We narrowed to 5,133 results for: codon optimized
-
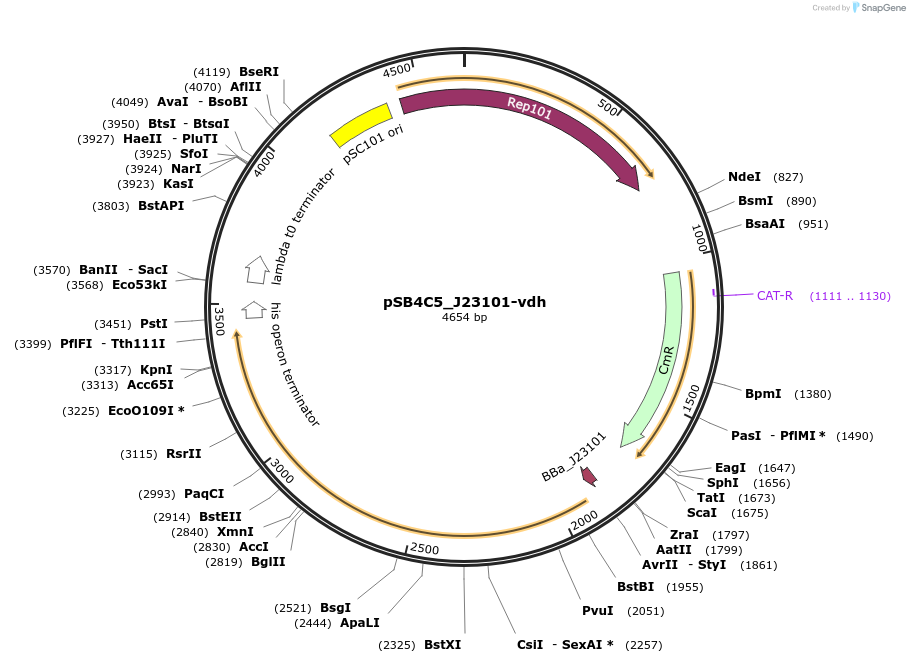
Plasmid#128130PurposeExpressing vdh gene (for the enzyme transforming benzaldehyde to benzoate) under control of the constitutive promoter J23101, and RBS B0032DepositorInsertvdh
UseSynthetic BiologyExpressionBacterialPromoterJ23101Available SinceNov. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
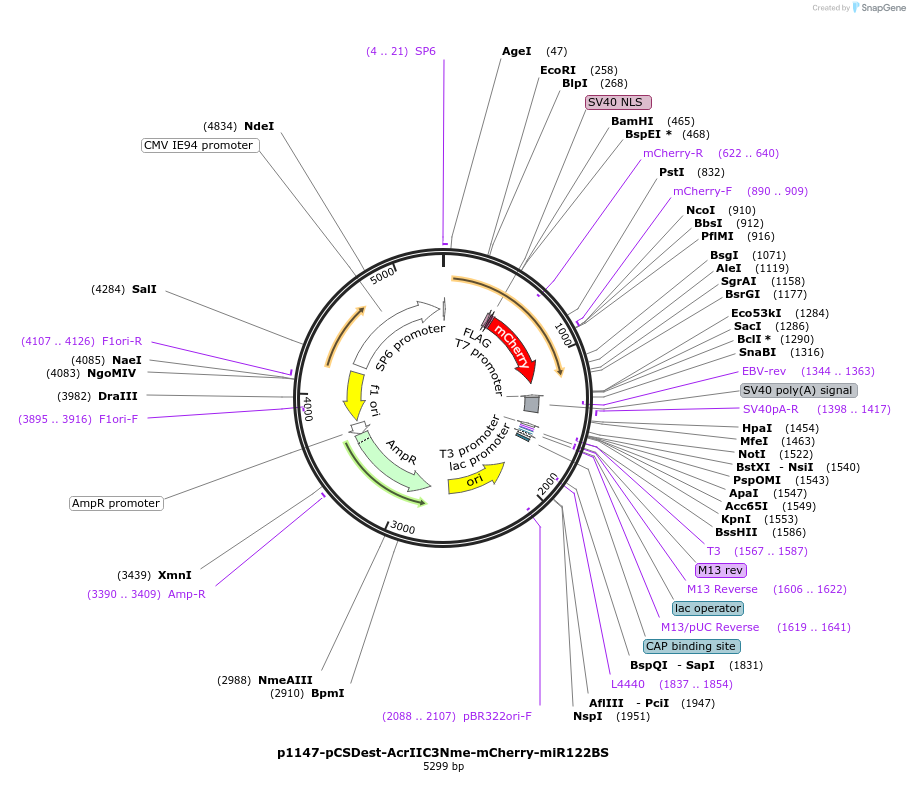
p1147-pCSDest-AcrIIC3Nme-mCherry-miR122BS
Plasmid#126203PurposeMammalian expression of AcrIIC3Nme fused to mCherry with 3x miR-122 binding sites in 3' UTRDepositorInsertcodon-optimized AcrIIC3Nme
TagsFLAG/NLS and mCherryExpressionMammalianPromoterCMVAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
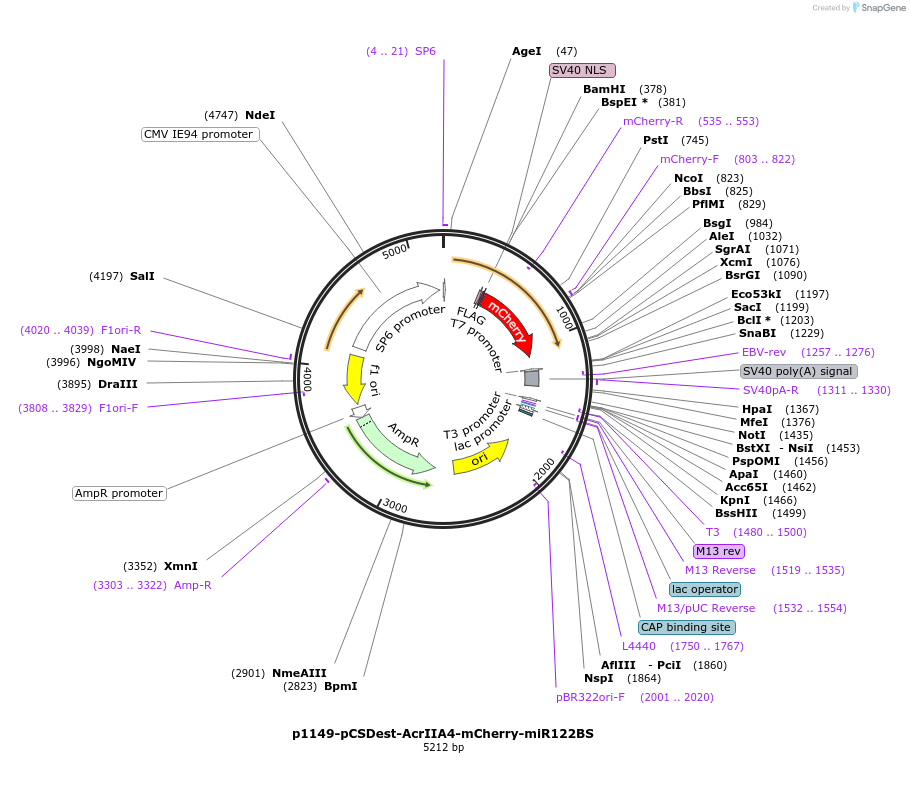
p1149-pCSDest-AcrIIA4-mCherry-miR122BS
Plasmid#126205PurposeMammalian expression of AcrIIA4 fused to mCherry with 3x miR-122 binding sites in 3' UTRDepositorInsertcodon-optimized AcrIIA4
TagsFLAG/NLS and mCherryExpressionMammalianPromoterCMVAvailable SinceSept. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
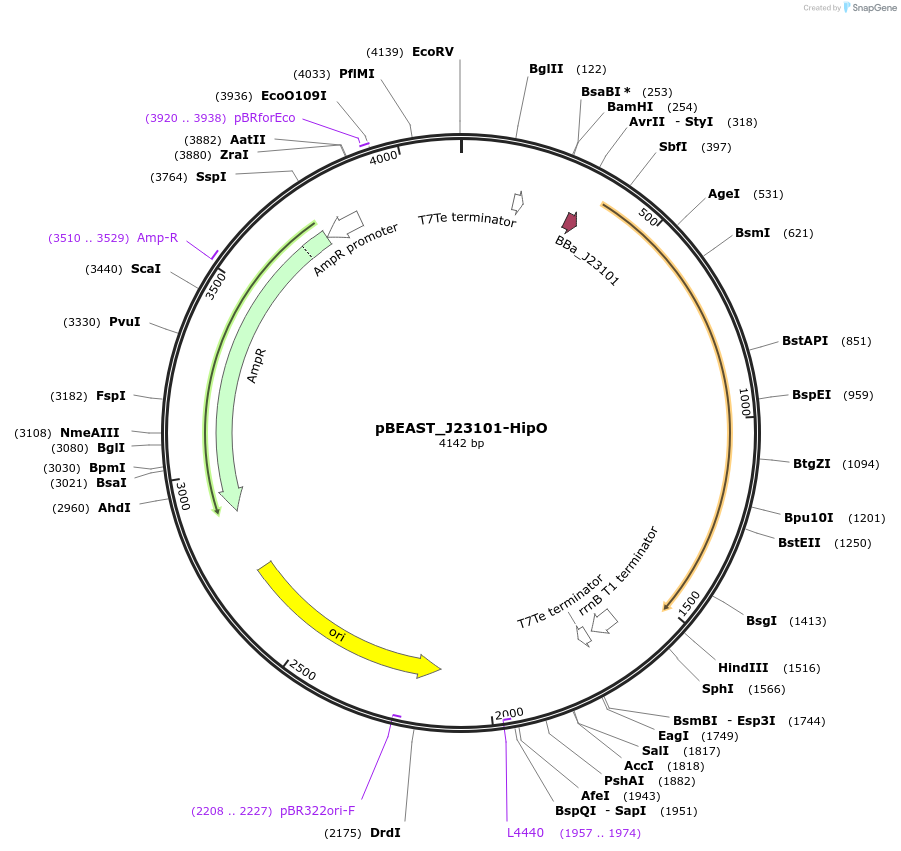
pBEAST_J23101-HipO
Plasmid#128133PurposeThe cell-free adapted backbone, pBEAST, expressing HipO (the enzyme converting hippurate to benzoate) under control of the constitutive promoter J23101, and RBS B0032DepositorInsertHipO
UseSynthetic BiologyExpressionBacterialPromoterJ23101Available SinceAug. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
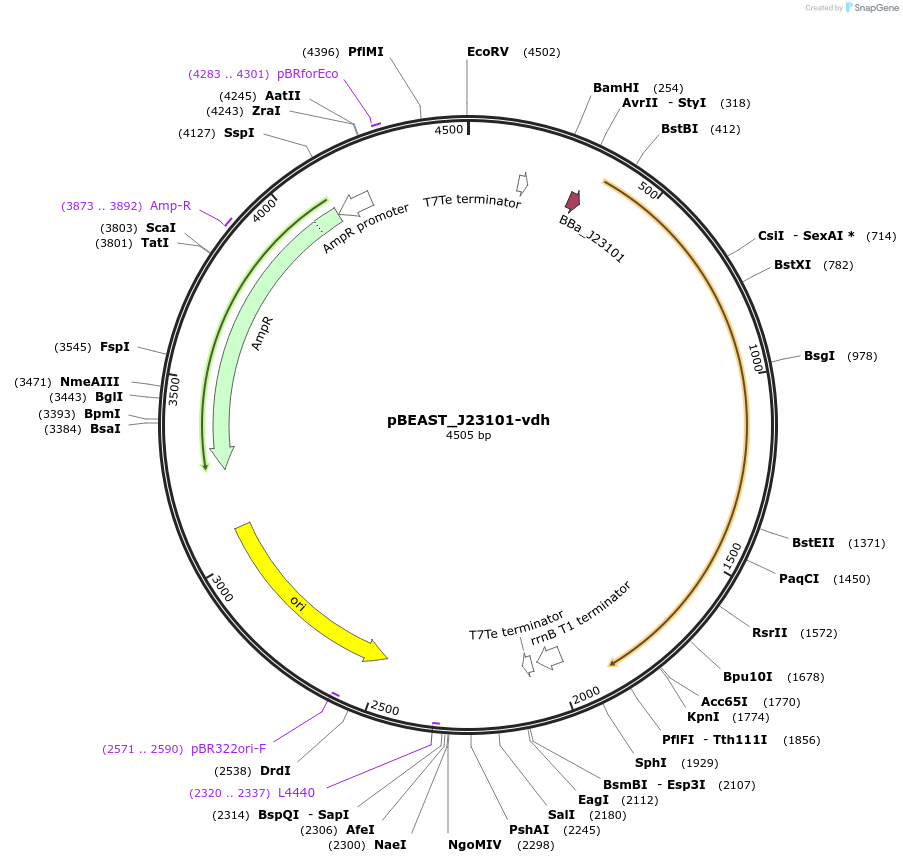
pBEAST_J23101-vdh
Plasmid#128134PurposeThe cell-free adapted backbone, pBEAST, expressing vdh (the enzyme converting benzaldehyde to benzoate) under control of the constitutive promoter J23101, and RBS B0032DepositorInsertvdh
UseSynthetic BiologyExpressionBacterialPromoterJ23101Available SinceAug. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
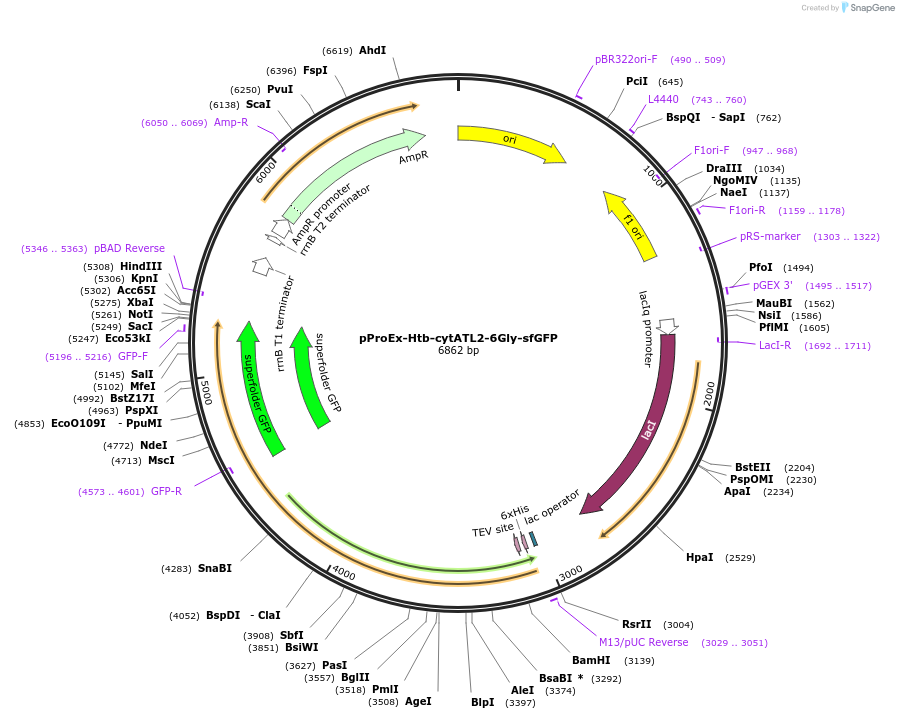
pProEx-Htb-cytATL2-6Gly-sfGFP
Plasmid#124065PurposeE. coli. exp. and purif. of X. laevis cytATL fragment fused to GFP in c-terminus.DepositorInsertcytATL-sfGFP
TagssfGFPExpressionBacterialPromotertrcAvailable SinceMay 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
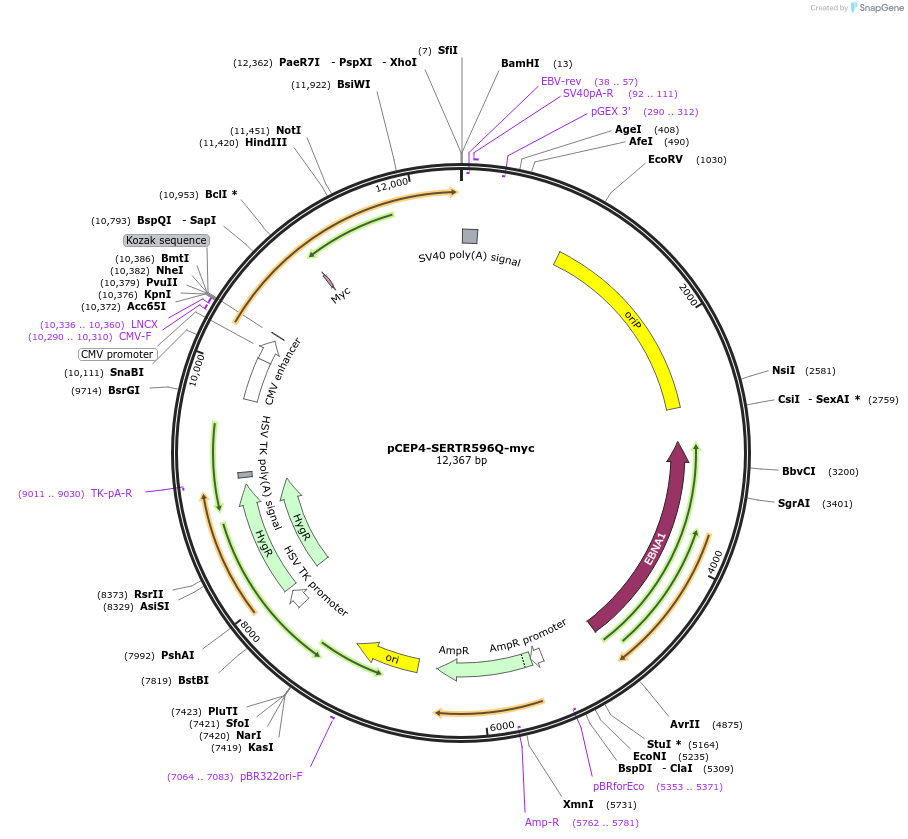
pCEP4-SERTR596Q-myc
Plasmid#107483PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation R596Q conferring gain of function phenotypeDepositorAvailable SinceMay 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
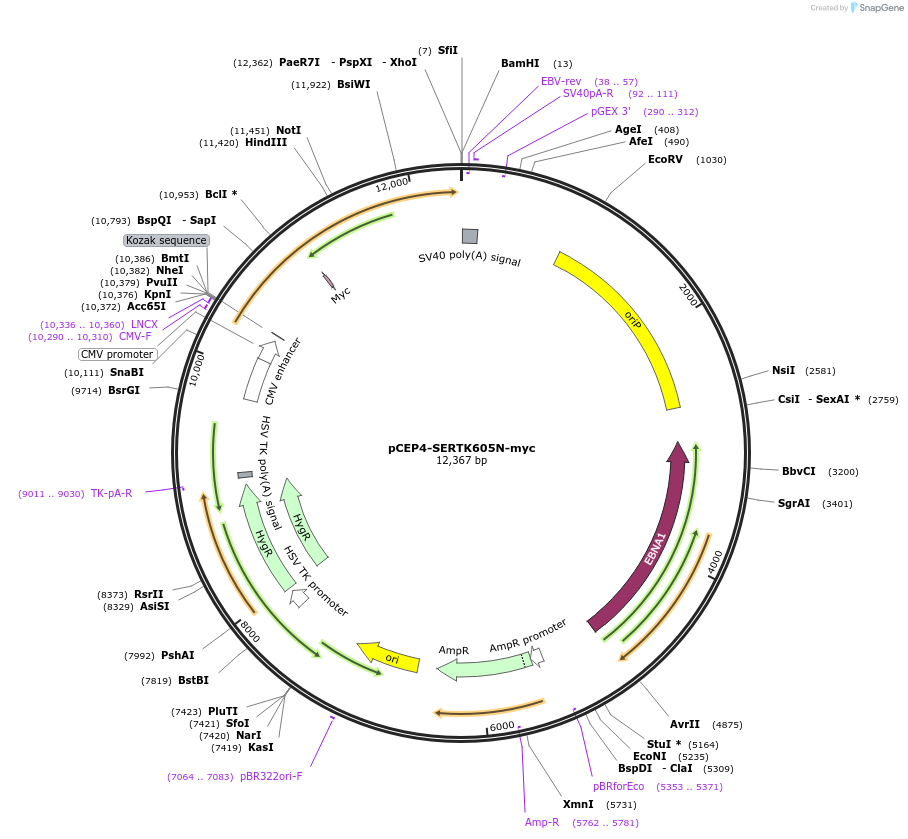
pCEP4-SERTK605N-myc
Plasmid#107475PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation K605N approxmately wildtype activity, decreased surface expressionDepositorAvailable SinceMarch 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
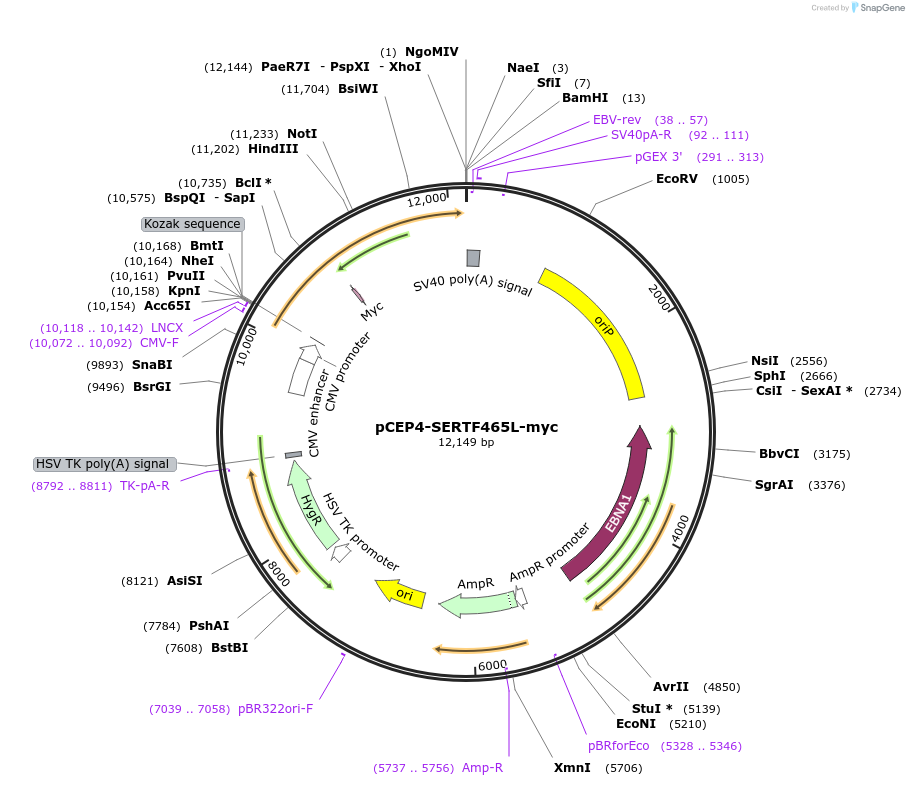
pCEP4-SERTF465L-myc
Plasmid#107469PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation F465L conferring gain of function phenotypeDepositorAvailable SinceMarch 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
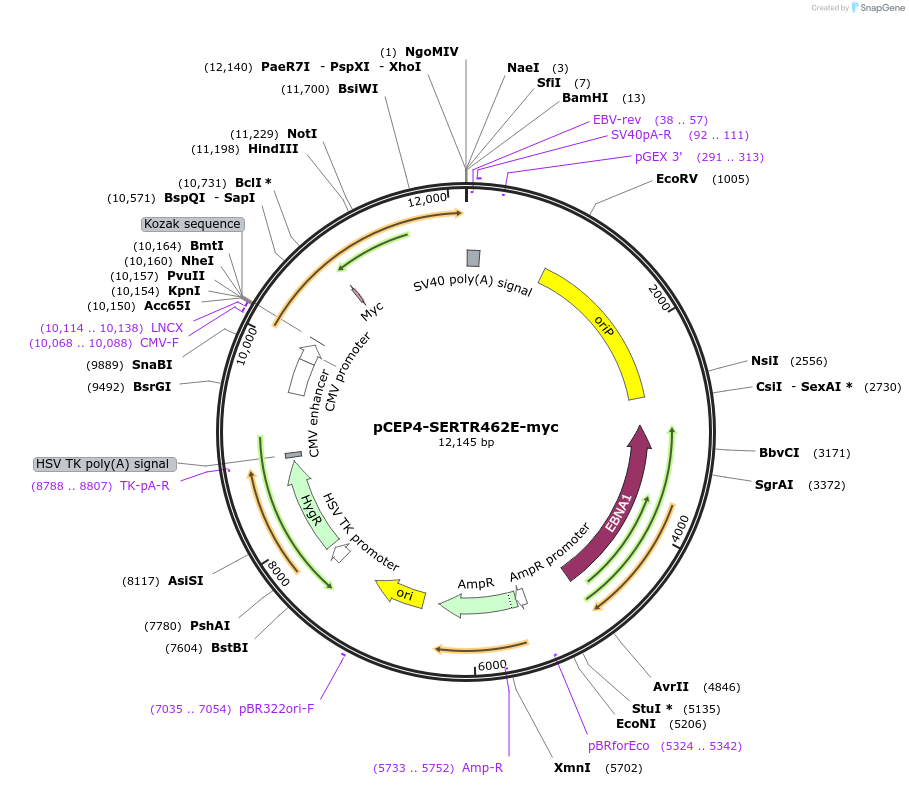
pCEP4-SERTR462E-myc
Plasmid#107489PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation R462E with approximately wildtype activity, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
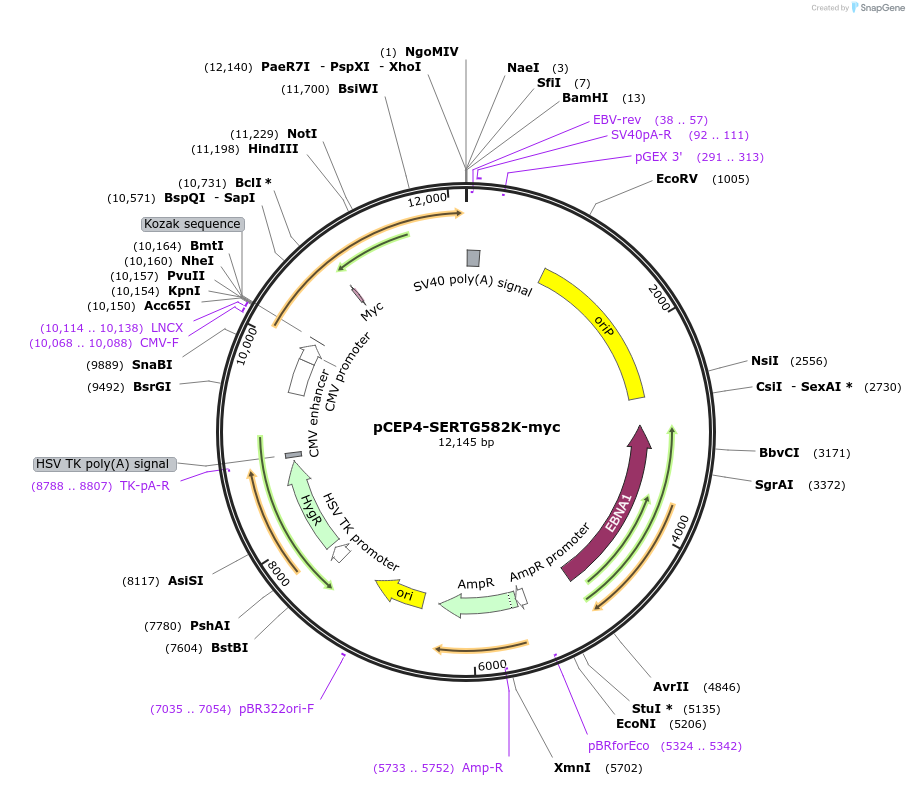
pCEP4-SERTG582K-myc
Plasmid#107497PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation G582K conferring gain of function phenotype, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
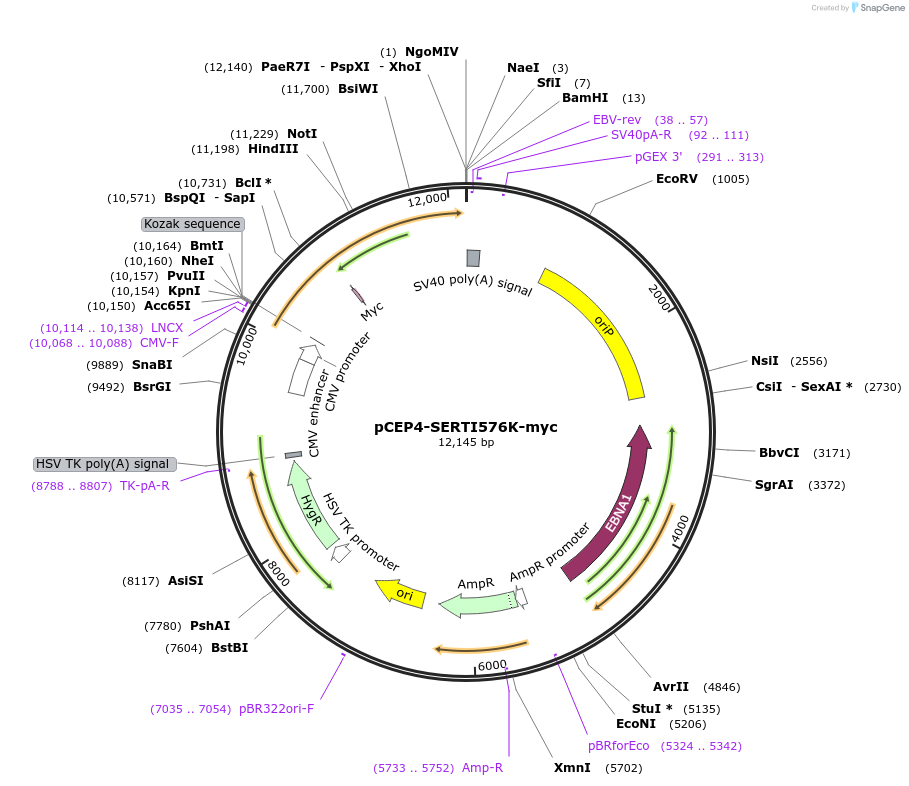
pCEP4-SERTI576K-myc
Plasmid#107496PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation I576K with approximately wildtype activity, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
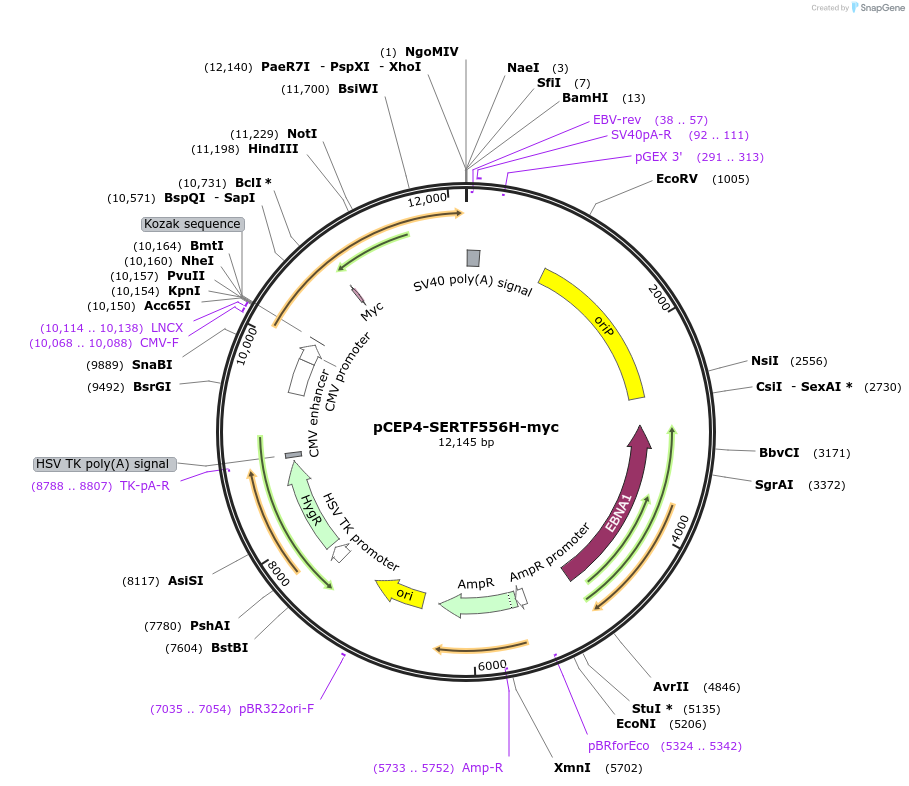
pCEP4-SERTF556H-myc
Plasmid#107495PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation F556H conferring gain of function phenotypeDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
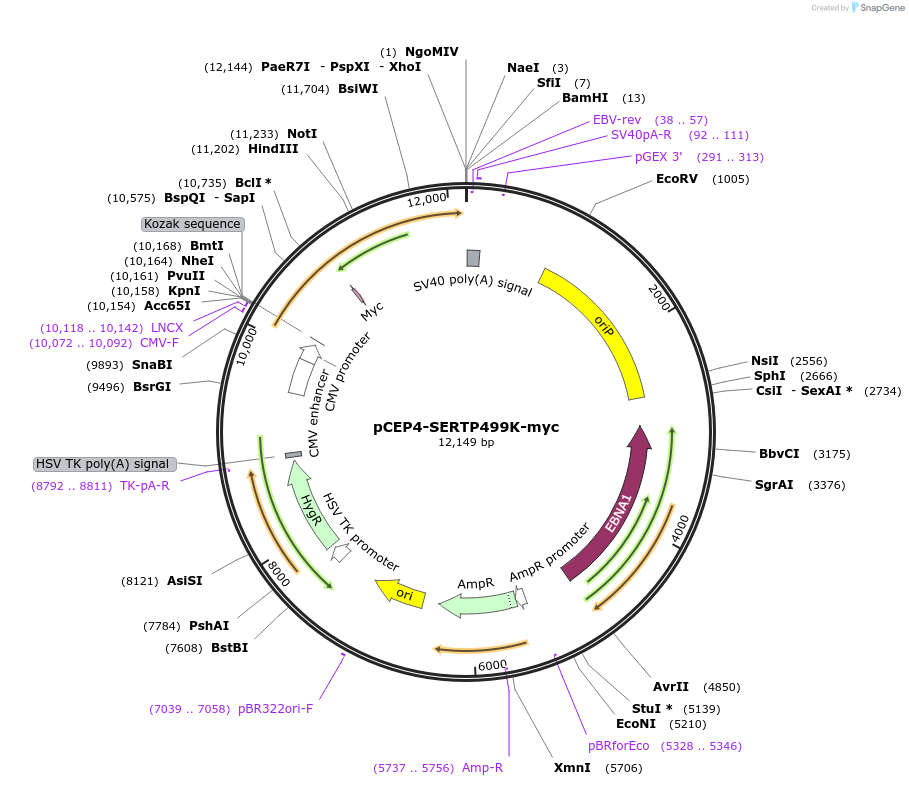
pCEP4-SERTP499K-myc
Plasmid#107494PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation P499K conferring gain of function phenotypeDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
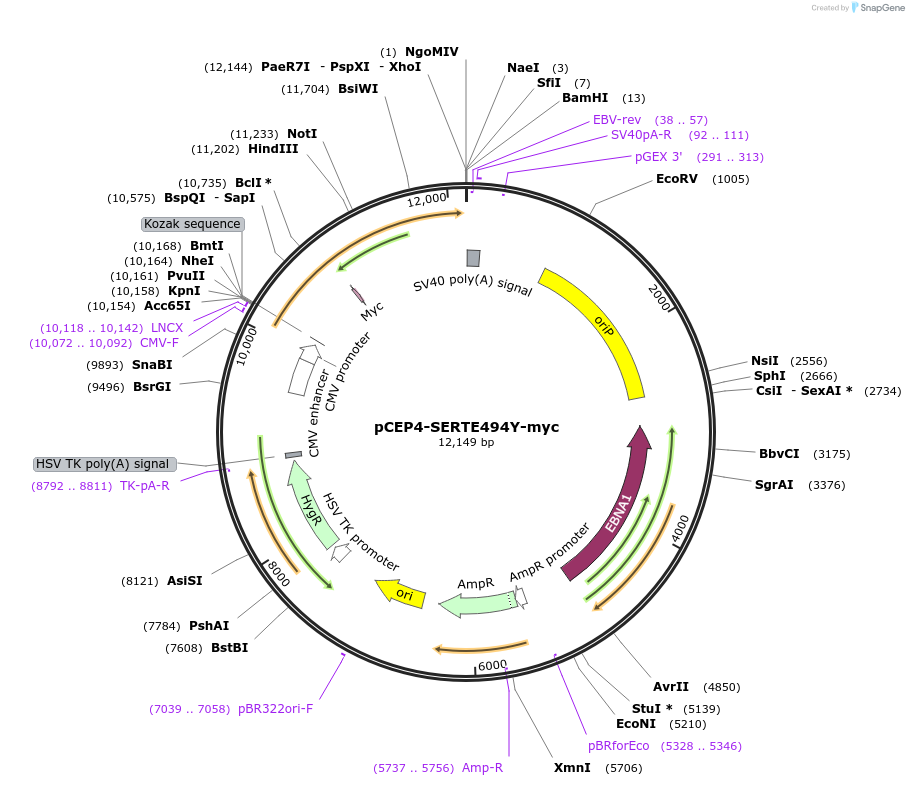
pCEP4-SERTE494Y-myc
Plasmid#107493PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation E494Y conferring gain of function phenotypeDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
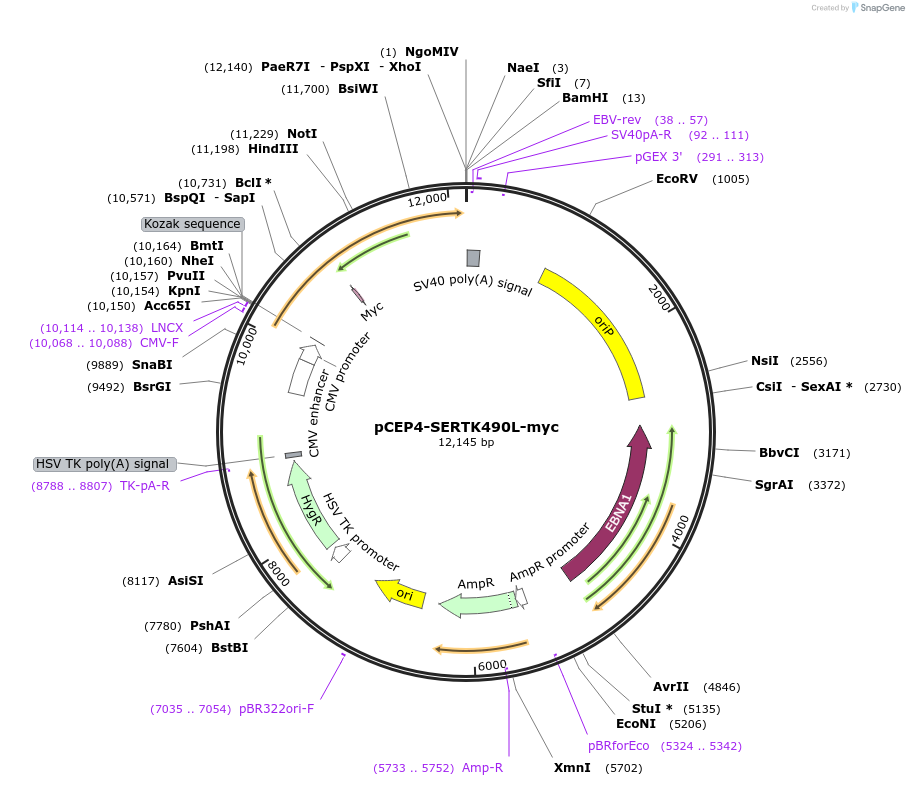
pCEP4-SERTK490L-myc
Plasmid#107492PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation K490L conferring gain of function phenotype, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
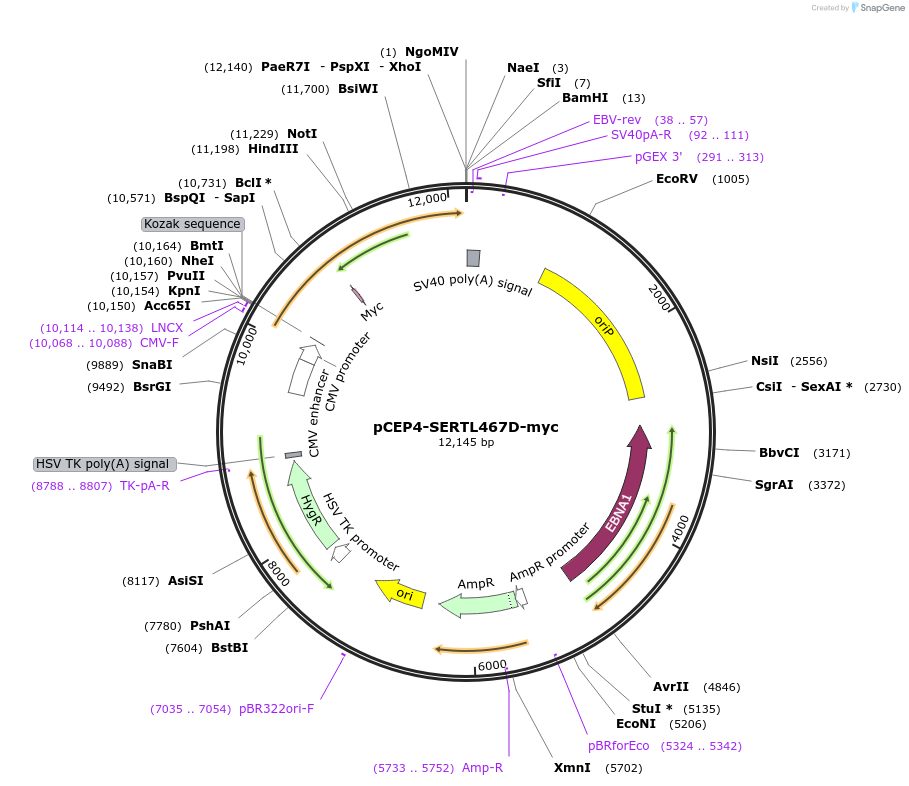
pCEP4-SERTL467D-myc
Plasmid#107490PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation L467D with approximately wildtype activity, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
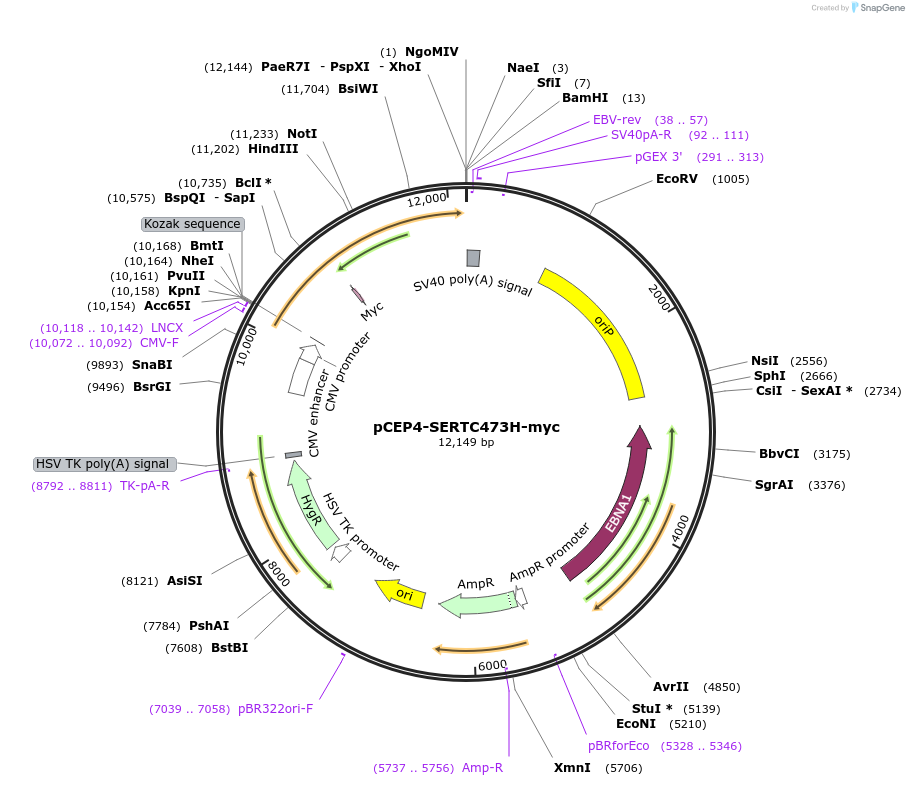
pCEP4-SERTC473H-myc
Plasmid#107491PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation C473H conferring gain of function phenotypeDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
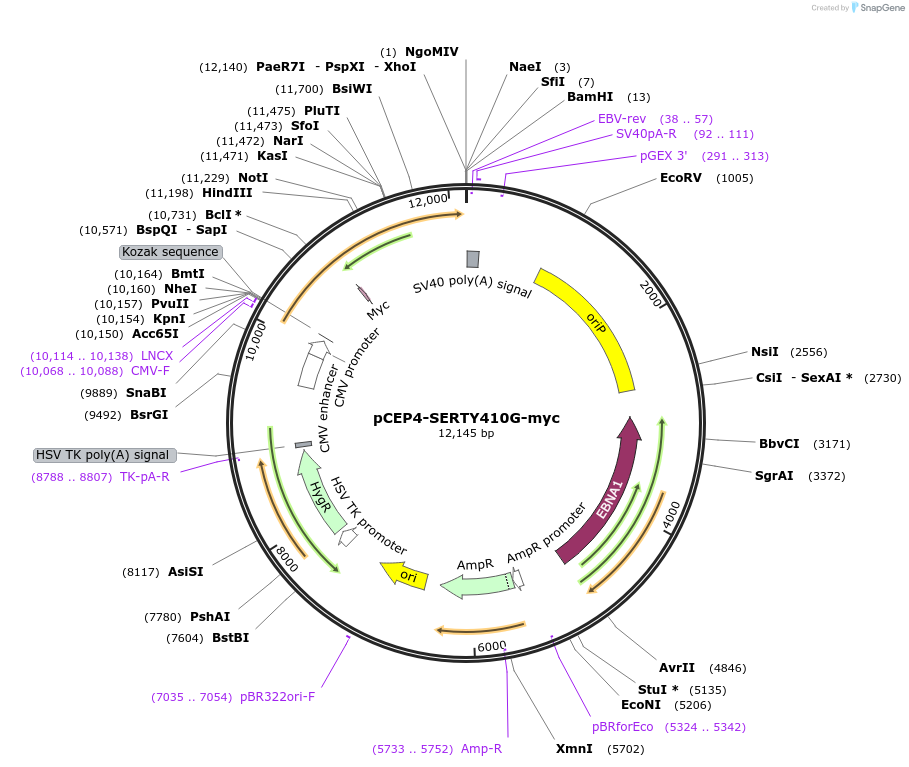
pCEP4-SERTY410G-myc
Plasmid#107488PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation Y410G with approximately wildtype activity, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
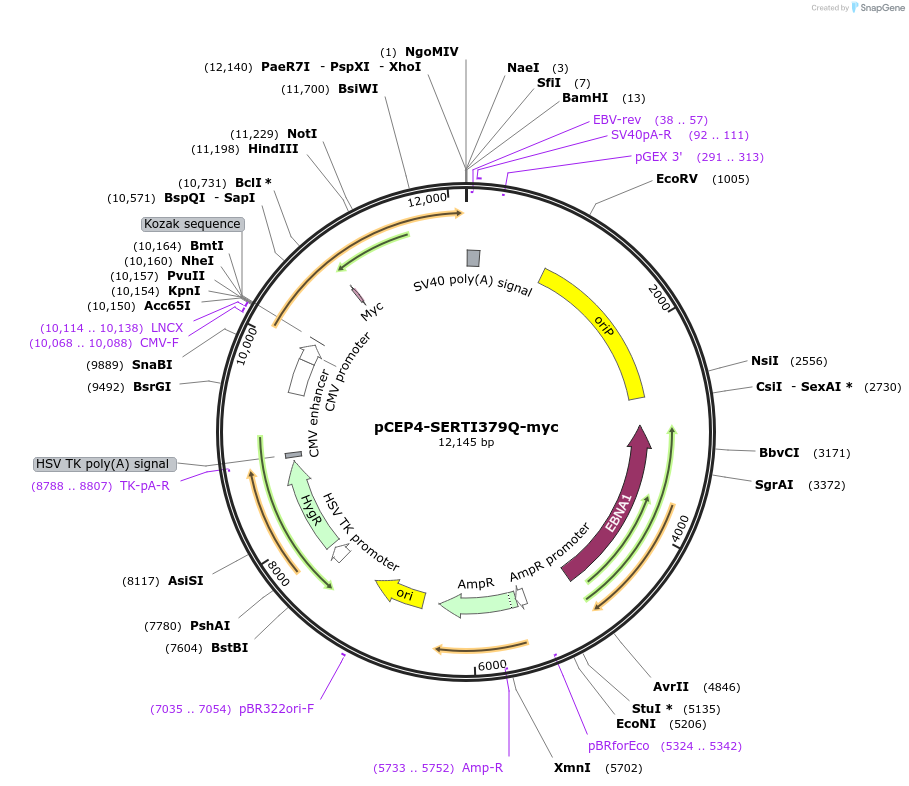
pCEP4-SERTI379Q-myc
Plasmid#107485PurposeMammalian expression plasmid for codon optimized mutant hSERT with internal MYC tag inserted in ECL2, mutation I379Q conferring gain of function phenotype, decreased surface expressionDepositorAvailable SinceMarch 15, 2018AvailabilityAcademic Institutions and Nonprofits only