We narrowed to 8,706 results for: PAN
-
Plasmid#207595PurposeGFP-BAB expressionDepositorInsertGFP-BAB
ExpressionBacterialAvailable SinceNov. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
pJS109
Plasmid#207590PurposeGFP-ABBA expressionDepositorInsertGFP-ABBA
ExpressionBacterialAvailable SinceNov. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB430
Plasmid#203621PurposeTU for the expression of phleomycin resistance.DepositorInsertPpcbC:ble:TamdS
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
FB431
Plasmid#203622PurposeTU for the expression of terbinafine resistance.DepositorInsertPgpdA:ergA:TamdS
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
pGL0_37 [tetA pSC101]
Plasmid#198950PurposeLevel 0 partDepositorInserttetA pSC101
UseSynthetic BiologyAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pGL0_36 [PtetA pSC101]
Plasmid#198949PurposeLevel 0 partDepositorInsertPtetA pSC101
UseSynthetic BiologyAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
PMT-mKate2_2
Plasmid#203786PurposeBrightness calibrationDepositorInsertPMT-mKate2 dimer
ExpressionMammalianAvailable SinceJuly 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
PMT-mCherry2_2
Plasmid#203784PurposeBrightness calibrationDepositorInsertPMT-mCherry2 dimer
ExpressionMammalianAvailable SinceJuly 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
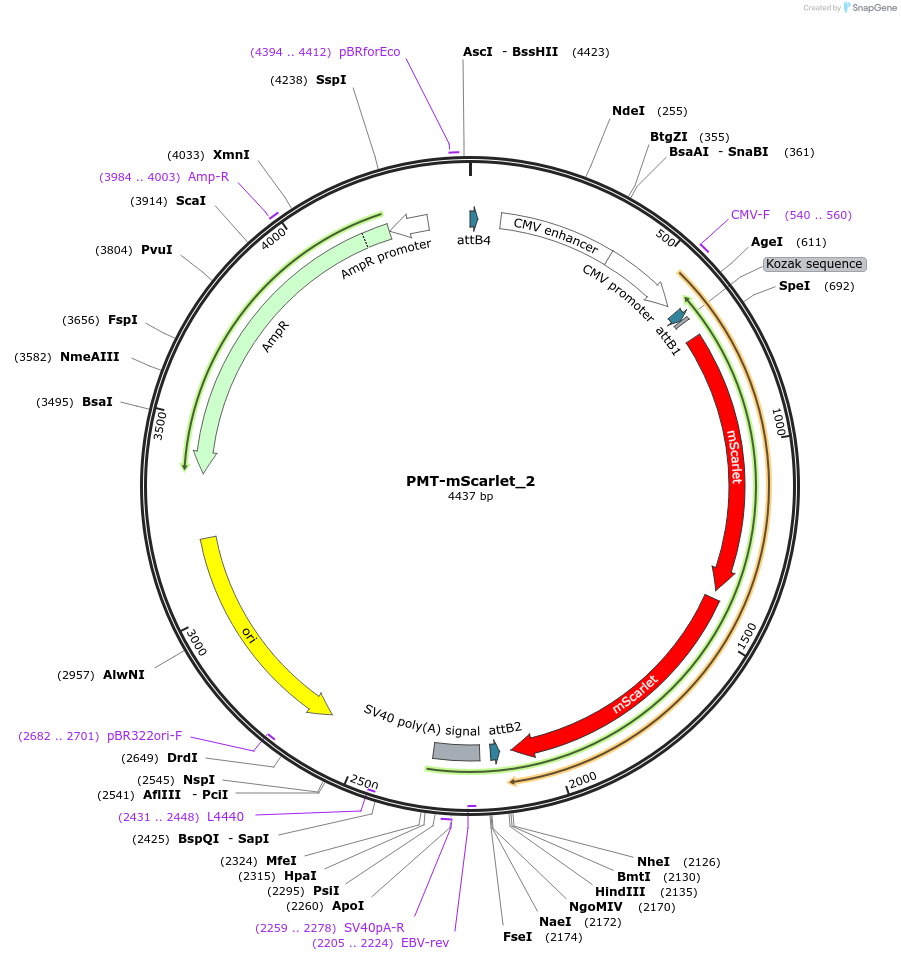
PMT-mScarlet_2
Plasmid#203780PurposeBrightness calibrationDepositorInsertPMT-mScarlet dimer
ExpressionMammalianAvailable SinceJuly 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
PMT-mCherry_2
Plasmid#203782PurposeBrightness calibrationDepositorInsertPMT-mCherry dimer
ExpressionMammalianAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
pCxcl1-Firefly-1-Tbx5-site
Plasmid#177832PurposeLuciferase reporter for mouse Cxcl1 with one Tbx5 consensus binding siteDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCxcl1-Firefly-2-Tbx5-sites
Plasmid#177833PurposeLuciferase reporter for mouse Cxcl1 with two Tbx5 consensus binding sitesDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
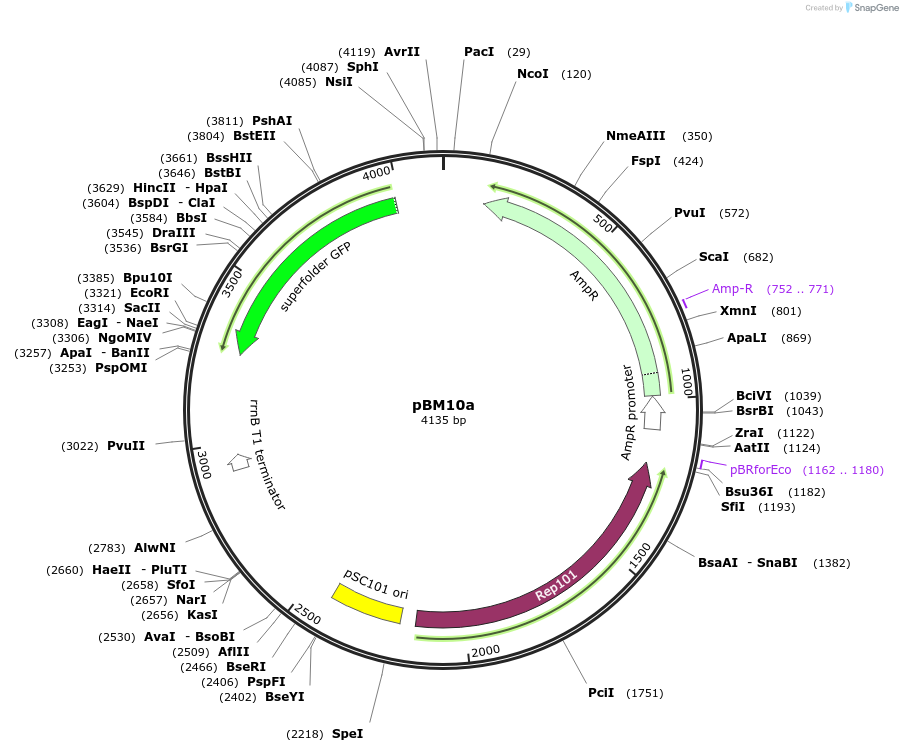
pBM10a
Plasmid#179688PurposeTarget plasmid for context profilingDepositorInsertACA target
ExpressionBacterialAvailable SinceApril 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRS426-PGAL7-A_GFP_TCYC1
Plasmid#177162PurposeExpresses yeGFP under the control of a synthetic promoter (PGAL7-A)DepositorInsertyeGFP
ExpressionYeastPromoterSynthetic promoter (PGAL7-A)Available SinceJan. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRS426-PGAL7-B_GFP_TCYC1
Plasmid#177163PurposeExpresses yeGFP under the control of a synthetic promoter (PGAL7-B)DepositorInsertyeGFP
ExpressionYeastPromoterSynthetic promoter (PGAL7-B)Available SinceJan. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRS426-PGAL7-C_RFP_TCYC1
Plasmid#177164PurposeExpresses yeRFP under the control of a synthetic promoter (PGAL7-C)DepositorInsertyeCherry
ExpressionYeastPromoterSynthetic promoter (PGAL7-C)Available SinceJan. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRS426-PGAL7-D_RFP_TCYC1
Plasmid#177165PurposeExpresses yeRFP under the control of a synthetic promoter (PGAL7-D)DepositorInsertyeCherry
ExpressionYeastPromoterSynthetic promoter (PGAL7-D)Available SinceJan. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pIRENE-div-Firefly
Plasmid#170661PurposeLuciferase reporter for mouse IRENE-div lncRNADepositorInsertIRENE-div
UseLuciferasePromoterIRENE-divAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCxcl1-Firefly-Chantico-Tdel
Plasmid#163900PurposeLuciferase reporter for mouse Cxcl1 with Chantico promoter Tbx5 site deletionDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
pChantico-Tdel-Firefly
Plasmid#163902PurposeLuciferase reporter for Chantico lncRNA with Tbx5 site deletionDepositorInsertLuciferase
UseLuciferasePromoterChantico with Tbx5 consensus binding site deletionAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
pChantico-pAS-Firefly
Plasmid#163903PurposeLuciferase reporter for Chantico lncRNA with triple pAS insertion downstream of the promoterDepositorInsertLuciferase
UseLuciferasePromoterChanticoAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
pG-02
Plasmid#166933PurposeTarget AD1.A5 part (G)DepositorInsertStar sense AD1.S5 +RBS - Block G- Flanked by BsaI restriction sites
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
pC-03
Plasmid#166927PurposeSTAR-AD1.A6 part (C)DepositorInsertStar Antisense AD1.A6 - Block C - Flanked by BsaI sites
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
pC-02
Plasmid#166926PurposeSTAR-AD1.A5 part (C)DepositorInsertStar Antisense AD1.A5- Block C- Flanked by BsaI restriction sites
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
pMV23
Plasmid#128887PurposePosition 4 module- STOP (stop codon)DepositorInsertSTOP
UseSynthetic BiologyExpressionYeastAvailable SinceFeb. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
C-13-BE
Plasmid#135041PurposeC-T base editing using LpCDA1L1_4 fused to C terminal of nCas9DepositorInsertLpCDA1L1_4
UseCRISPRExpressionMammalianPromoterU6 with EF-1alpha intronAvailable SinceFeb. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
C-7-BE
Plasmid#135035PurposeC-T base editing using LpCDA1L1_1 fused to C terminal of nCas9DepositorInsertLpCDA1L1_1
UseCRISPRExpressionMammalianPromoterU6 with EF-1alpha intronAvailable SinceFeb. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
C-6-BE
Plasmid#135034PurposeC-T base editing using LjDA1L2_2 fused to C terminal of nCas9DepositorInsertLjDA1L2_2
UseCRISPRExpressionMammalianPromoterU6 with EF-1alpha intronAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
C-11-BE
Plasmid#135039PurposeC-T base editing using LpCDA1L2_2 fused to C terminal of nCas9DepositorInsertLpCDA1L2_2
UseCRISPRExpressionMammalianPromoterU6 with EF-1alpha intronAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
pMV10
Plasmid#128892PurposePosition 6 module- URA3DepositorAvailable SinceAug. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMV5
Plasmid#128886PurposePosition 4 module- CLN2-PEST (Degron)DepositorInsertCLN2-PEST
UseSynthetic BiologyExpressionYeastAvailable SinceAug. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
-
pCfB2229
Plasmid#67537PurposeEasyClone system-based yeast integrative vector carrying loxP-flanked CaLYS5 marker, integration into S. cerevisiae XI-5 chromosomal location, USER site for cloning, amp resistanceDepositorTypeEmpty backboneUseCre/Lox; IntegrativeExpressionYeastAvailable SinceNov. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
pattB-synaptobrevin-13-QFBDAD-G4MD257-761-hsp70
Plasmid#46122DepositorInsertQFBDAD-G4MD257-761
ExpressionInsectAvailable SinceAug. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pattB-synaptobrevin-12-QFBDAD-G4MD50-761-hsp70
Plasmid#46121DepositorInsertQFBDAD-G4MD50-761
ExpressionInsectAvailable SinceAug. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pLenti6.2_mNeonGreen2
Plasmid#113727PurposeExpresses the fluorescent protein mNeonGreen2 in a lentiviral backboneDepositorInsertmNeonGreen2
UseLentiviralExpressionMammalianPromoterCMVAvailable SinceMay 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
pRSF-1b-S2
Plasmid#128590PurposeExpresses tagless S2 ribosomal protein in E.coliDepositorAvailable SinceMay 25, 2022AvailabilityAcademic Institutions and Nonprofits only