8,173 results
-
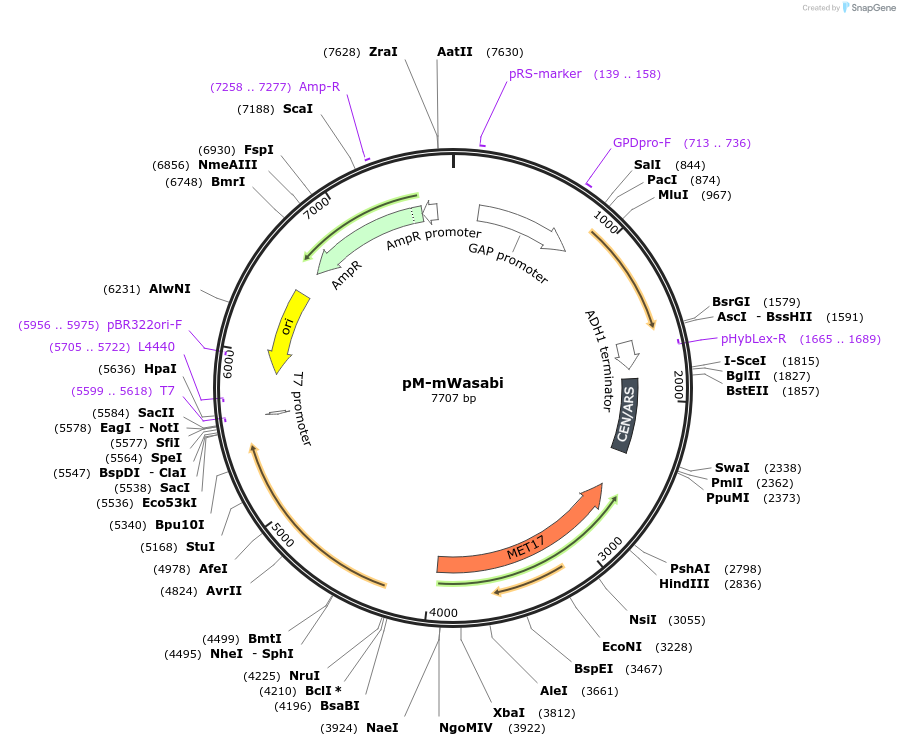
Plasmid#176554PurposeExpression of mWasabi for auxotrophic selection in the absence of methionineDepositorInsertmWasabi
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
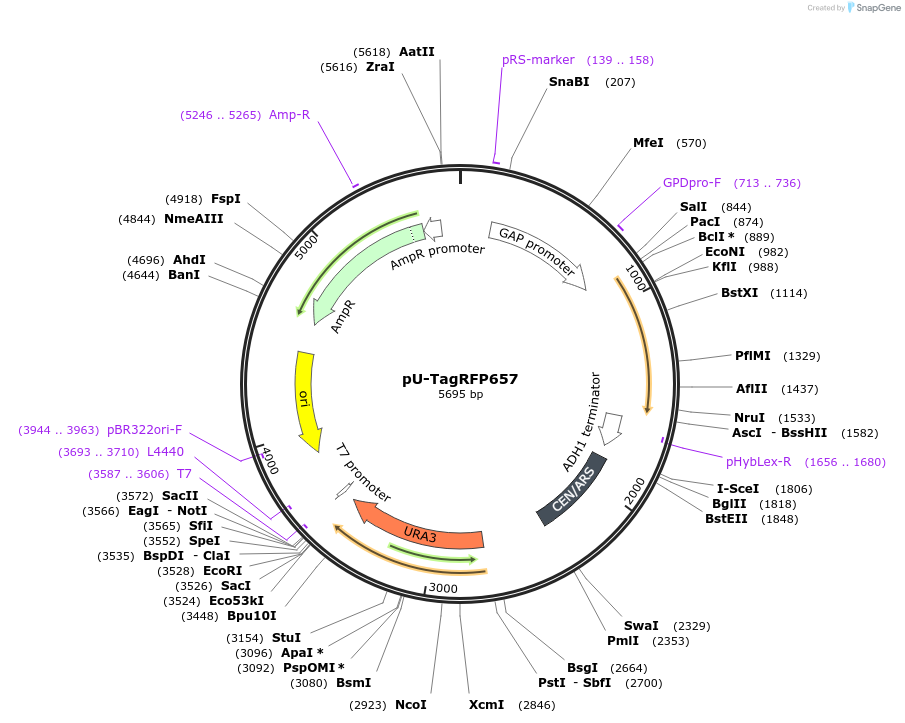
pU-TagRFP657
Plasmid#176562PurposeExpression of TagRFP657 for auxotrophic selection in the absence of uracilDepositorInsertTagRFP657
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
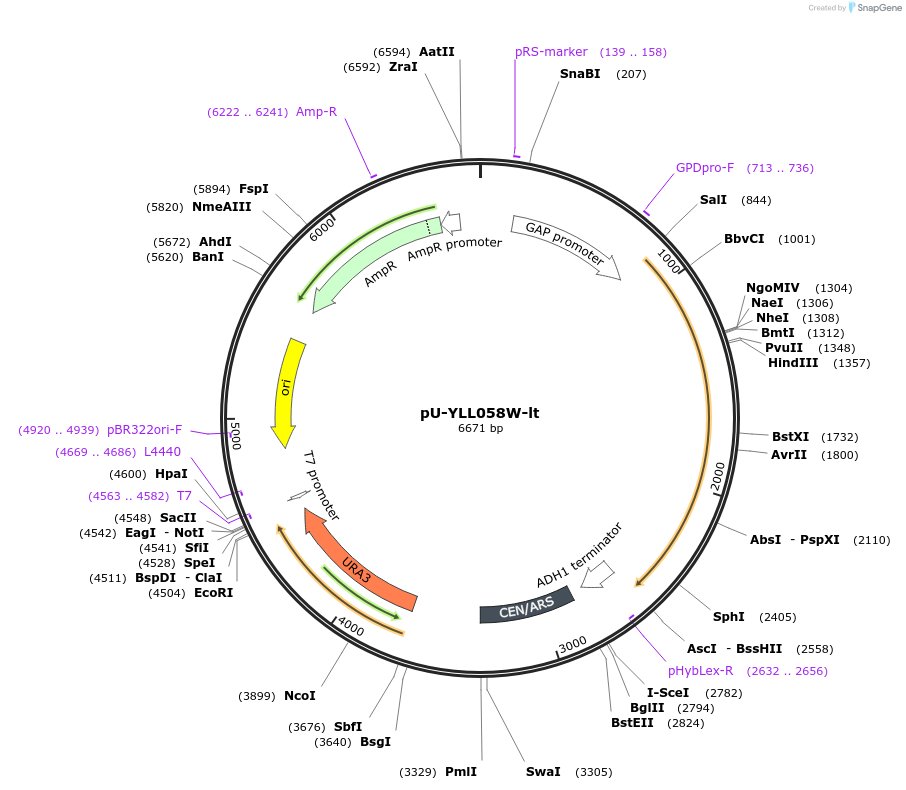
pU-YLL058W-lt
Plasmid#176564PurposeExpression of YLL058W for Lanchea thermotolerans for auxotrophic selection in the absence of uracilDepositorInsertYLL058W-lt
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
pU-YLL058W-zr
Plasmid#176565PurposeExpression of YLL058W from Zygosaccharomyces rouxii for auxotrophic selection in the absence of uracilDepositorInsertYLL058W-zr
ExpressionYeastMutationCodon optimised for expression in S.cerevisaePromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
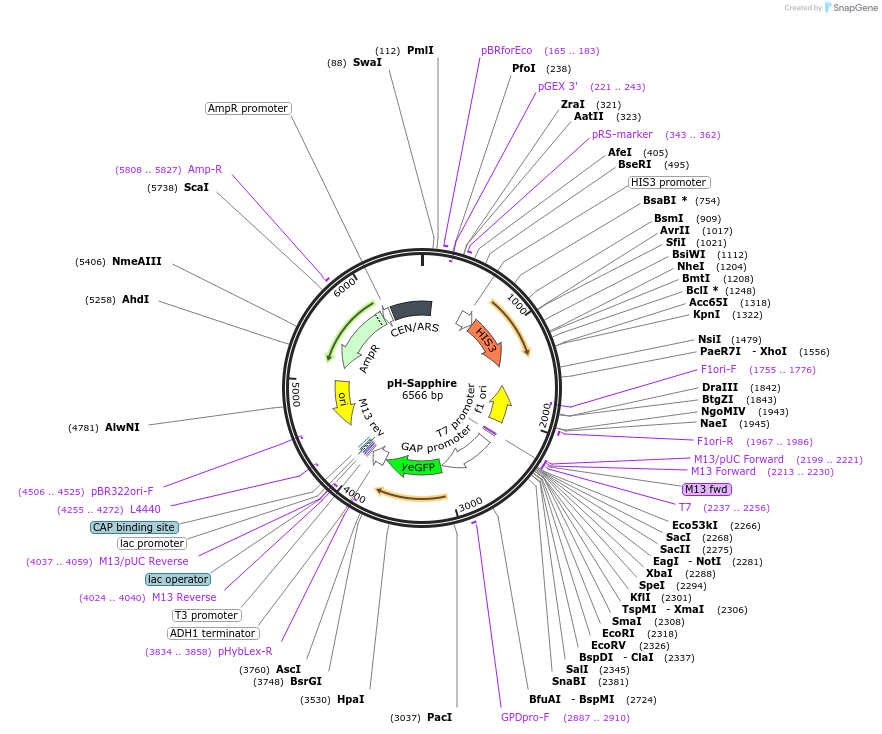
pH-Sapphire
Plasmid#176566PurposeExpression of Sapphire for auxotrophic selection in the absence of lysineDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
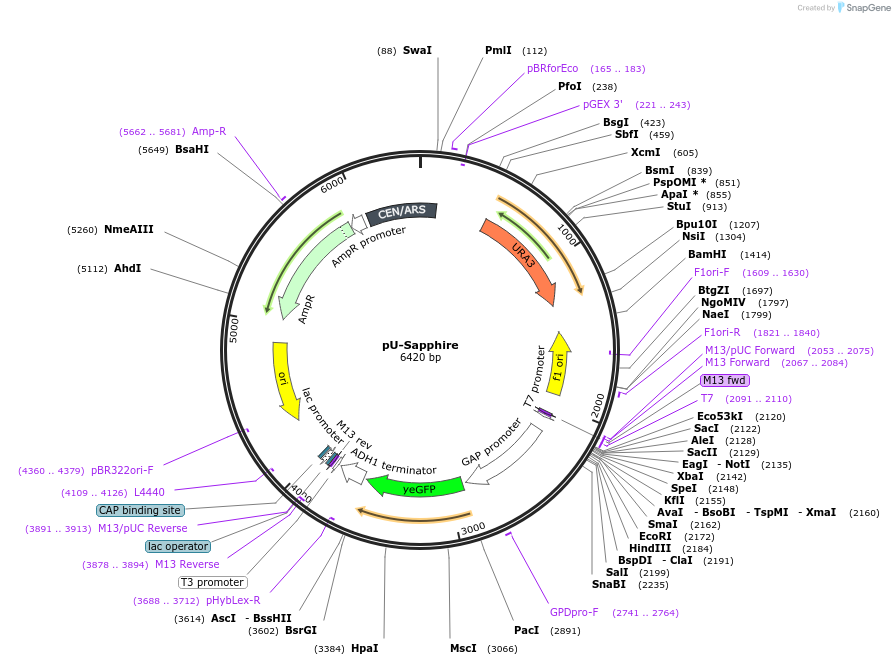
pU-Sapphire
Plasmid#176567PurposeExpression of Sapphire for auxotrophic selection in the absence of uracilDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
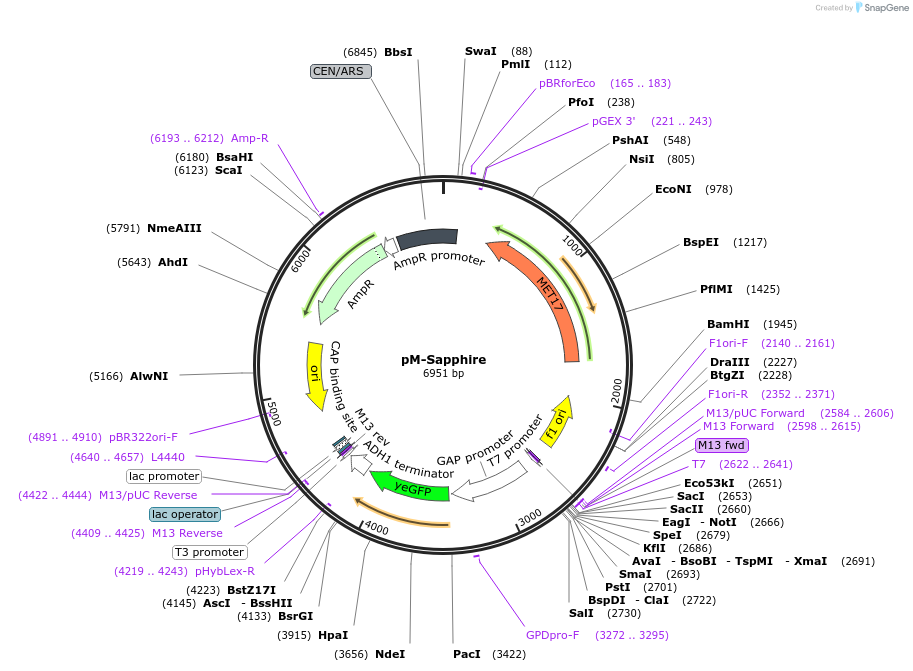
pM-Sapphire
Plasmid#176569PurposeExpression of Sapphire for auxotrophic selection in the absence of methionineDepositorInsertyETSapphire
ExpressionYeastPromoterTDH1(GAP)Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
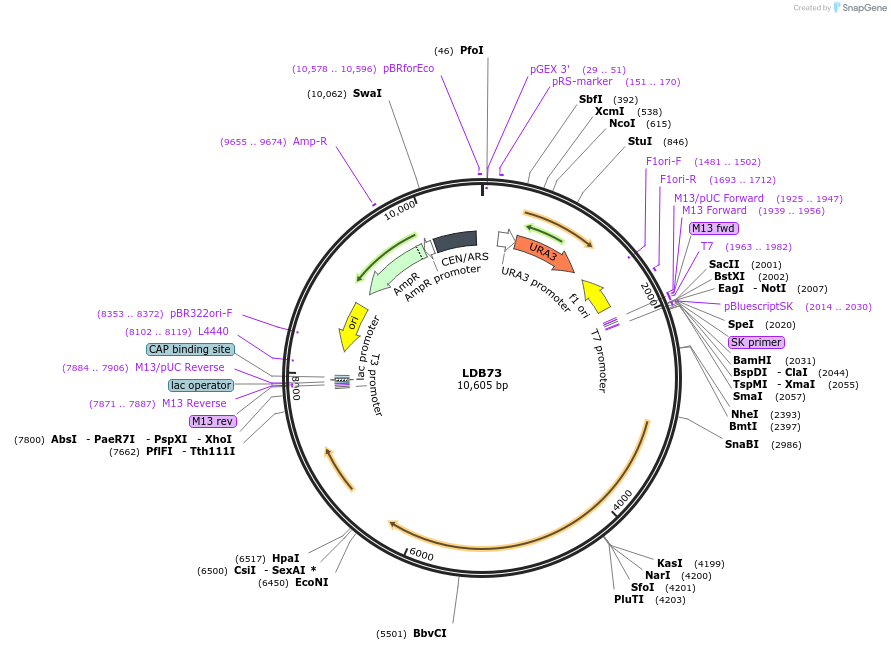
LDB73
Plasmid#185437PurposeNUP1 CEN URA3 vector for synthetic lethal sectoring assays and screeningDepositorInsertNUP1
ExpressionYeastAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
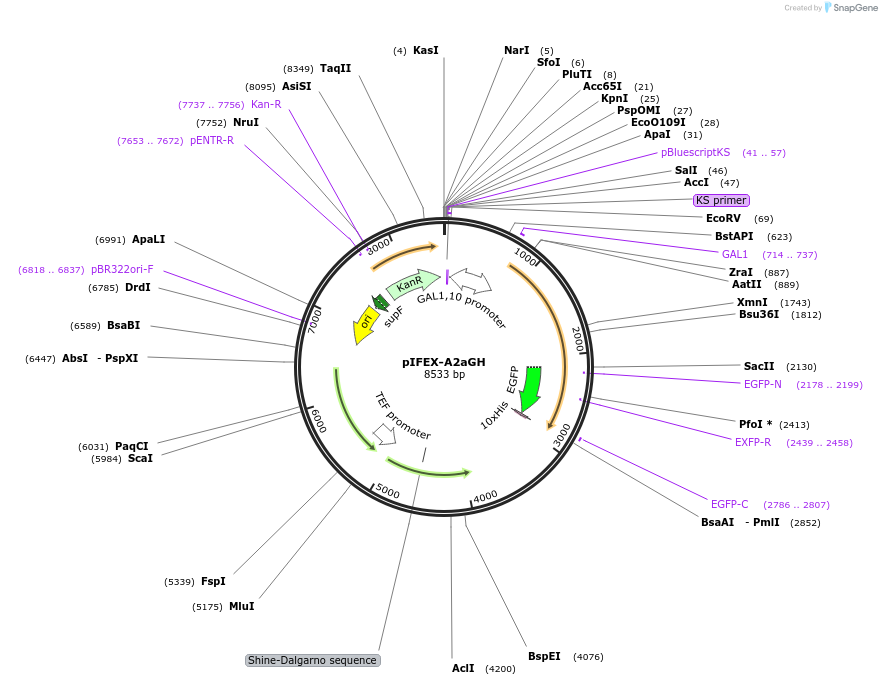
pIFEX-A2aGH
Plasmid#160542PurposepITY4 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
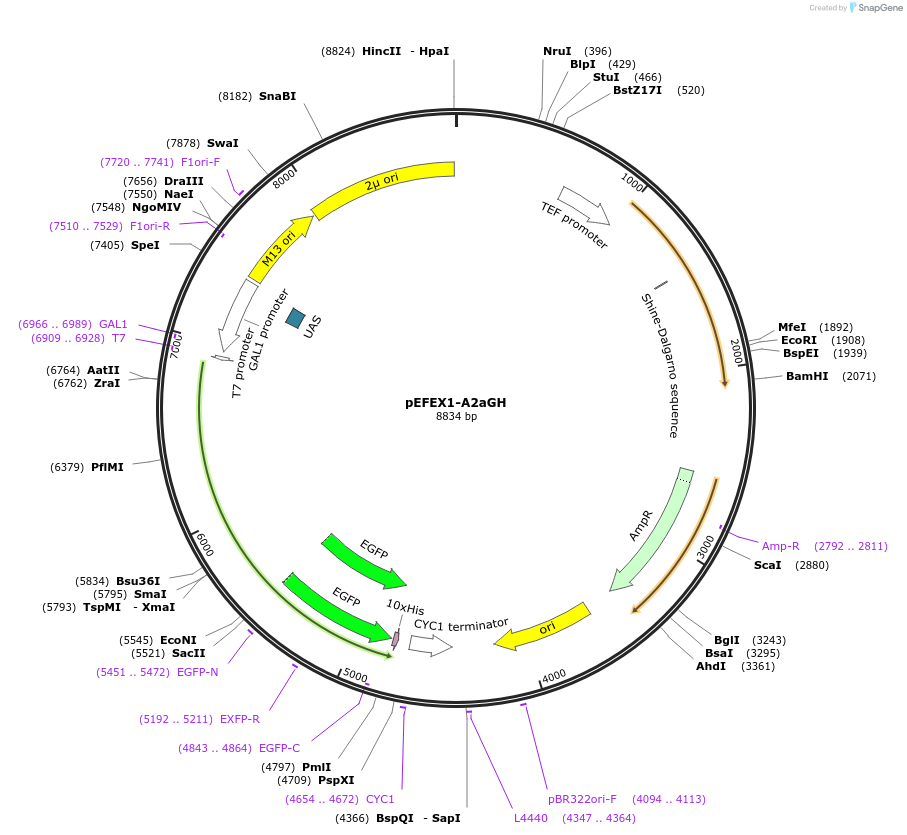
pEFEX1-A2aGH
Plasmid#160543PurposepYES2 derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
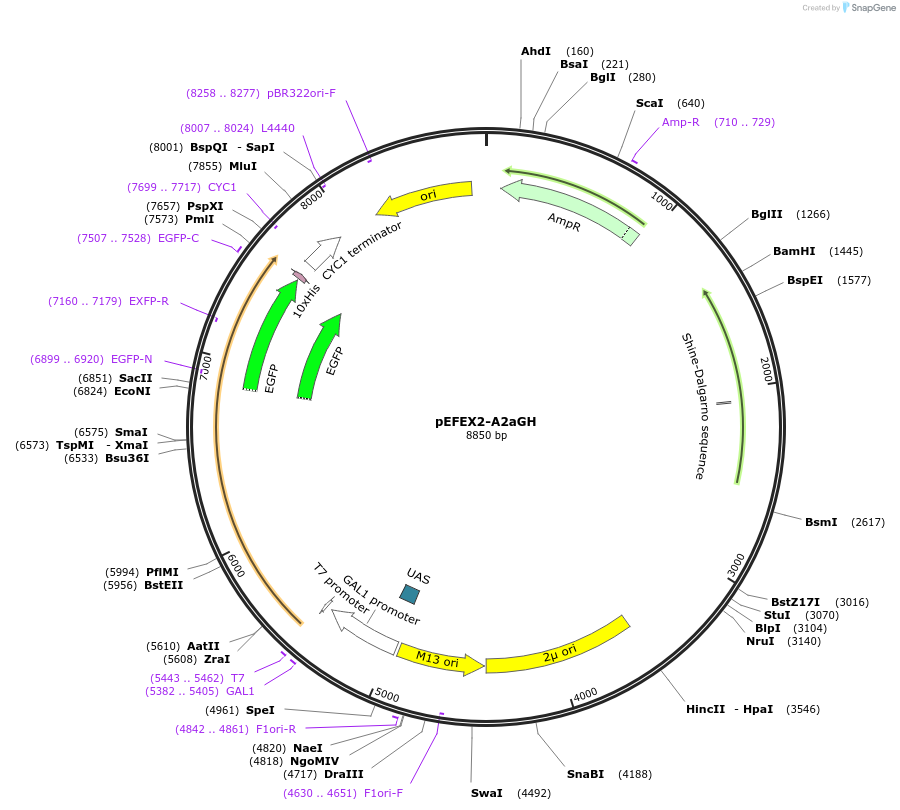
pEFEX2-A2aGH
Plasmid#160544PurposepYES2 derivative reporter plasmid encoding PPGI1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
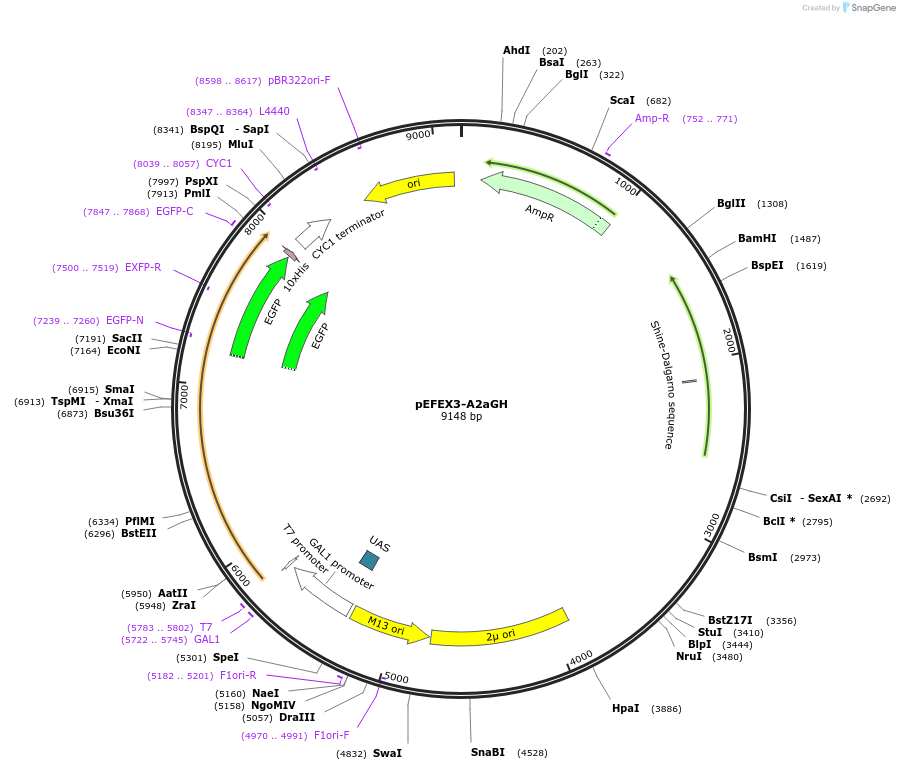
pEFEX3-A2aGH
Plasmid#160545PurposepYES2 derivative reporter plasmid encoding PREV1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
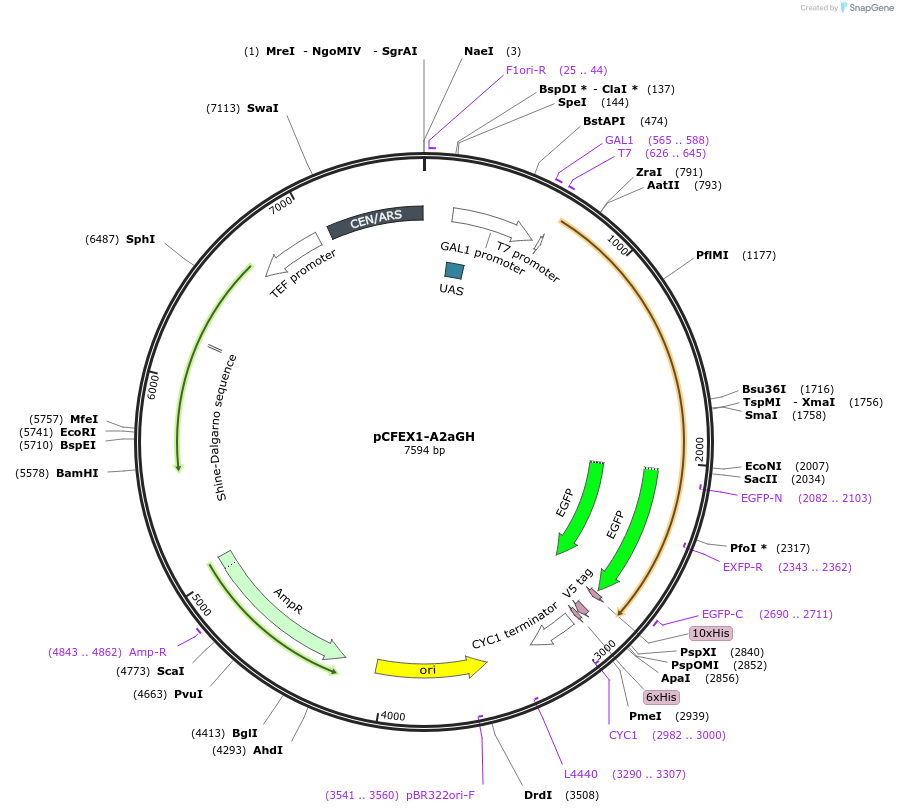
pCFEX1-A2aGH
Plasmid#160546PurposepYC2/CT derivative reporter plasmid encoding Ashbya gossypii PTEF1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
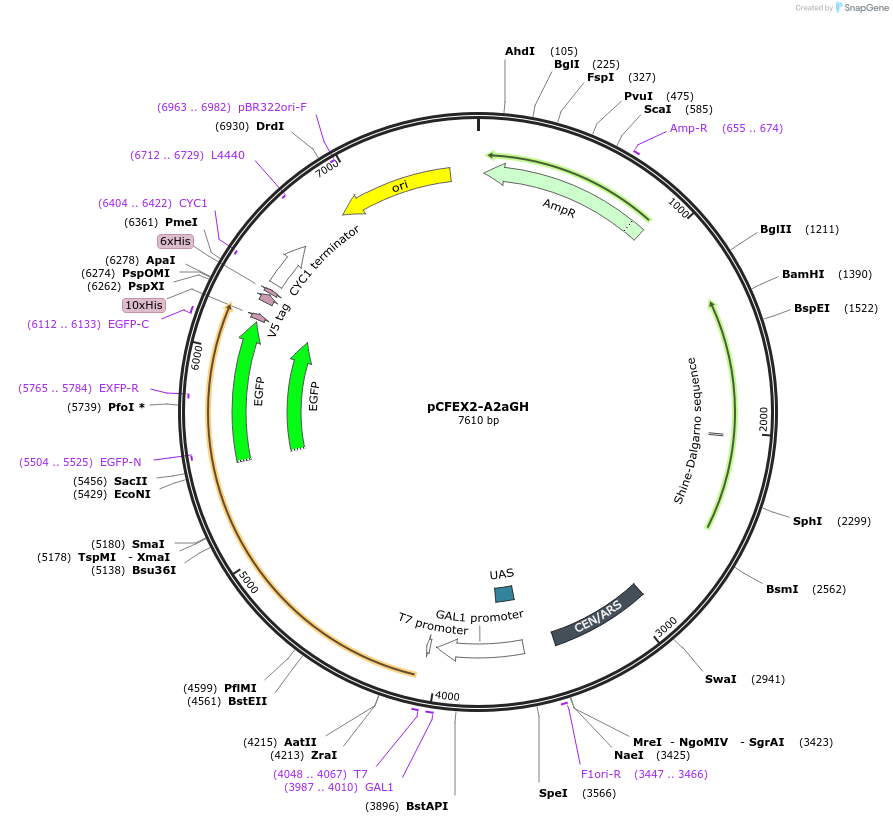
pCFEX2-A2aGH
Plasmid#160547PurposepYC2/CT derivative reporter plasmid encoding PPGI1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
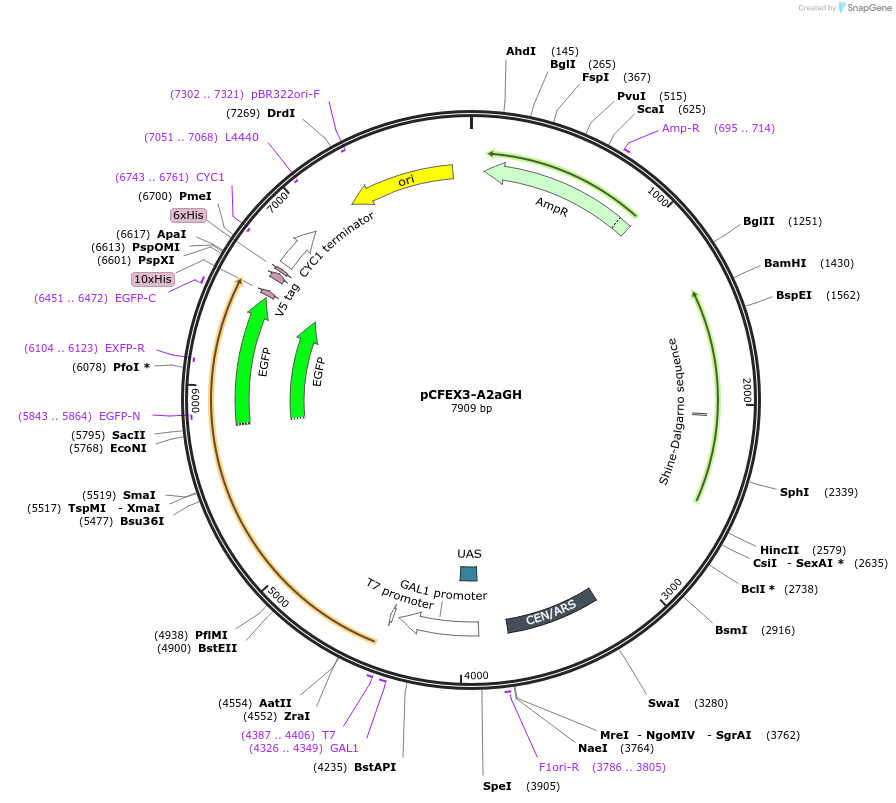
pCFEX3-A2aGH
Plasmid#160548PurposepYC2/CT derivative reporter plasmid encoding PREV1-Saccharomyces cerevisiae FEX1-MFaT; as well as PGAL1-Homo sapiens ADORA2A-EGFP-10HIS-MFaT.DepositorInsertExpressionYeastMutationN/AAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
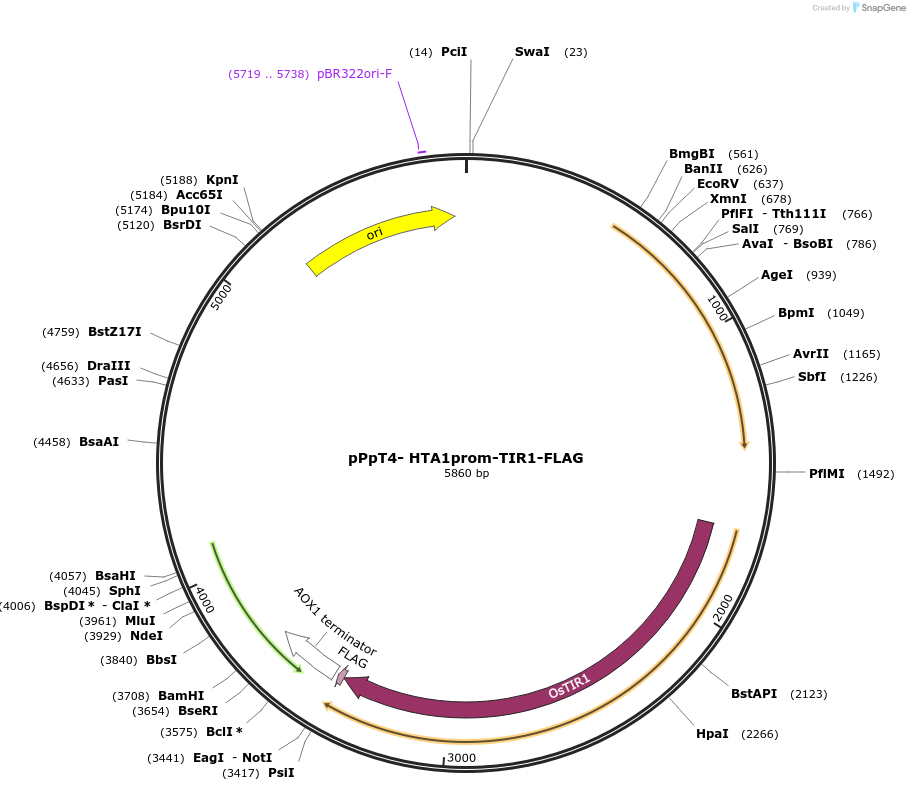
pPpT4- HTA1prom-TIR1-FLAG
Plasmid#189725PurposeEnables expression of OsTIR1 from the HTA1 promoter, used to implement the AID system in Komagataella phaffii (original name pLLpLL001)DepositorInsertOsTIR1
TagsFLAGExpressionYeastPromoterHTA1Available SinceSept. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
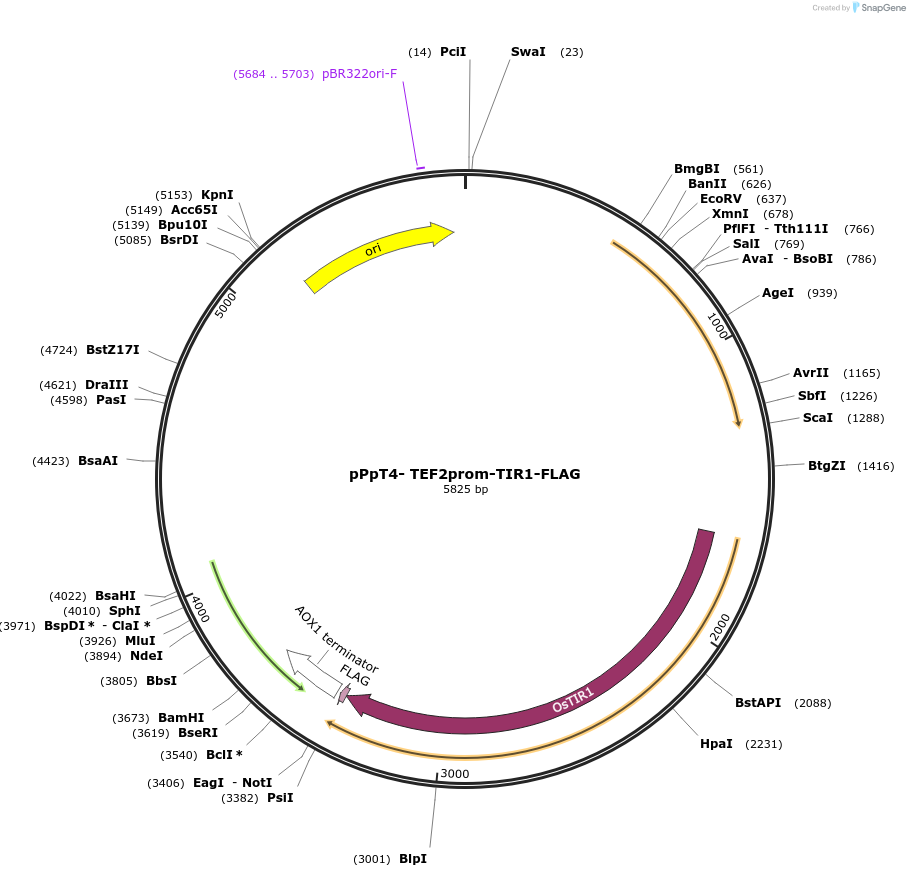
pPpT4- TEF2prom-TIR1-FLAG
Plasmid#189726PurposeEnables expression of OsTIR1 from the TEF2 promoter, used to implement the AID system in Komagataella phaffii (original name pLL002)DepositorInsertOsTIR1
TagsFLAGExpressionYeastPromoterTEF2Available SinceSept. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
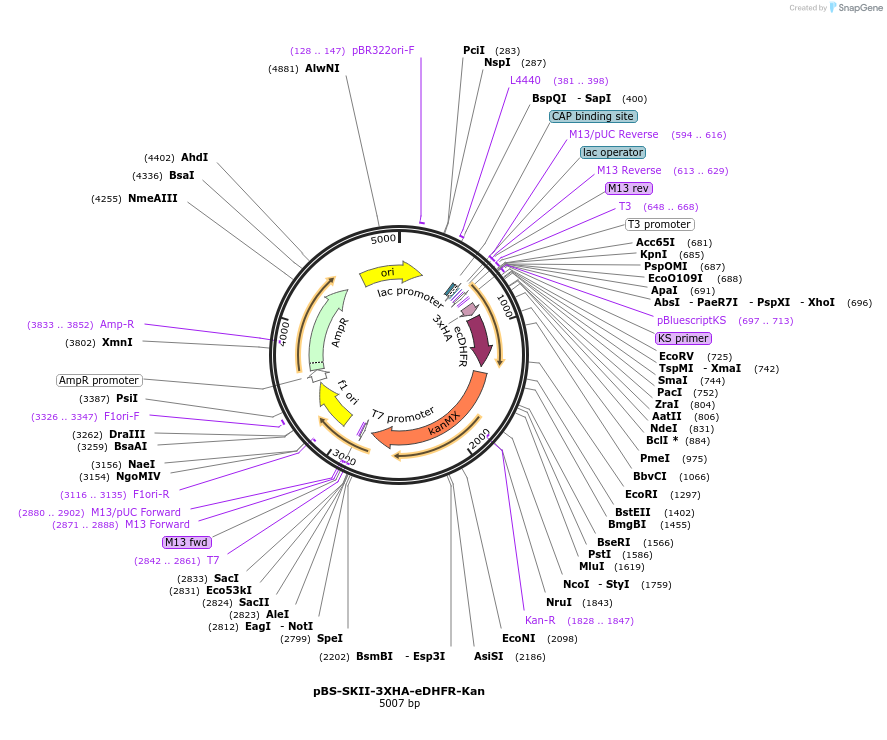
pBS-SKII-3XHA-eDHFR-Kan
Plasmid#188928PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-eDHFR-Kan
ExpressionYeastAvailable SinceSept. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
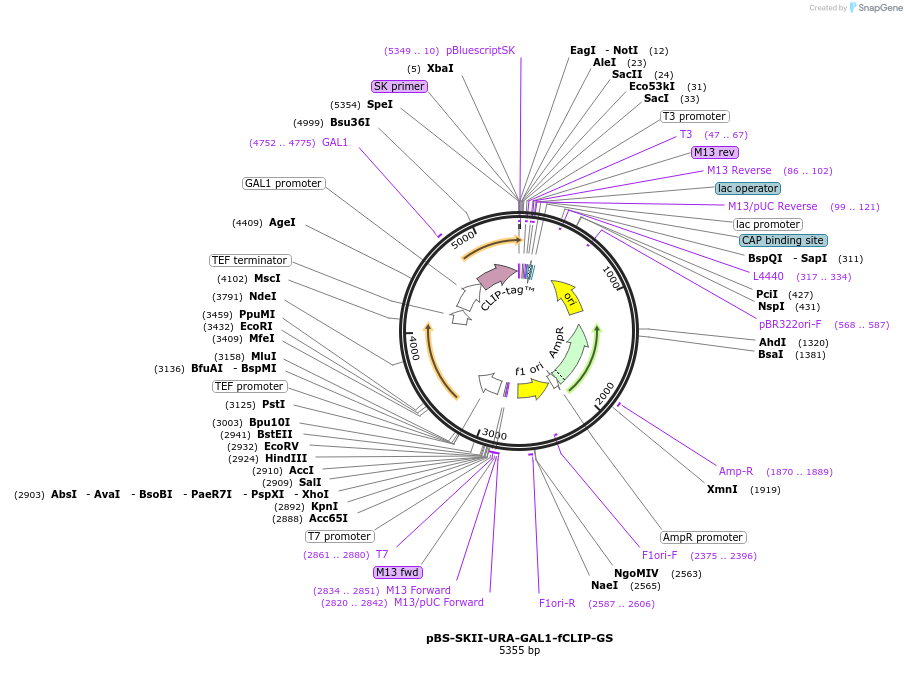
pBS-SKII-URA-GAL1-fCLIP-GS
Plasmid#188939PurposeIntegration cassette to create N-terminal fusions for fluorescent taggingDepositorInsertURA-GAL1-fCLIP-GS
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
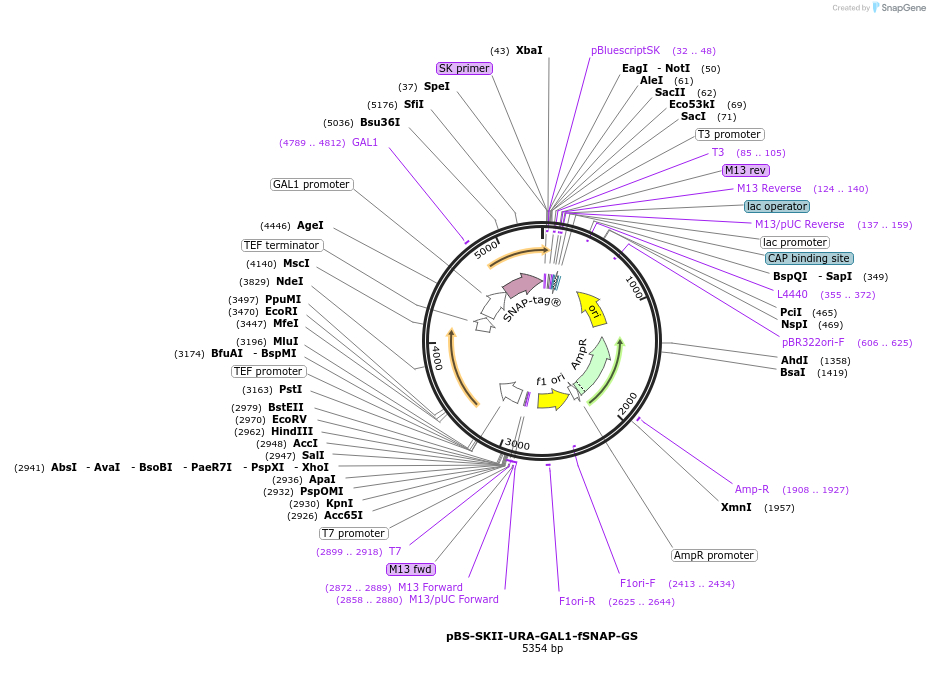
pBS-SKII-URA-GAL1-fSNAP-GS
Plasmid#188938PurposeIntegration cassette to create N-terminal fusions for fluorescent taggingDepositorInsertURA-GAL1-fSNAP-GS
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
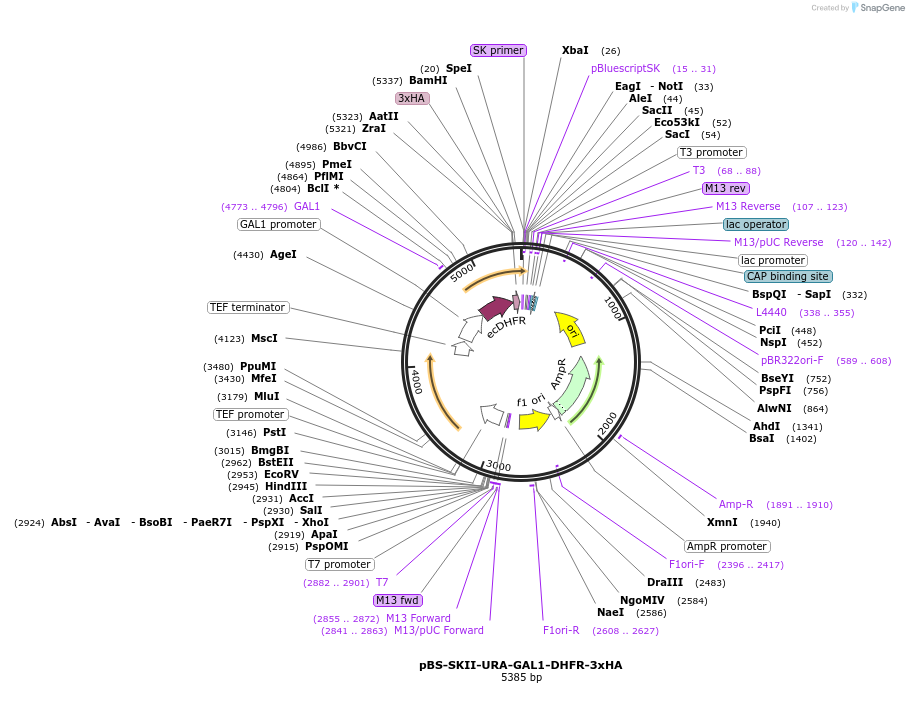
pBS-SKII-URA-GAL1-DHFR-3xHA
Plasmid#188934PurposeIntegration cassette to create N-terminal fusions for fluorescent taggingDepositorInsertURA-GAL1-DHFR-3xHA
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
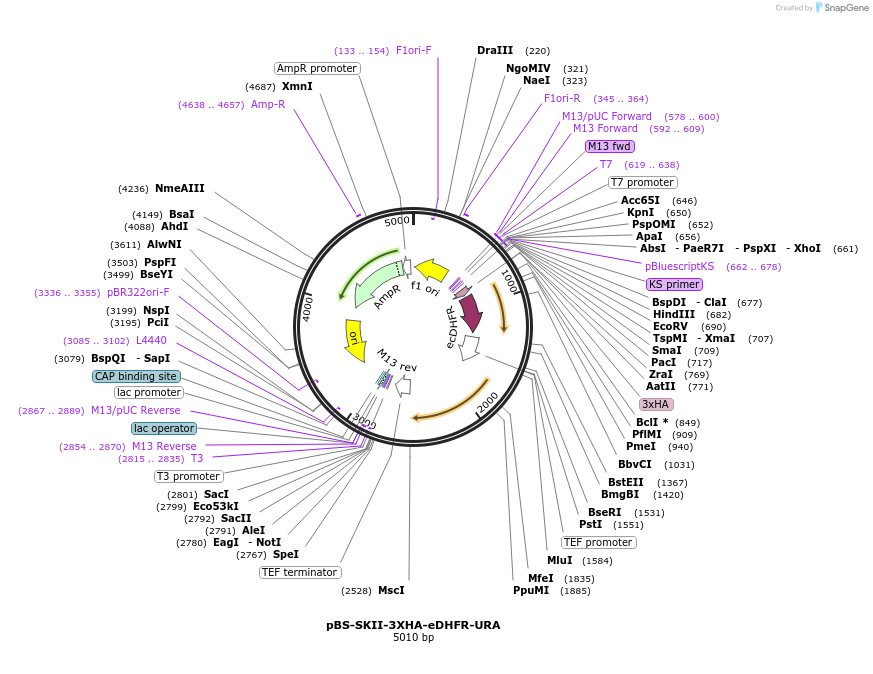
pBS-SKII-3XHA-eDHFR-URA
Plasmid#188927PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-eDHFR-URA
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
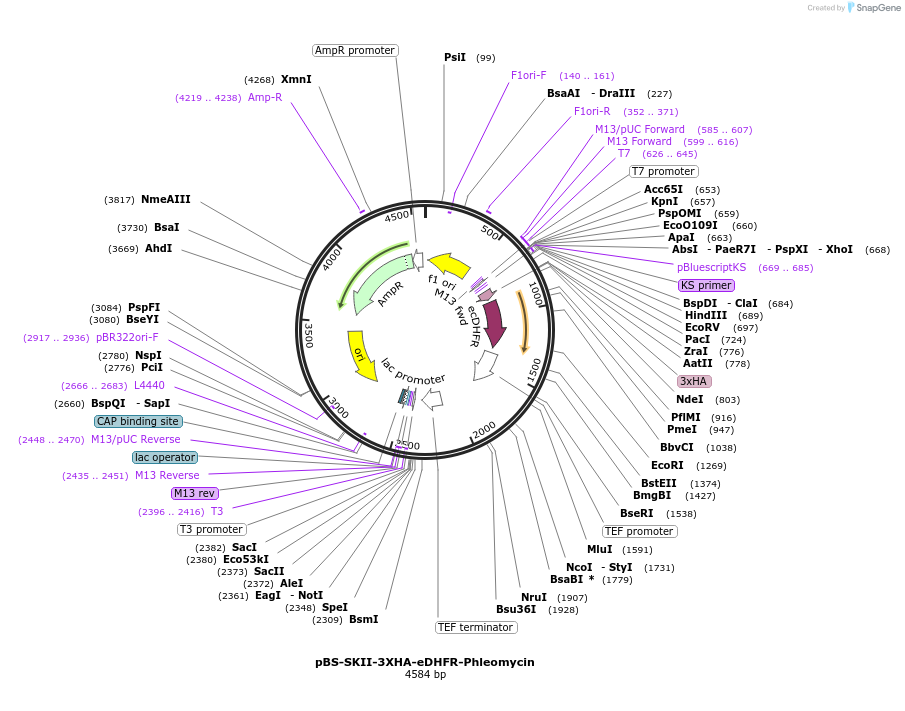
pBS-SKII-3XHA-eDHFR-Phleomycin
Plasmid#188925PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-eDHFR-Phleomycin
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
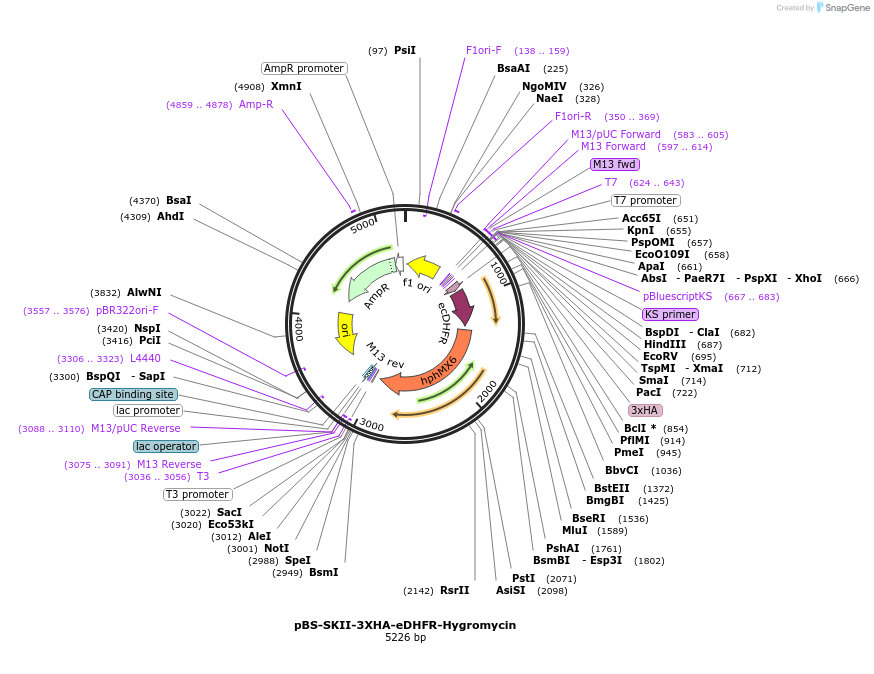
pBS-SKII-3XHA-eDHFR-Hygromycin
Plasmid#188924PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-eDHFR-Hygromycin
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
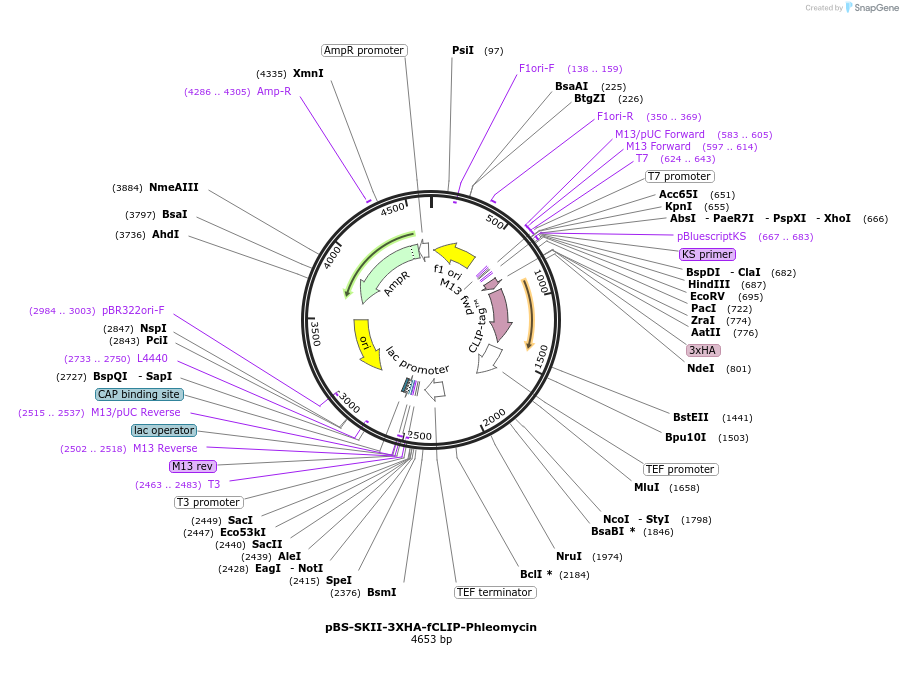
pBS-SKII-3XHA-fCLIP-Phleomycin
Plasmid#188920PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-fCLIP-Phleomycin
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
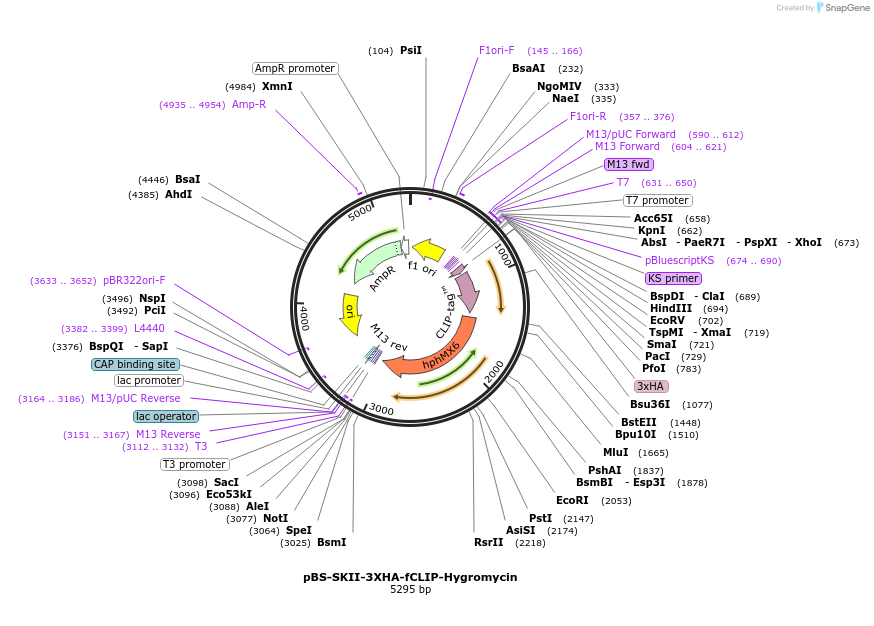
pBS-SKII-3XHA-fCLIP-Hygromycin
Plasmid#188919PurposeIntegration cassette to create C-terminal fusions for fluorescent taggingDepositorInsert3XHA-fCLIP-Hygromycin
ExpressionYeastAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
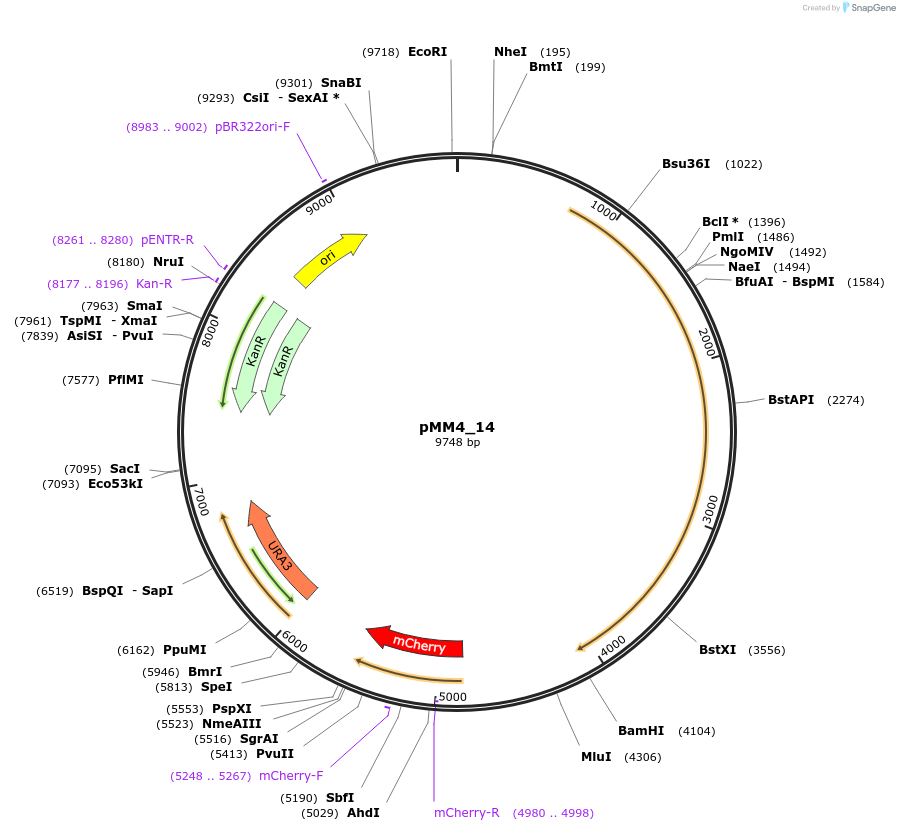
pMM4_14
Plasmid#183062PurposePlasmid expressing a biosensor for acetic acid: BM3R1-Haa1-mTurquoise2 under RET2p and mCherry under HREsYGP1p with flanks for genome integration into HO locusDepositorInsertsmCherry
BM3R1-HAA1-mTurquoise2
UseSynthetic BiologyTagsmCherry and mTurquoise2ExpressionYeastPromoterHREsYGP1p and RET2Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
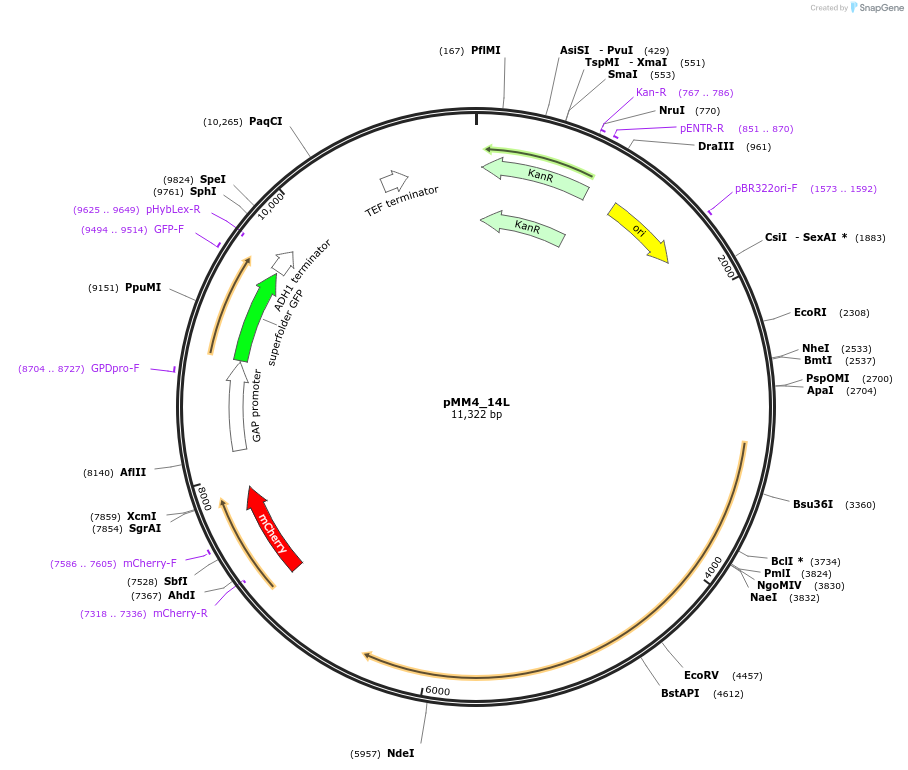
pMM4_14L
Plasmid#183063PurposePlasmid expressing a biosensor for acetic acid and pHluorin: BM3R1-Haa1-mTurquoise2 under RET2p, mCherry under HREsYGP1p and sfpHluorin under TDH3p with flanks for genome integration into HO locusDepositorInsertsmCherry
sfpHluorin
BM3R1-HAA1-mTurquoise2
UseSynthetic BiologyTagsmCherry and mTurquoise2ExpressionYeastPromoterRET2, Synthetic promoter, and TDH3Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
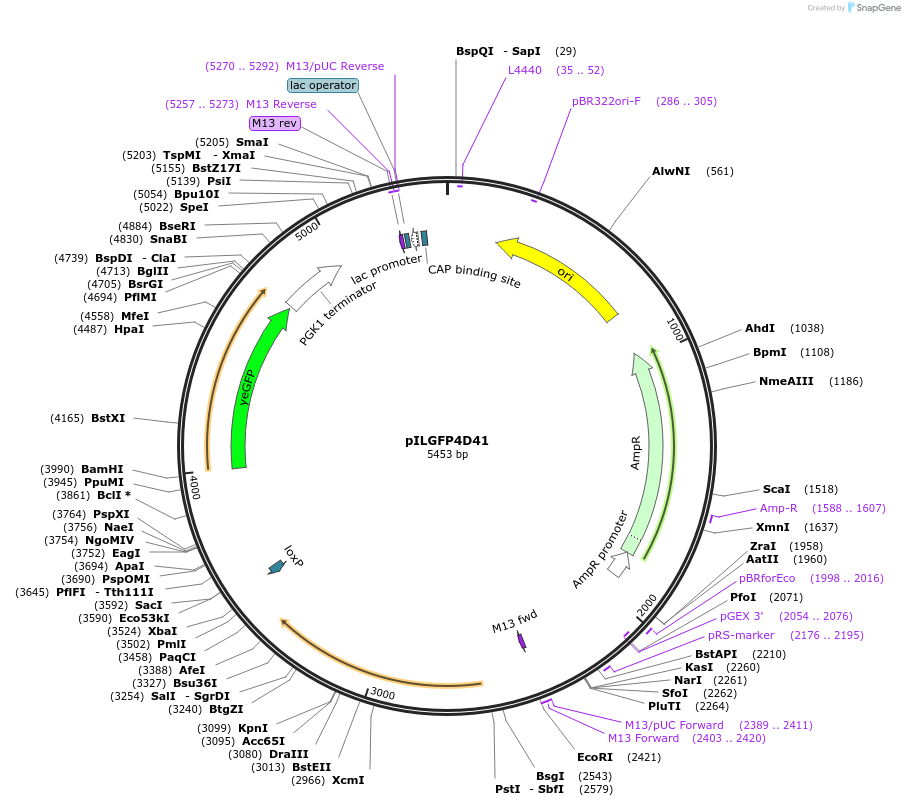
pILGFP4D41
Plasmid#185841PurposeTesting the CYC1 core promoter inserted with 4 Z268 elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PCYC1+[Z268]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
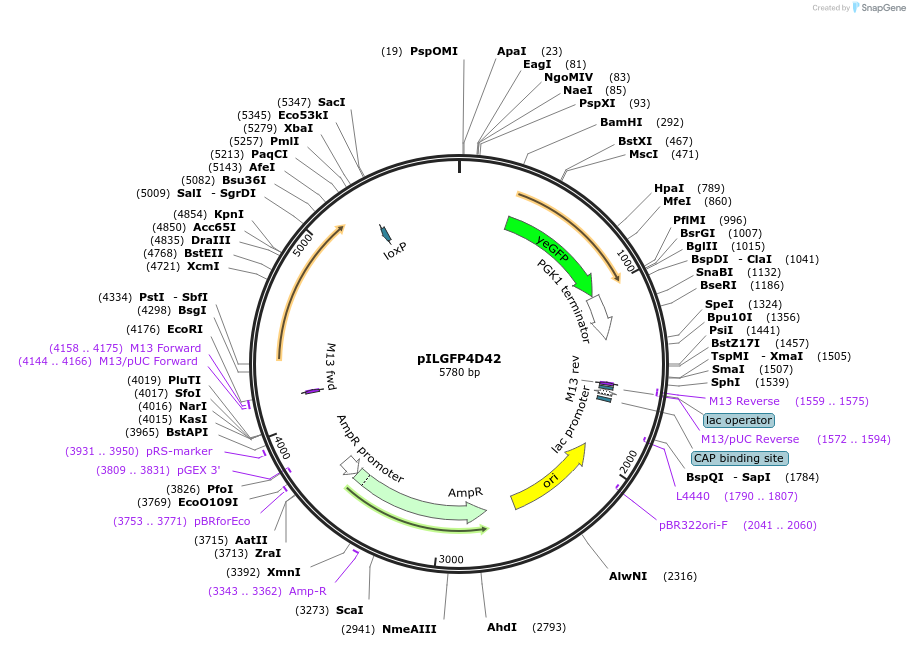
pILGFP4D42
Plasmid#185840PurposeTesting the TEF1 promoter inserted with 4 Z268 elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1+[Z268]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
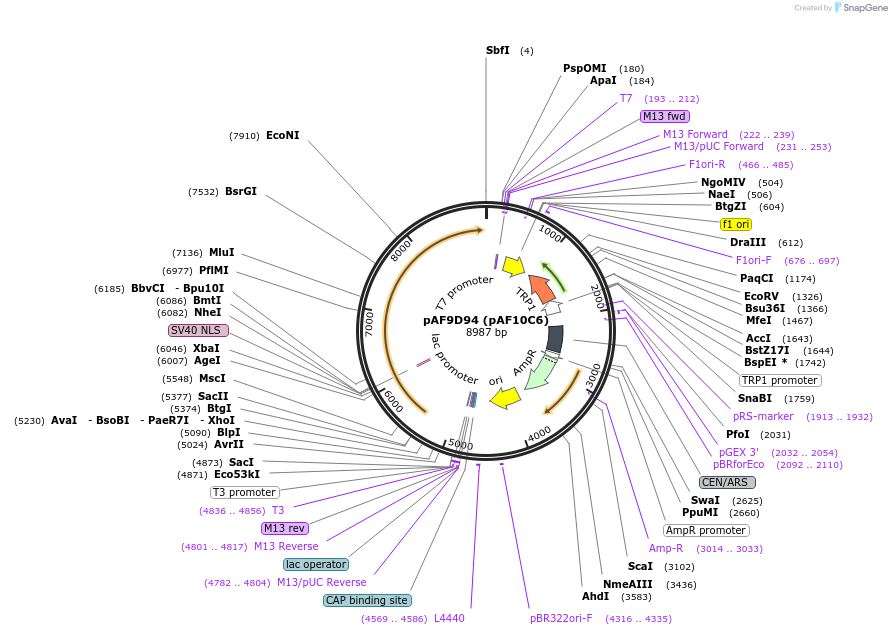
pAF9D94 (pAF10C6)
Plasmid#185848PurposeExpressing artificial trans-repressor in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>CYC8>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
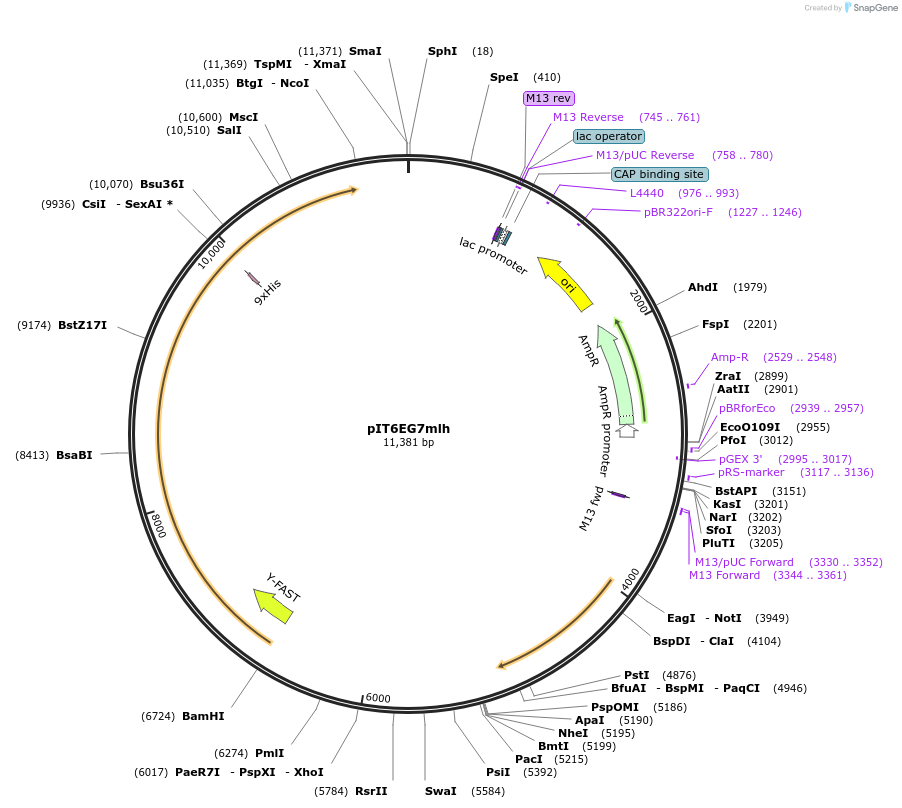
pIT6EG7mlh
Plasmid#185887PurposeIn vivo gene amplification (HamAmp) of limonene synthetic module at RPL25 locus (23 copy).DepositorInsertsee comments
ExpressionYeastMutationERG20 F96W N127WAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
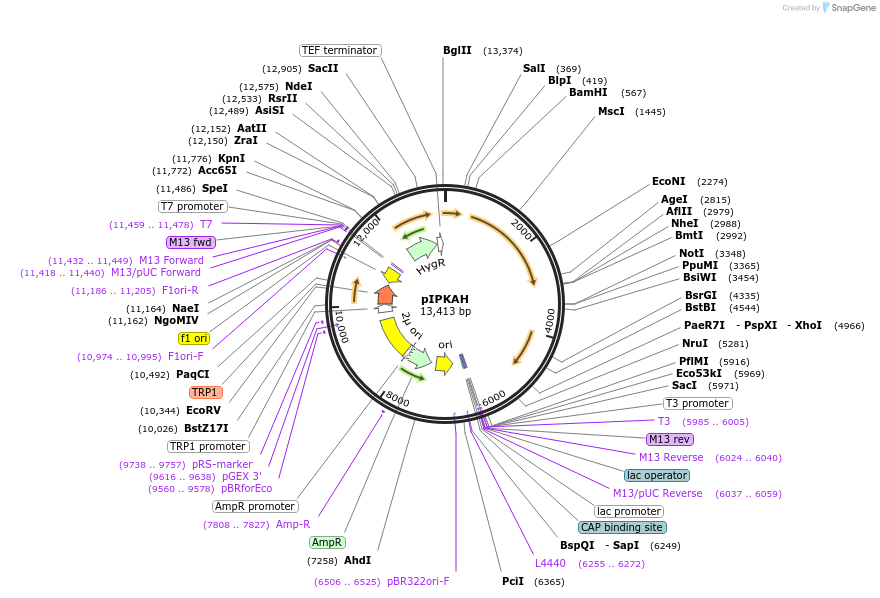
pIPKAH
Plasmid#185869PurposeKnocking out GPP1 and expressing a phosphoketolase pathway (Lactobacillus reuteri xfp and Methanosarcina barkeri pta)DepositorInsertGPP1(-220,80)> PAg.TEF1>hphMX6>TAg.TEF1-PENO2> Lr.xfp>T_TPI1-PTDH3>Mb.pta>TFBA1-GPP1(754, 1400)
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
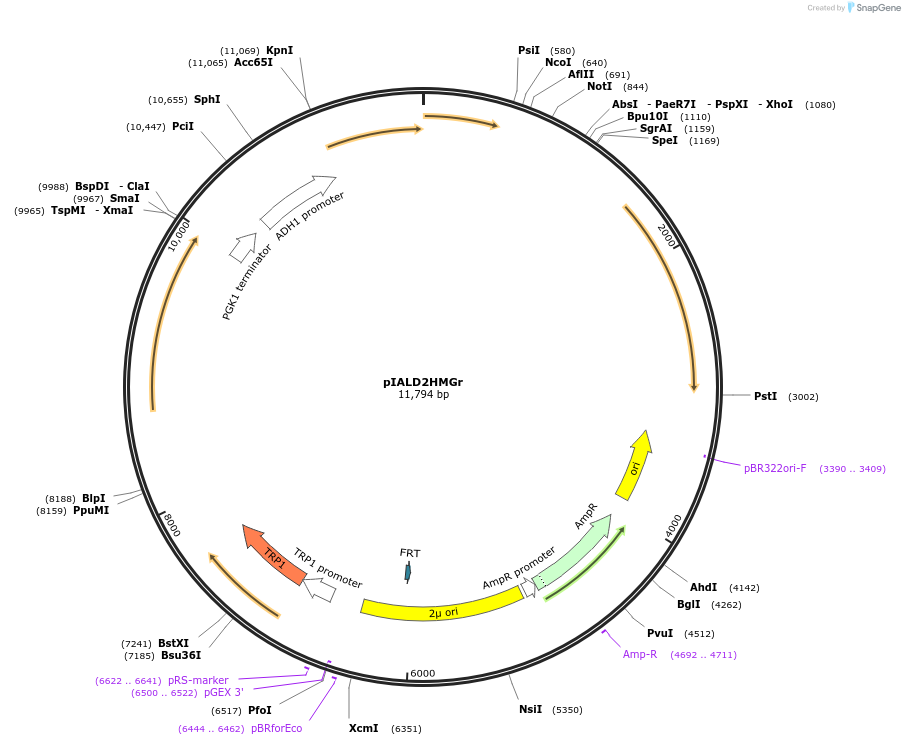
pIALD2HMGr
Plasmid#185867PurposeKnocking out ALD6; expressing acetylating aldehyde dehydrogenase (Lactobacillus reuteri pduP and L. reuteri EutE) and an NADH-preferring HMG-CoA reductase (Delftia acidovorans mvaA) in S. cerevisiaeDepositorInsertALD6(-125, 40)> PSk.GAL1>Da.mvaA>TPGK1-PADH1> Lr.pduP> TPDC1-PGAL2>Lr.EutE-ALD6(1054, 1749)
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
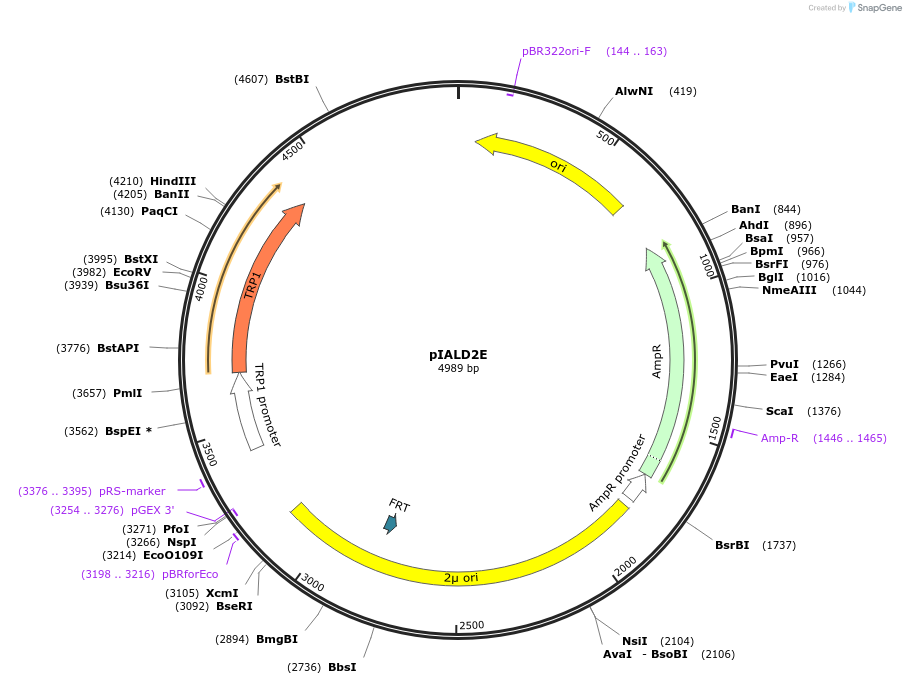
pIALD2E
Plasmid#185866PurposeKnocking out ALD6DepositorInsertALD(-125, 40)- ALD6(1054, 1749)
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
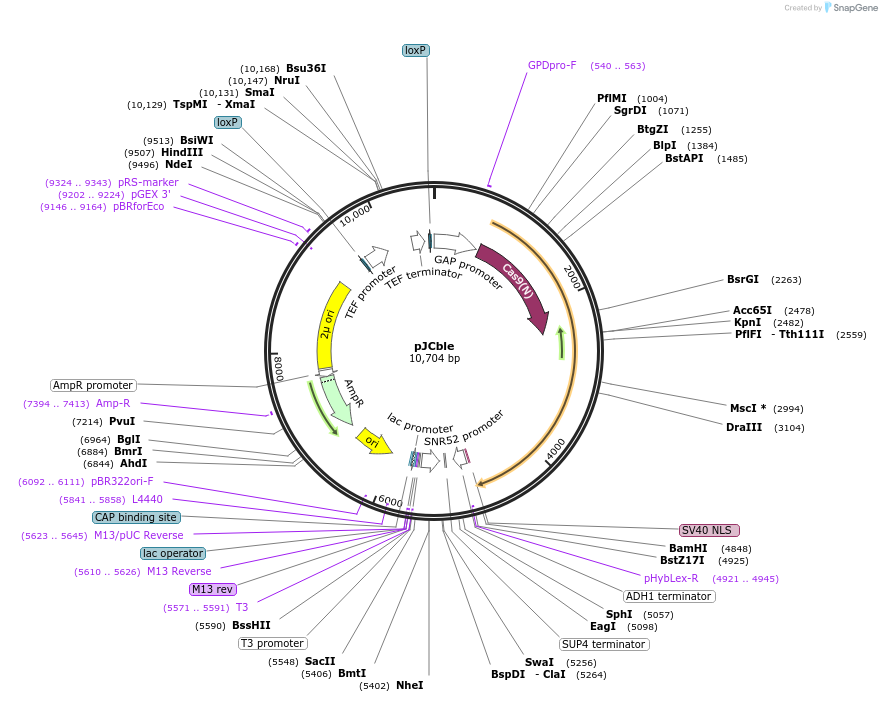
pJCble
Plasmid#185861PurposeThe CRISPR/Cas9-and-gRNA-expressing plasmid with a phleomycin-resistent selectable markerDepositorInsertPTDH3-Cas9(N); PSNR52-srRNA-TSUP4; ble
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
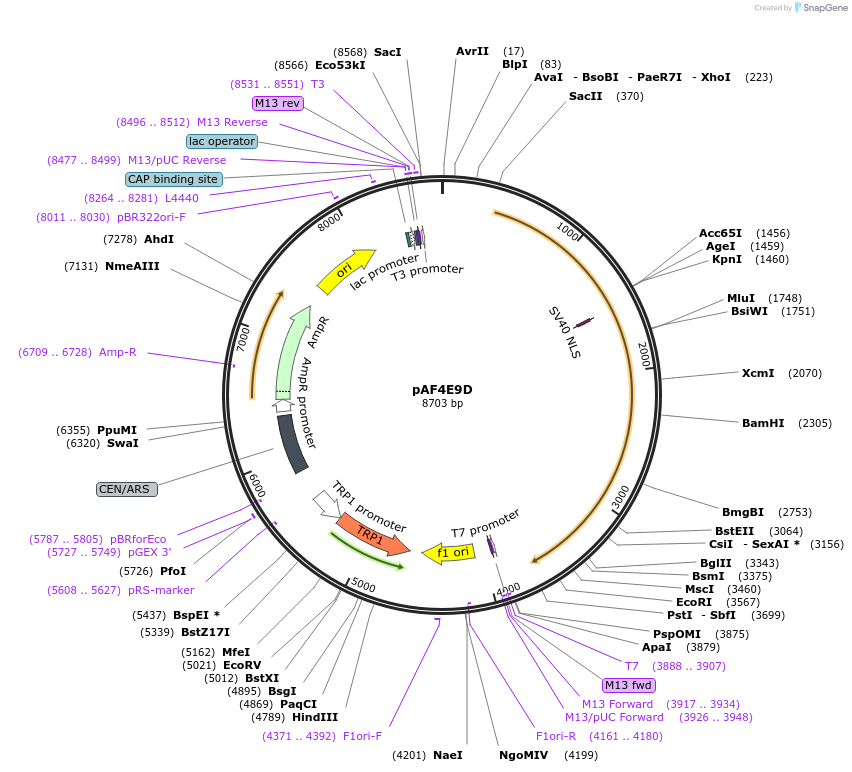
pAF4E9D
Plasmid#185844PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>LacI>TUP1>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
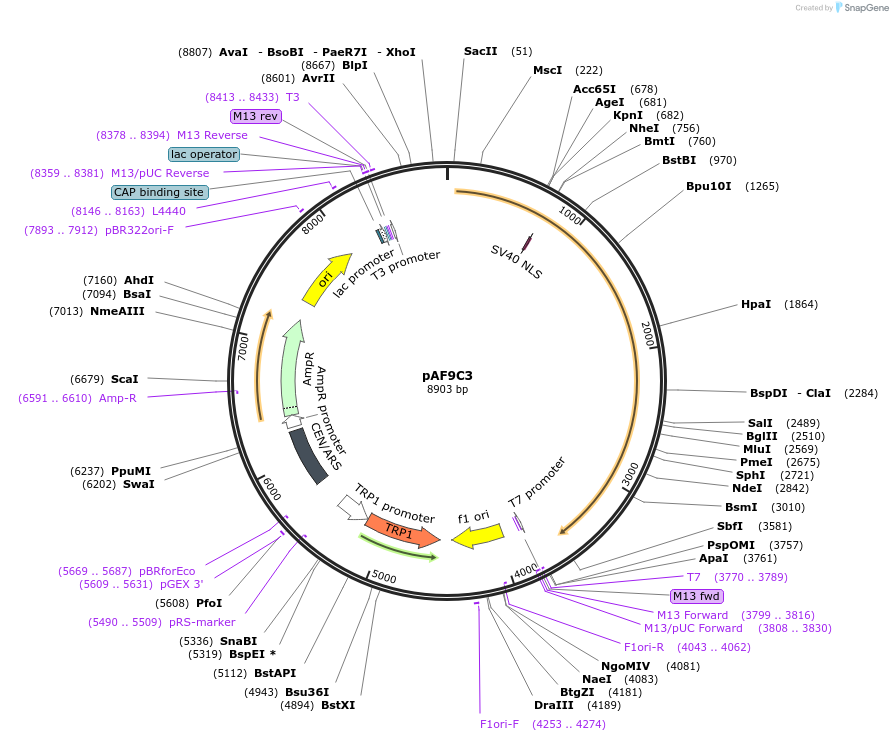
pAF9C3
Plasmid#185843PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>RevTetR_r1.7>SIN3C>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
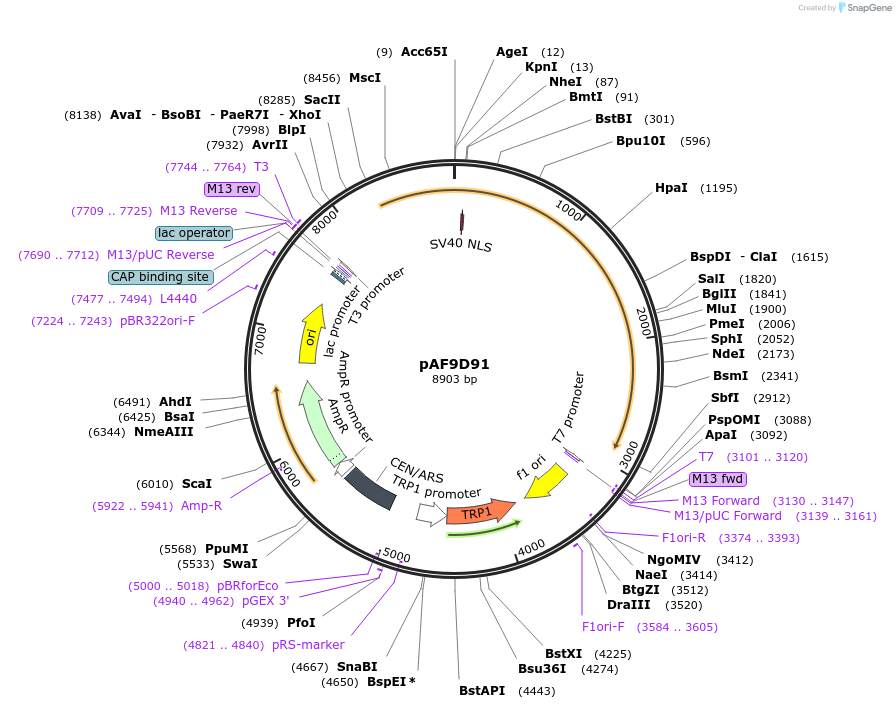
pAF9D91
Plasmid#185845PurposeExpressing artificial trans-activator in Saccharomyces cerevisiaeDepositorInsertPHAC1>TetR>SIN3C>PABF1
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
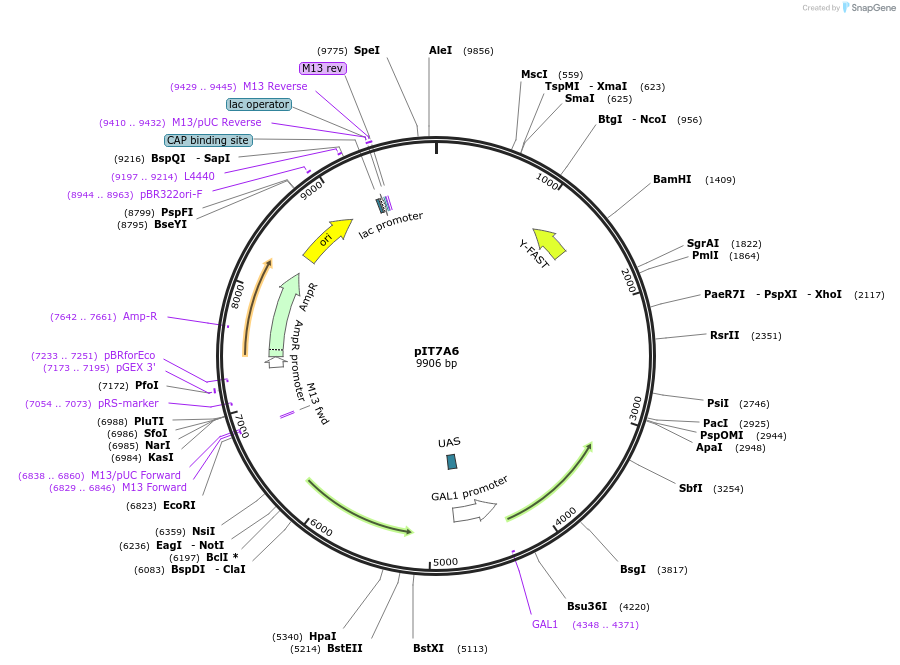
pIT7A6
Plasmid#185896PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (15 copy).DepositorInsertPRPL25(Arm 1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm 3)-ARS305-PSk.GAL2>Y.FAST-EVBR1.2A>TRPL41B-PPDA1>RPL25(partial;Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only