor vector that could be used for expression of sgRNAs, contains restriction enzyme sites for golden gate assembly using BsaIDepositorInsertlacZ
UseCRISPR and Synthetic BiologyAvailable SinceAug. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
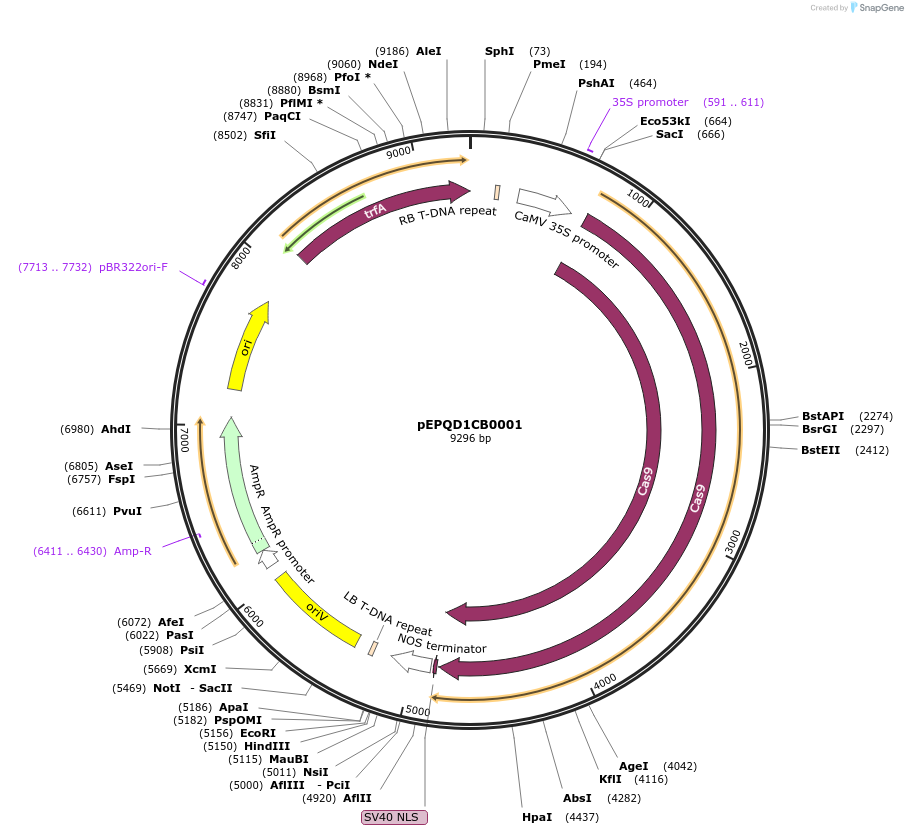
pEPQD1CB0001
Plasmid#185625PurposeMoClo Level 1, position 2 reverse, transcriptional unit for expression of plant codon optimized Cas9 from Streptococcus pyogenes driven by 35S promoterDepositorInsertSpCas9
UseCRISPR and Synthetic BiologyAvailable SinceAug. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
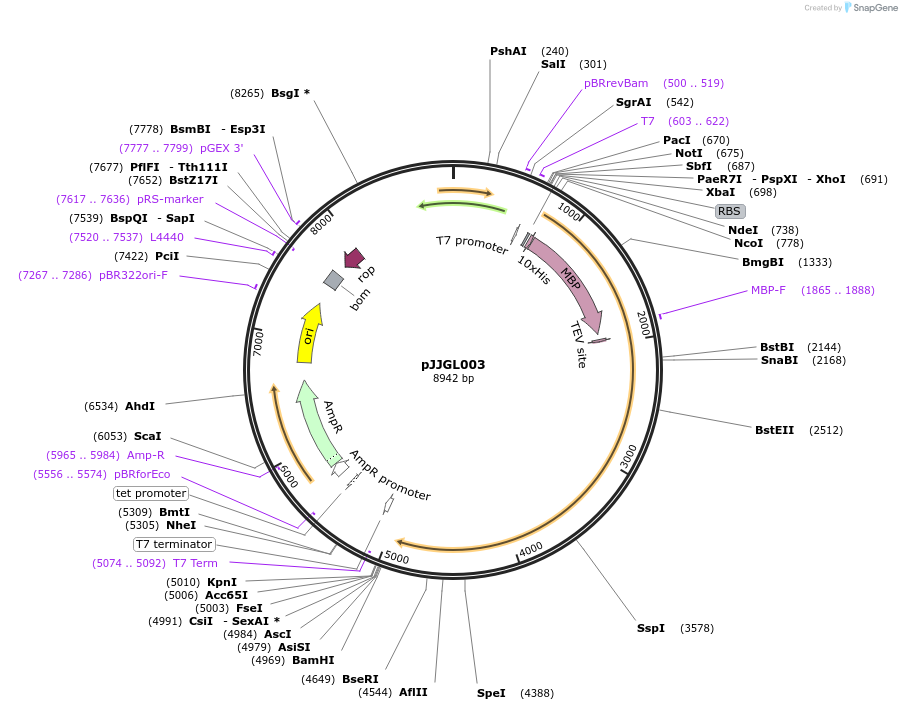
pJJGL003
Plasmid#180607PurposePlasmid expressing E. coli codon optimized His-MBP-TEV-DpbCasX+Region3 insertionDepositorInsertDpbCasX with R3 loop
UseCRISPRTags10x His and MBPExpressionBacterialPromoterT7Available SinceApril 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
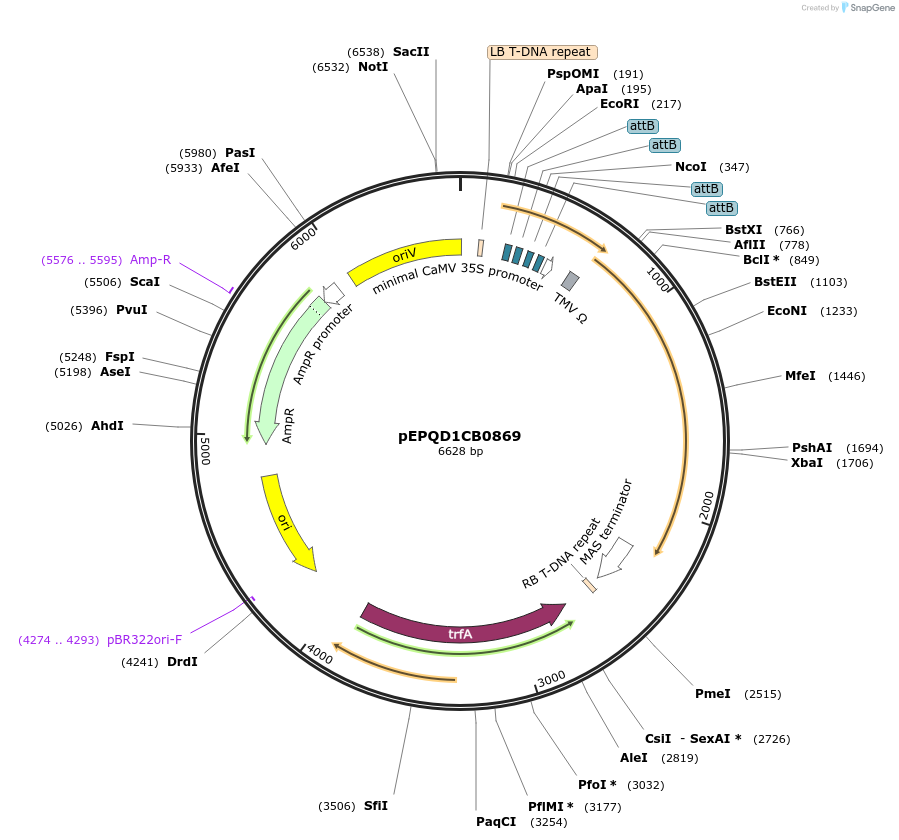
pEPQD1CB0869
Plasmid#177086PurposeMoClo Level 1, position 5, transcriptional unit for transient expression of 7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsert7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
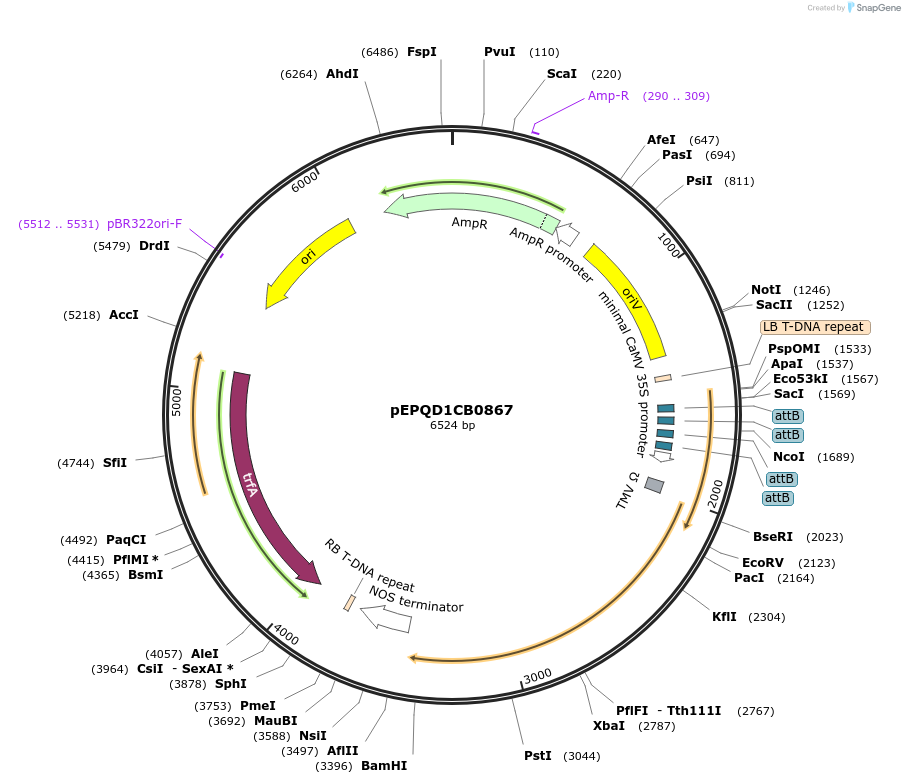
pEPQD1CB0867
Plasmid#177085PurposeMoClo Level 1, position 3, transcriptional unit for transient expression of 7-deoxyloganetic acid glucosyl transferase (Cr7-DLGT) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsert7-deoxyloganetic acid glucosyl transferase (Cr7-DLGT) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only