We narrowed to 29,785 results for: des.2
-
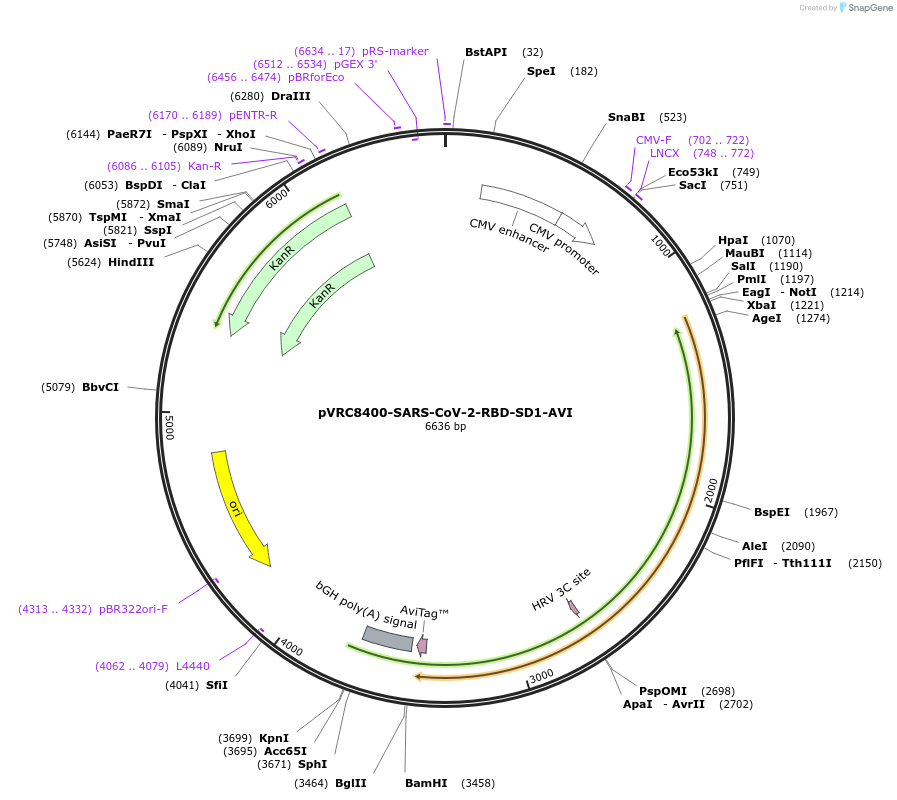
Plasmid#160477PurposeExpresses SARS-CoV-2 RBD-SD1 domain with single chain Fc, HRV3C protease cleavage site, and AVI tagDepositorInsertSARS-CoV-2-RBD-SD1
TagsAVI, HRV3C, and scFcExpressionMammalianAvailable SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
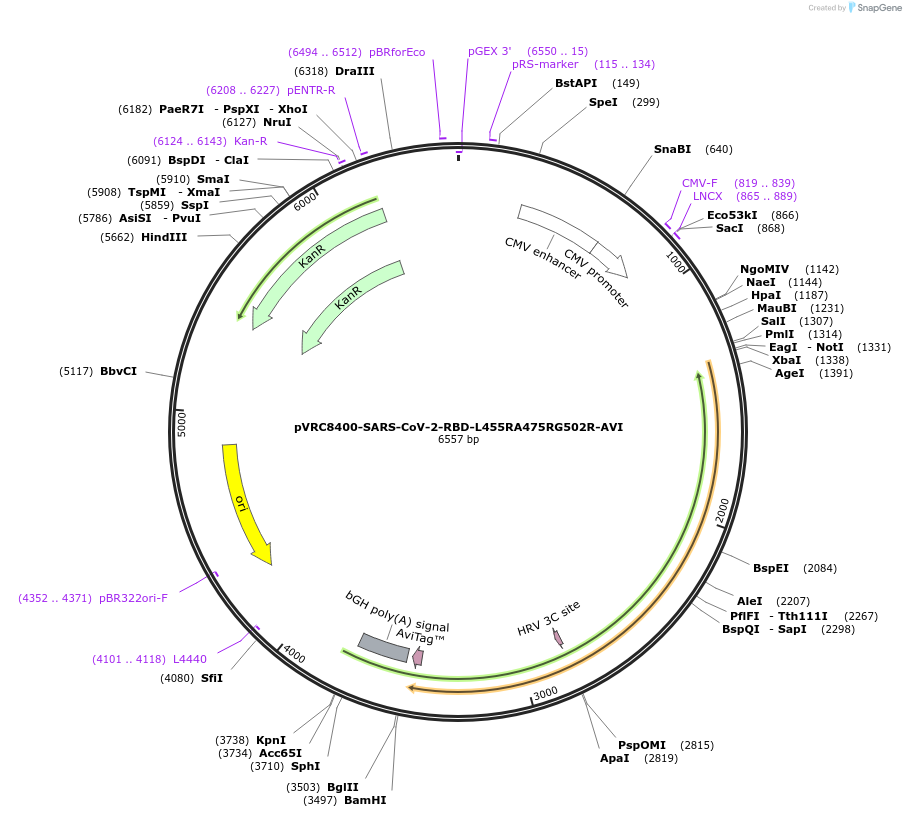
pVRC8400-SARS-CoV-2-RBD-L455RA475RG502R-AVI
Plasmid#160480PurposeExpresses SARS-CoV-2 RBD-L455RA475RG502R domain with single chain Fc, HRV3C protease cleavage site, and AVI tagDepositorInsertSARS-CoV-2-RBD-L455RA475RG502R
TagsAVI, HRV3C, and scFcExpressionMammalianMutationchanged Leucine 455 to Arginine, Alanine 475 to A…Available SinceOct. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
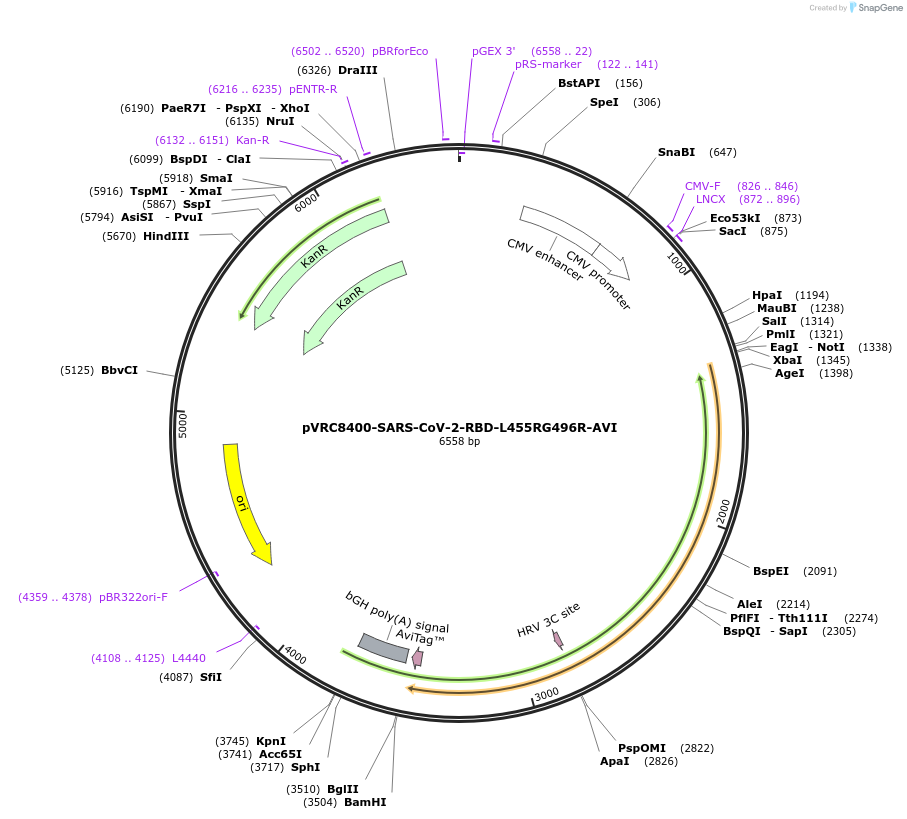
pVRC8400-SARS-CoV-2-RBD-L455RG496R-AVI
Plasmid#160479PurposeExpresses SARS-CoV-2 RBD-L455RG496R domain with single chain Fc, HRV3C protease cleavage site, and AVI tagDepositorInsertSARS-CoV-2-RBD-L455RG496R
TagsAVI, HRV3C, and scFcExpressionMammalianMutationchanged Leucine 455 to Arginine and changed Glyci…Available SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
pVRC8400-SARS-CoV-2-RBD-L455RA475R-AVI
Plasmid#160478PurposeExpresses SARS-CoV-2 RBD-L455RA475R domain with single chain Fc, HRV3C protease cleavage site, and AVI tagDepositorInsertSARS-CoV-2-RBD-L455RA475R
TagsAVI, HRV3C, and scFcExpressionMammalianMutationchanged Leucine 455 to Arginine and changed Alani…Available SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
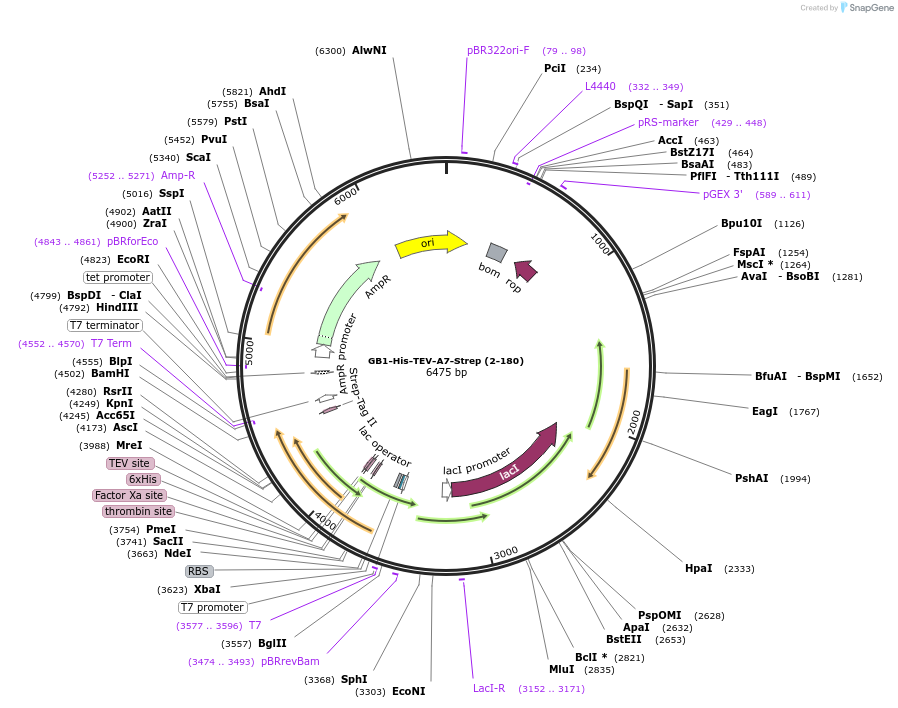
GB1-His-TEV-A7-Strep (2-180)
Plasmid#190067PurposeCodon-optimized bacterial expression plasmid. Expresses Annexin A7 proline rich domain with strep.DepositorInsertAnnexin A7 PRD (ANXA7 Human)
TagsGB1-His-TEV and StrepExpressionBacterialMutationPRD only, residues 2-180Available SinceSept. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
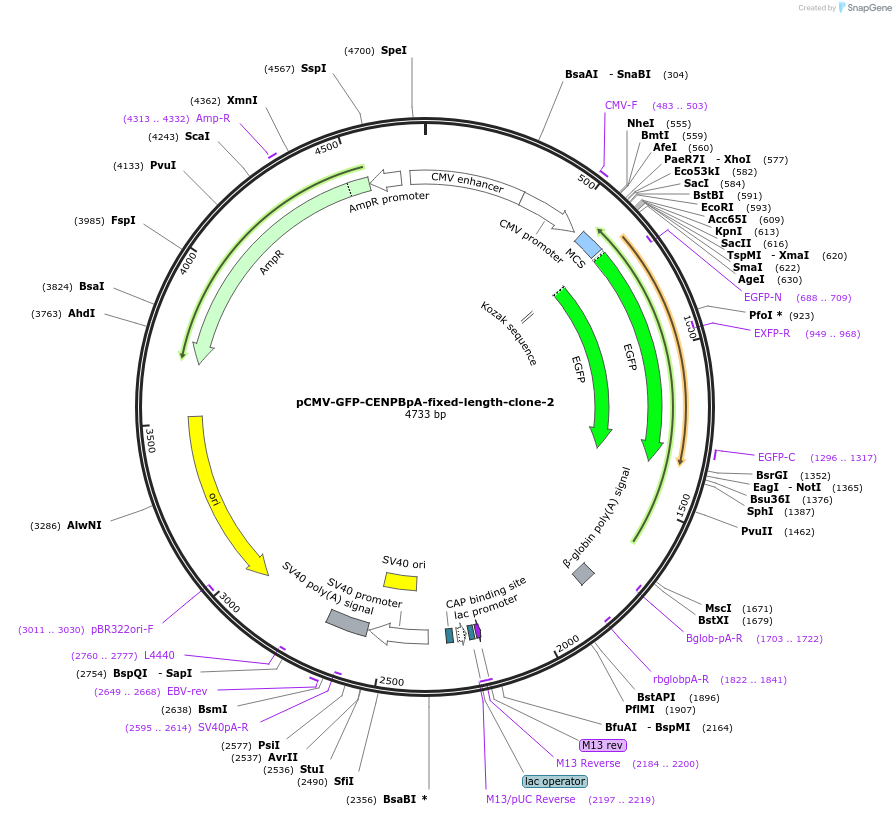
pCMV-GFP-CENPBpA-fixed-length-clone-2
Plasmid#120180PurposeRandomly picked clone 2 of the pCMV-GFP-CENPBpA-fixed-length libraryDepositorInsertCENPB poly(A) site
ExpressionMammalianAvailable SinceApril 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
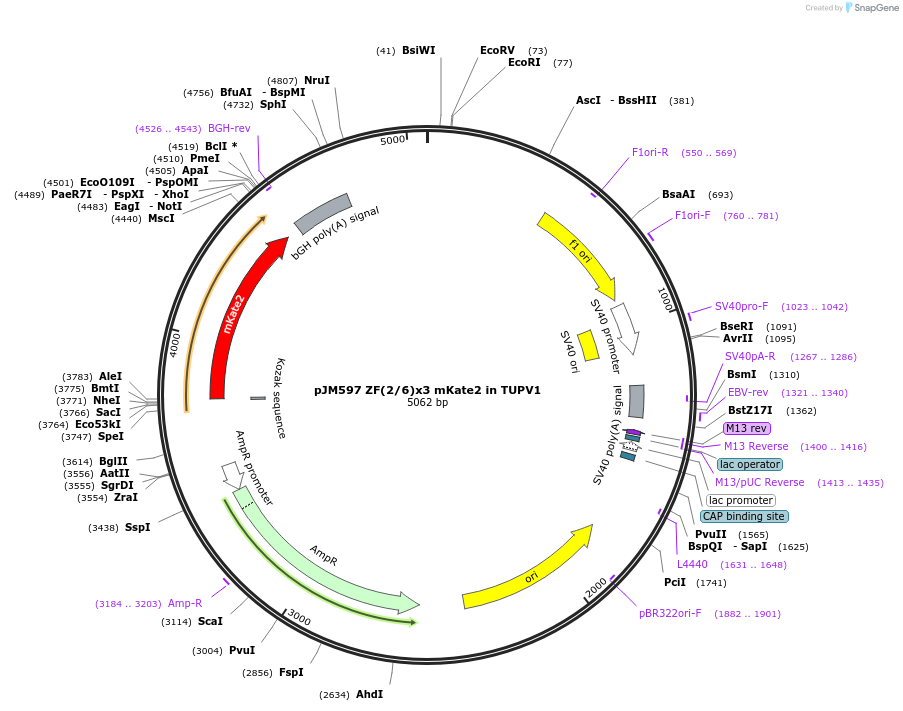
pJM597 ZF(2/6)x3 mKate2 in TUPV1
Plasmid#161531PurposeInducible expression of mKate2 under the ZF(2/6)x3 promoterDepositorInsertmKate2
UseSynthetic BiologyExpressionMammalianPromoterZF(2/6)x3Available SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
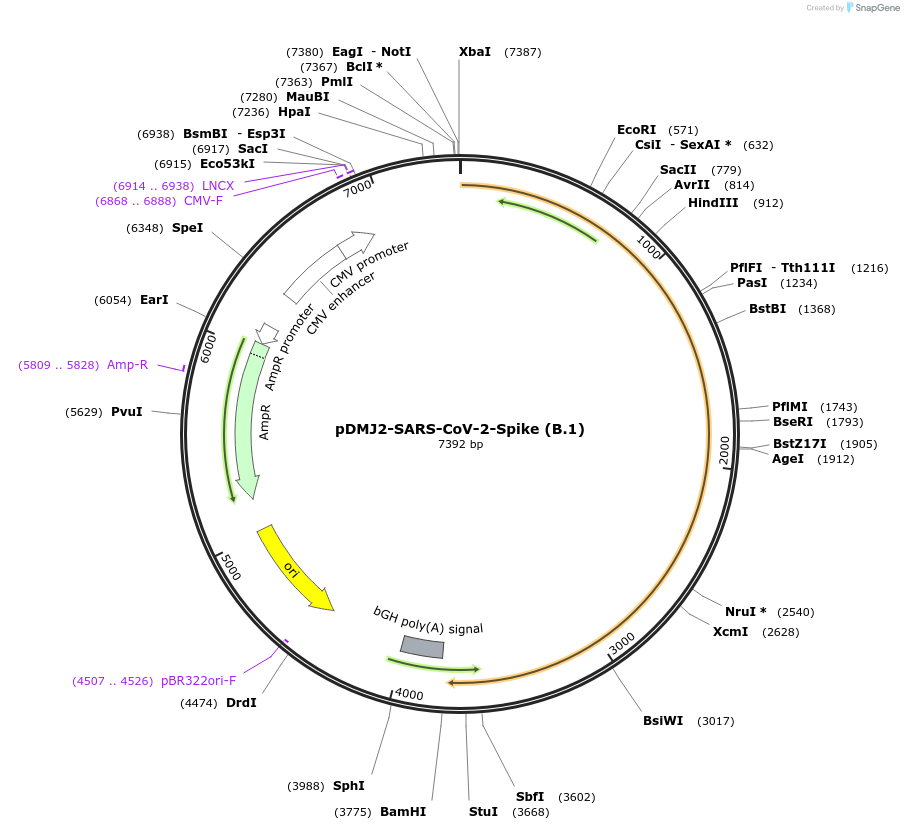
pDMJ2-SARS-CoV-2-Spike (B.1)
Plasmid#241033PurposeExpresses SARS-CoV-2 spike (B.1) protein for creating pseudotyped virusDepositorAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
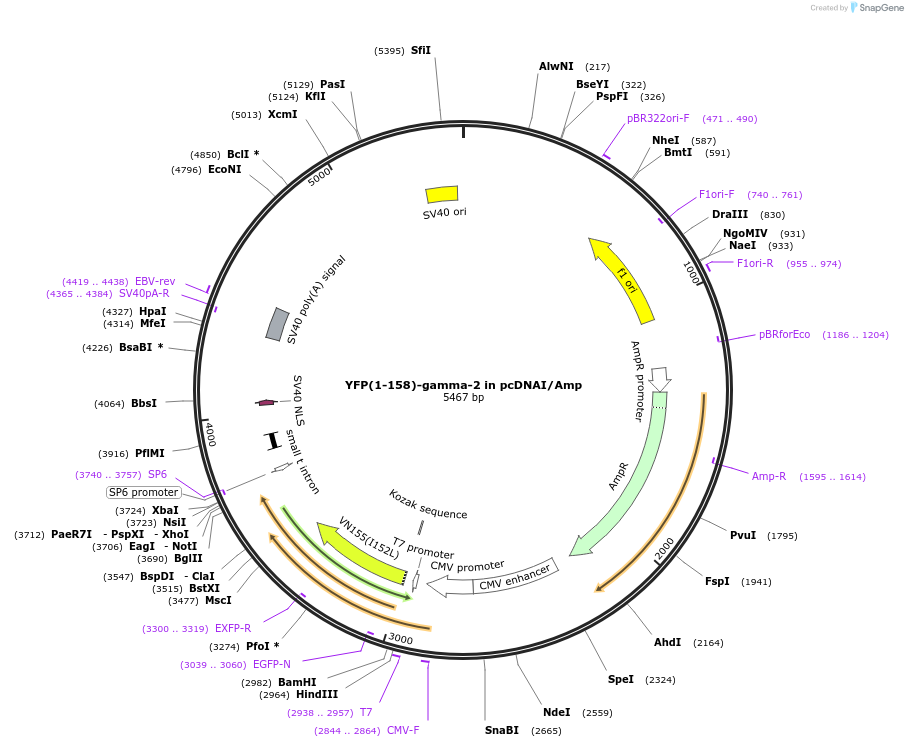
YFP(1-158)-gamma-2 in pcDNAI/Amp
Plasmid#55762PurposeAn amino-terminal YFP fragment was fused to Ggamma2. When co-expressed with a carboxyl terminal YFP or CFP fragment fused to a Gbeta subunit with which it interacts, a fluorescent signal is produced.DepositorInsertYFP(1-158)-gamma-2 (GNG2 Human, Aequorea victoria)
TagsYFP(1-158) was fused to the amino terminus of Gga…ExpressionMammalianMutationYFP (1-158) includes a substitution of Met for Gl…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
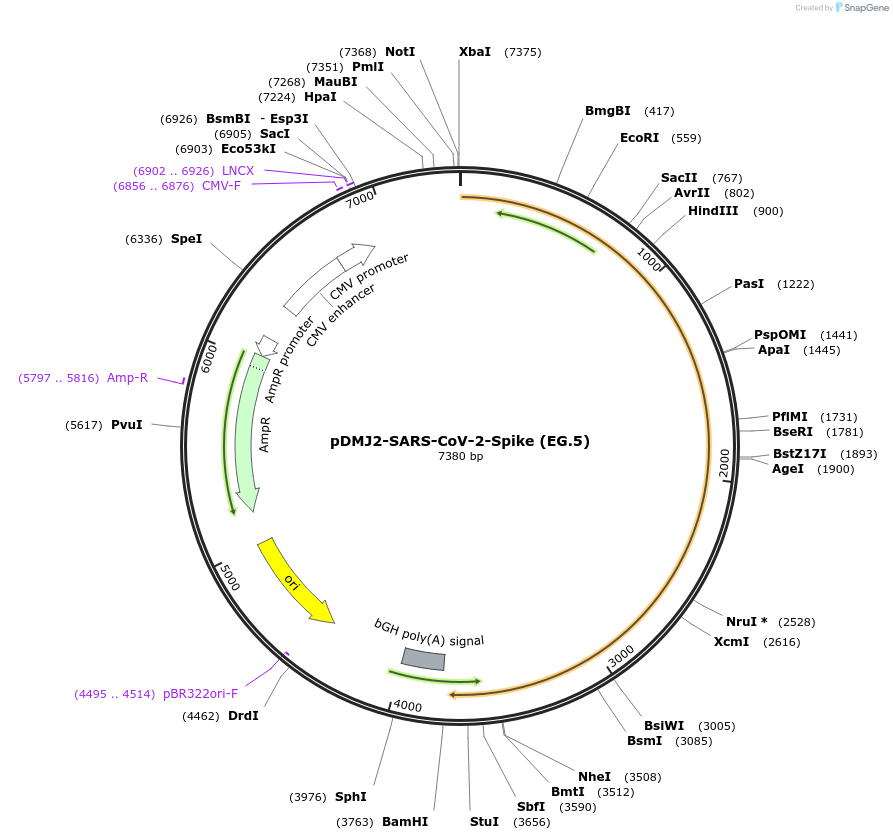
pDMJ2-SARS-CoV-2-Spike (EG.5)
Plasmid#241114PurposeExpresses SARS-CoV-2 spike (EG.5) protein for creating pseudotyped virusDepositorInsertSpike (EG.5) (S SARS-CoV-2)
ExpressionMammalianMutationT19I, L24del, P25del, P26del, A27S, V83A, G142D, …Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
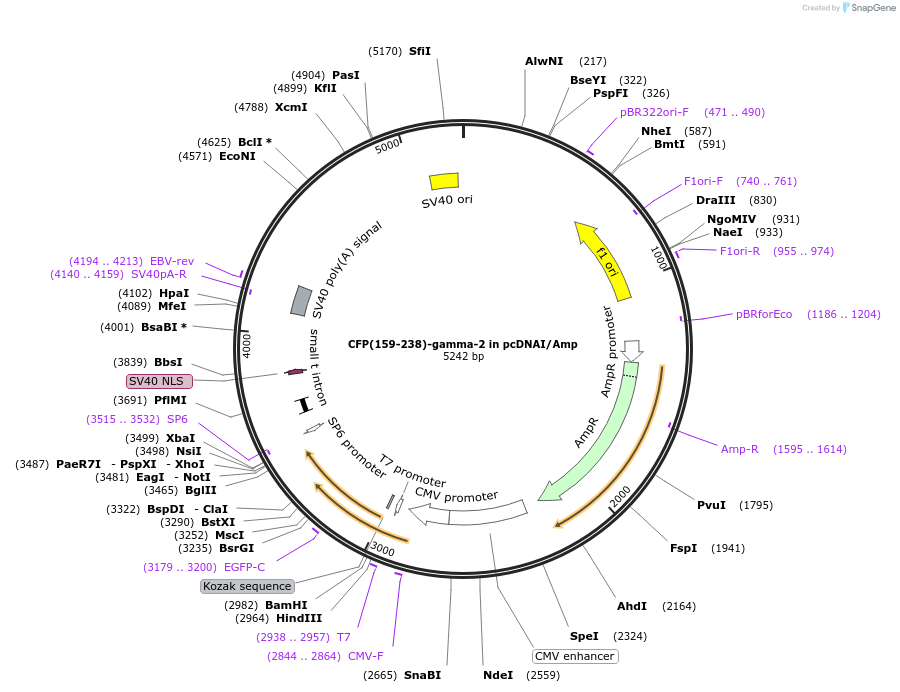
CFP(159-238)-gamma-2 in pcDNAI/Amp
Plasmid#55777PurposeA C-terminal CFP fragment was fused to Ggamma-2. When co-expressed with an N-terminal mCerulean, CFP, or YFP fragment fused to a Gbeta subunit with which it interacts a fluorescent signal is produced.DepositorInsertCFP(159-238)-gamma-2 (GNG2 Human, Aequorea victoria)
TagsCFP(159-238) was fused to the amino terminus of G…ExpressionMammalianMutationCFP(159-238) includes a substitution of His for A…PromoterCMVAvailable SinceJuly 30, 2015AvailabilityAcademic Institutions and Nonprofits only -
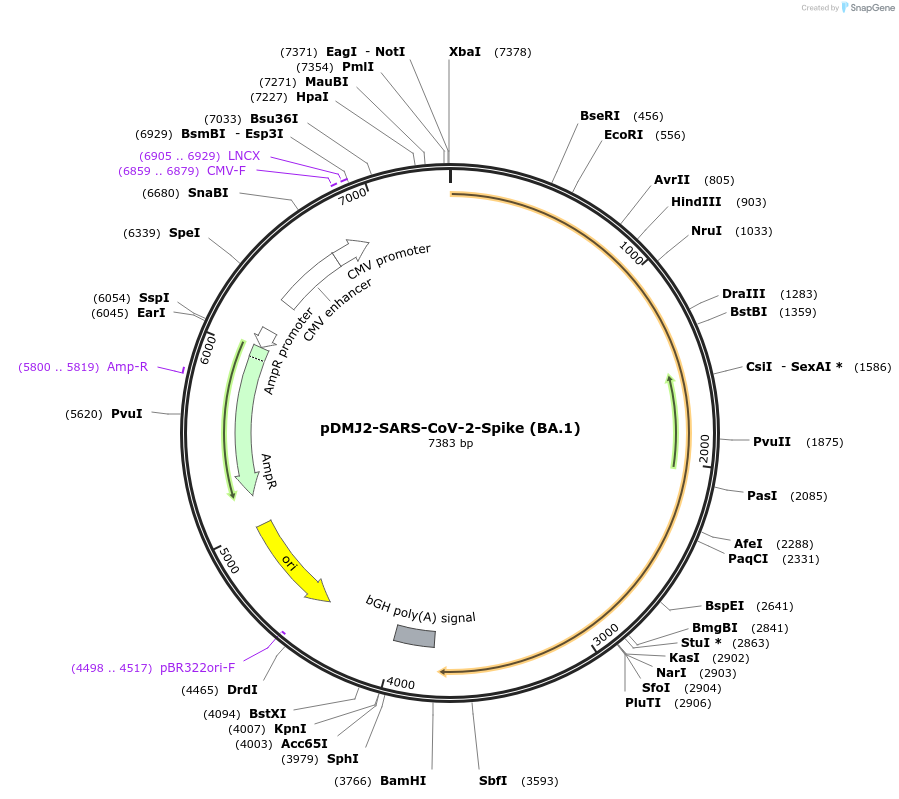
pDMJ2-SARS-CoV-2-Spike (BA.1)
Plasmid#241055PurposeExpresses SARS-CoV-2 spike (BA.1) protein for creating pseudotyped virusDepositorInsertSpike (BA.1) (S SARS-CoV-2)
ExpressionMammalianMutationA67V,H69del,V70del,T95I,G142D,V143del,Y144del,Y14…Available SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
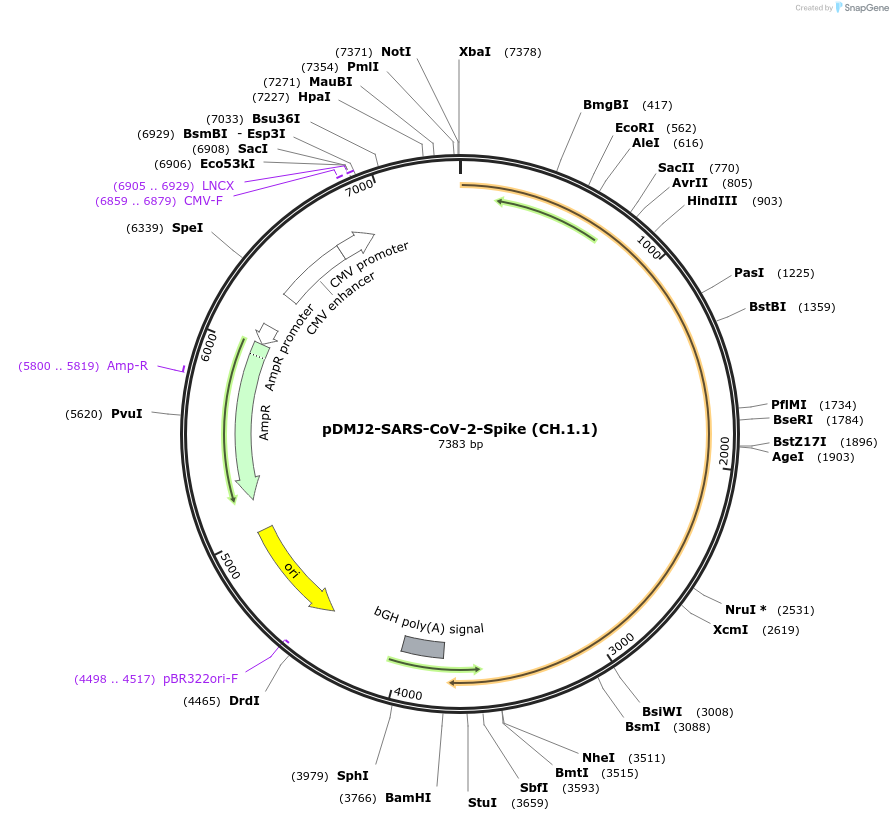
pDMJ2-SARS-CoV-2-Spike (CH.1.1)
Plasmid#241112PurposeExpresses SARS-CoV-2 spike (CH.1.1) protein for creating pseudotyped virusDepositorInsertSpike (CH.1.1) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,G142D,K147E,W152R,…Available SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
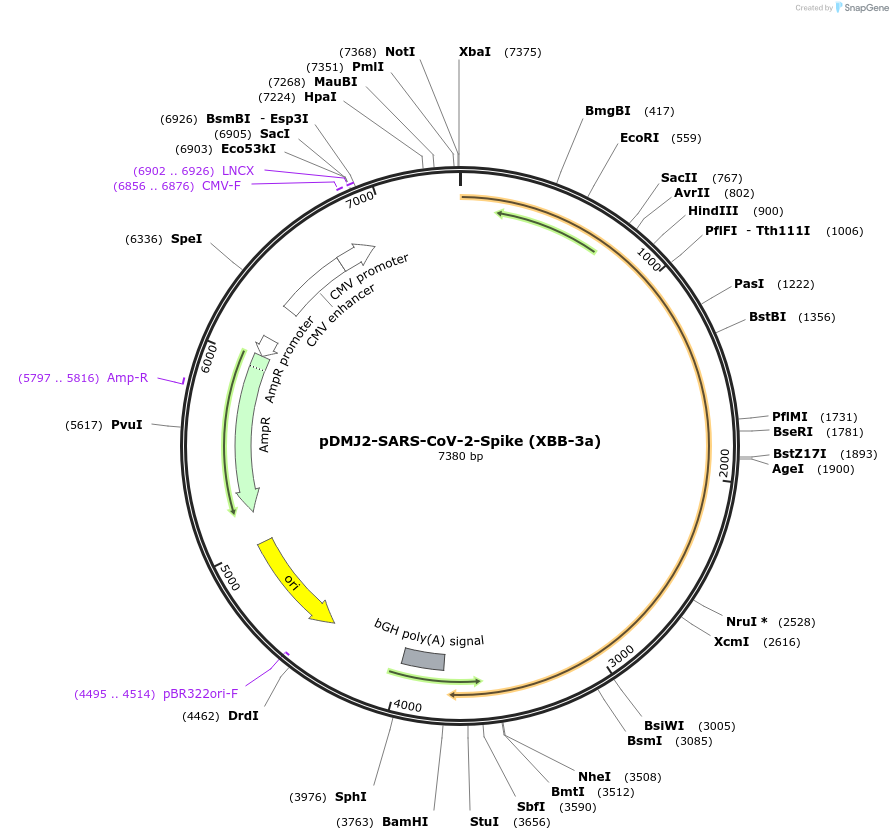
pDMJ2-SARS-CoV-2-Spike (XBB-3a)
Plasmid#241123PurposeExpresses SARS-CoV-2 spike (XBB-3a) protein for creating pseudotyped virusDepositorInsertSpike (XBB-3a) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,V83A,G142D,Y144del…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
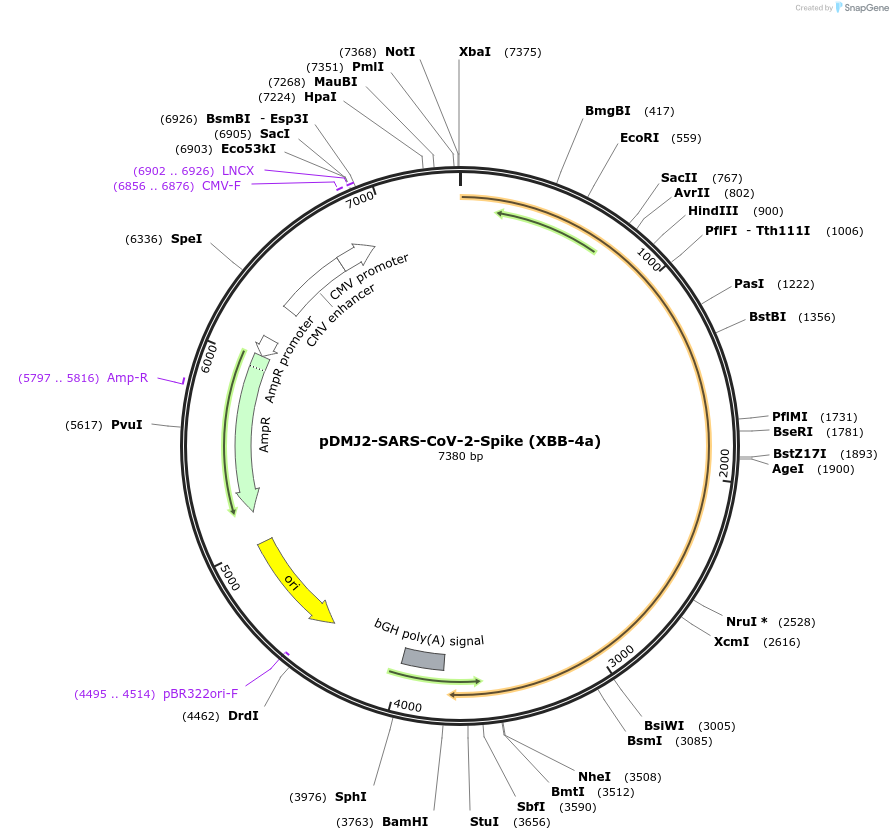
pDMJ2-SARS-CoV-2-Spike (XBB-4a)
Plasmid#241124PurposeExpresses SARS-CoV-2 spike (XBB-4a) protein for creating pseudotyped virusDepositorInsertSpike (XBB-4a) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,V83A,G142D,Y144del…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
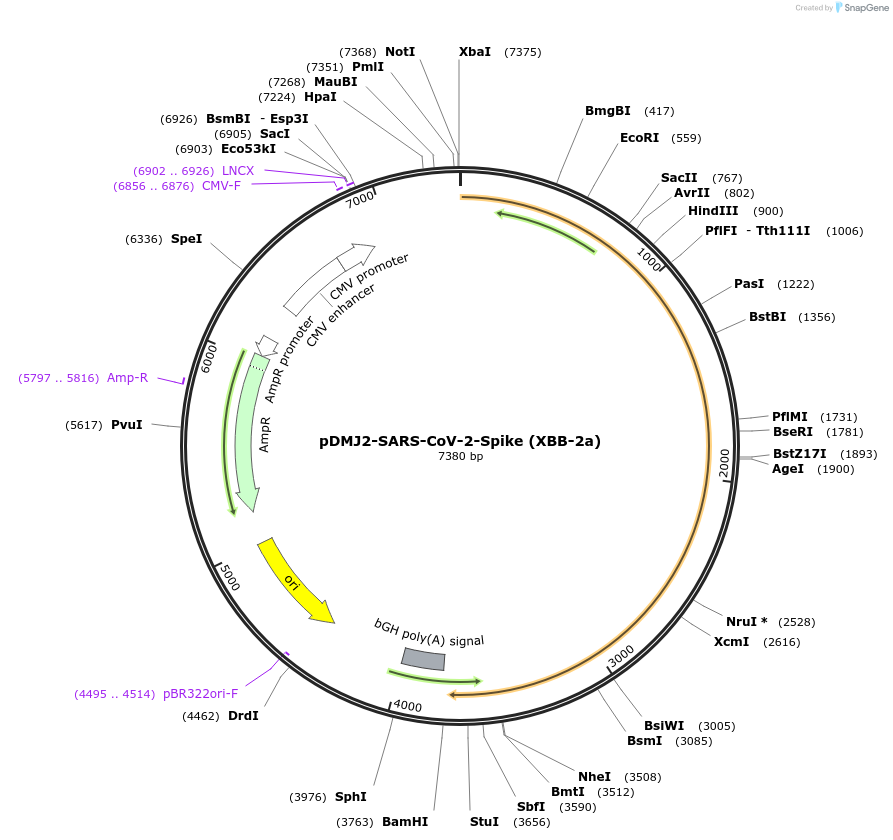
pDMJ2-SARS-CoV-2-Spike (XBB-2a)
Plasmid#241122PurposeExpresses SARS-CoV-2 spike (XBB-2a) protein for creating pseudotyped virusDepositorInsertSpike (XBB-2a) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,V83A,G142D,Y144del…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
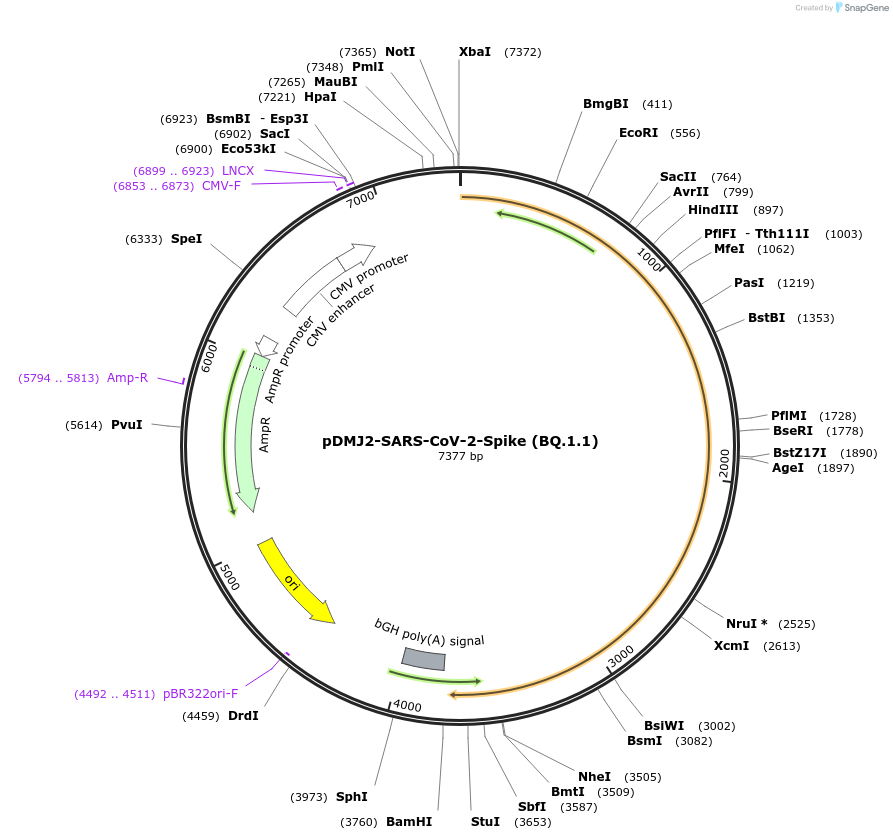
pDMJ2-SARS-CoV-2-Spike (BQ.1.1)
Plasmid#241111PurposeExpresses SARS-CoV-2 spike (BQ.1.1) protein for creating pseudotyped virusDepositorInsertSpike (BQ.1.1) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,H69del,V70del,G142…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
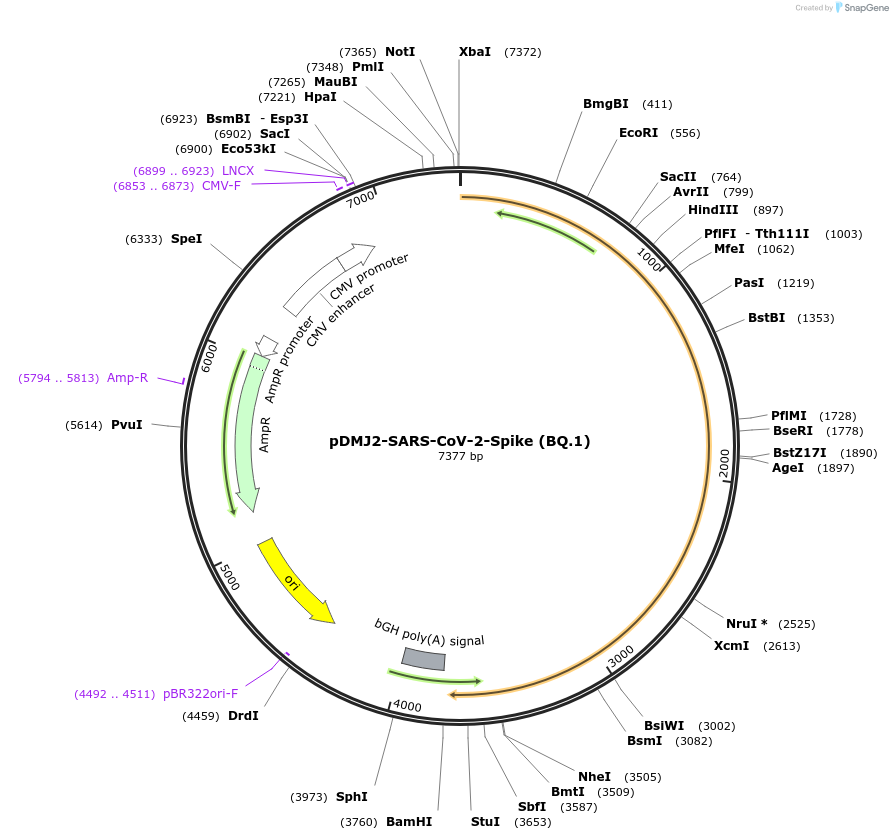
pDMJ2-SARS-CoV-2-Spike (BQ.1)
Plasmid#241110PurposeExpresses SARS-CoV-2 spike (BQ.1) protein for creating pseudotyped virusDepositorInsertSpike (BQ.1) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,H69del,V70del,G142…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
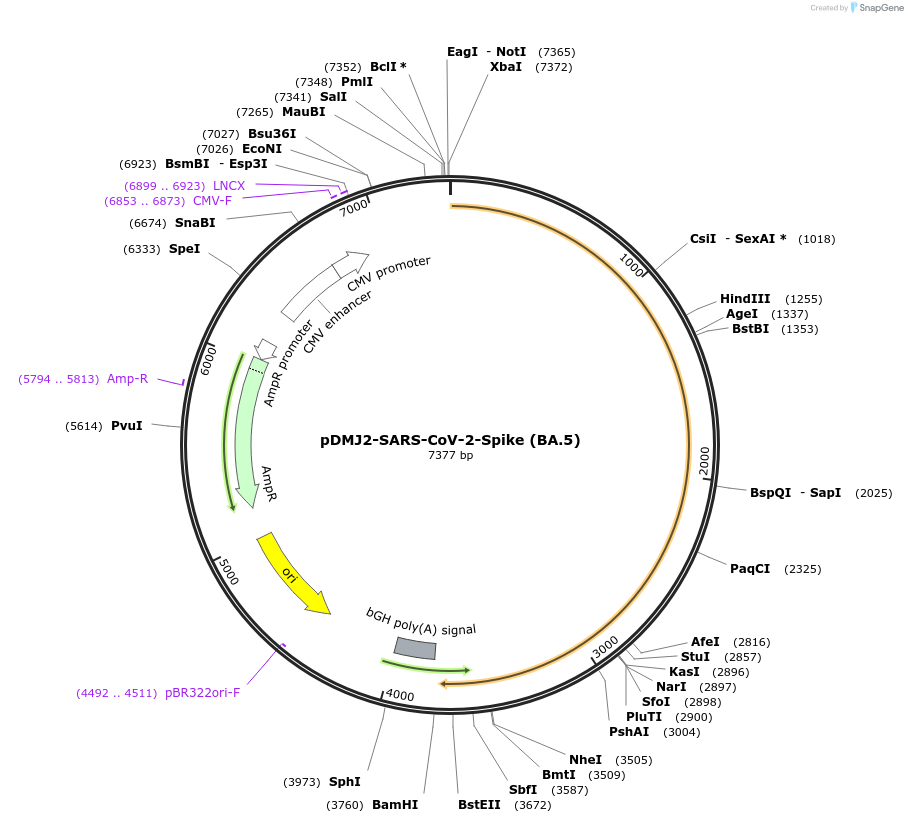
pDMJ2-SARS-CoV-2-Spike (BA.5)
Plasmid#241091PurposeExpresses SARS-CoV-2 spike (BA.5) protein for creating pseudotyped virusDepositorInsertSpike (BA.5) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,H69del,V70del,G142…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
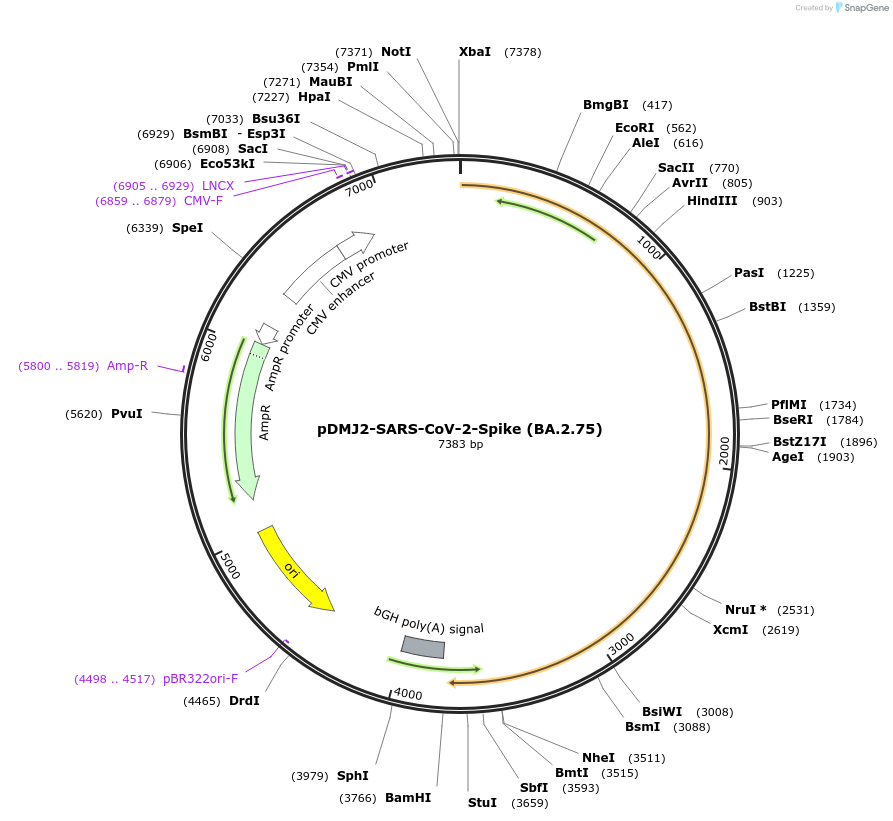
pDMJ2-SARS-CoV-2-Spike (BA.2.75)
Plasmid#241071PurposeExpresses SARS-CoV-2 spike (BA.2.75) protein for creating pseudotyped virusDepositorInsertSpike (BA.2.75) (S SARS-CoV-2)
ExpressionMammalianMutationT19I,L24del,P25del,P26del,A27S,G142D,K147E,W152R,…Available SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only