We narrowed to 15,575 results for: NOL
-
Plasmid#47196DepositorInsertZebrafishCommunity-klf8-right (si:ch211-208m1.2 Zebrafish)
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
pCellFree_G03 STAT6
Plasmid#67123PurposeStandard for cell-free expression benchmarking.DepositorAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCellFree_G03 TFDP3
Plasmid#67083PurposeStandard for cell-free expression benchmarking.DepositorAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCellFree_G03 TFDP1
Plasmid#67084PurposeStandard for cell-free expression benchmarking.DepositorAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCellFree_G03 TFDP2
Plasmid#67088PurposeStandard for cell-free expression benchmarking.DepositorAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
pCellFree_G03 TFF1
Plasmid#67047PurposeStandard for cell-free expression benchmarking.DepositorAvailable SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
mPlum-Alpha-Actinin-19
Plasmid#55949PurposeLocalization: alpha Actinin, Excitation: 590, Emission: 649DepositorAvailable SinceMarch 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
-
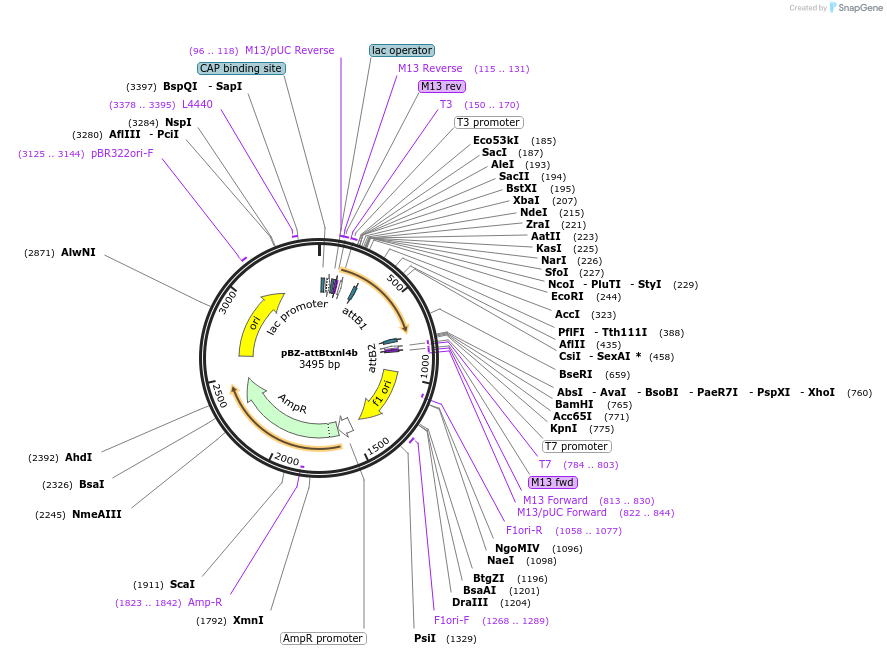
pBZ-attBtxnl4b
Plasmid#18667Purposecloning vector containing attB flanked TXNL4B useful for recombination based conversion of conventional vectors to Gateway compatible destination vectorsDepositorAvailable SinceJuly 26, 2008AvailabilityAcademic Institutions and Nonprofits only -
-
-
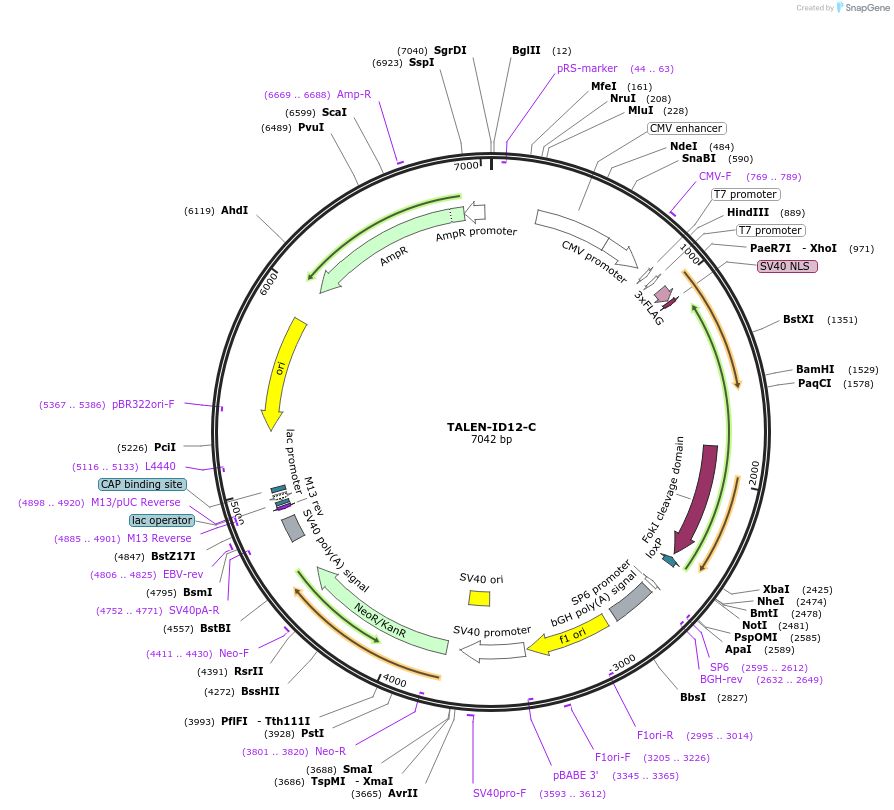
TALEN-ID12-C
Plasmid#41477DepositorInsertTALEN-FokI nuclease backbone
Tags3xFLAG and WT FokI nuclease domainExpressionMammalianPromoterCMVAvailable SinceJan. 2, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
-
-
TAL3577
Plasmid#47296DepositorInsertZebrafishCommunity-tetmethylcytosinedioxygenase3(previous si:ch211-220a11.1)-right
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 20, 2013AvailabilityAcademic Institutions and Nonprofits only -
TAL3576
Plasmid#47295DepositorInsertZebrafishCommunity-tetmethylcytosinedioxygenase3(previous si:ch211-220a11.1)-left
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 20, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
TAL3539
Plasmid#47258DepositorInsertZebrafishCommunity-si:dkey-208k22.3-right (si:dkey-208k22.3 Zebrafish)
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
TAL3538
Plasmid#47257DepositorInsertZebrafishCommunity-si:dkey-208k22.3-left (si:dkey-208k22.3 Zebrafish)
UseT7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceAug. 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
TAL3330
Plasmid#42793DepositorInsertZebrafishCommunity-oxtr-left (LOC100001530 Zebrafish)
Uset7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceMarch 22, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
-
TAL3331
Plasmid#42794DepositorInsertZebrafishCommunity-oxtr-right (LOC100001530 Zebrafish)
Uset7Tags3X Flag and WT FOKIExpressionMammalianPromoterCMVAvailable SinceMarch 19, 2013AvailabilityAcademic Institutions and Nonprofits only -