We narrowed to 2,834 results for: Star;
-
Plasmid#8546DepositorInsertHOXC4 (HOXC4 Human)
UseRetroviralExpressionMammalianMutationContains improved ribosomal binding site and NcoI…Available SinceMay 30, 2006AvailabilityAcademic Institutions and Nonprofits only -
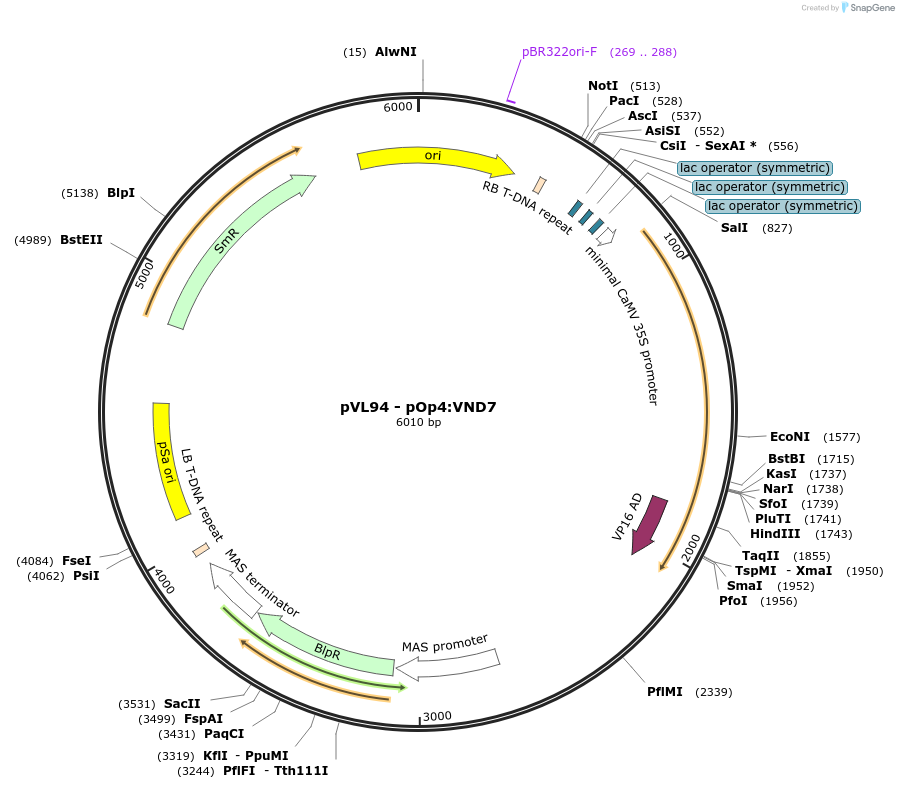
pVL94 - pOp4:VND7
Plasmid#116016Purposedestination vector for GreenGate cloning method, "Effector line", crossed with a "Driver line" the GOI, here VND7, will be expressed upon Dex-inductionDepositorInsertpOp4:B-dummy:VND7:VP16:tUBQ10::BastaR
ExpressionPlantAvailable SinceJuly 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
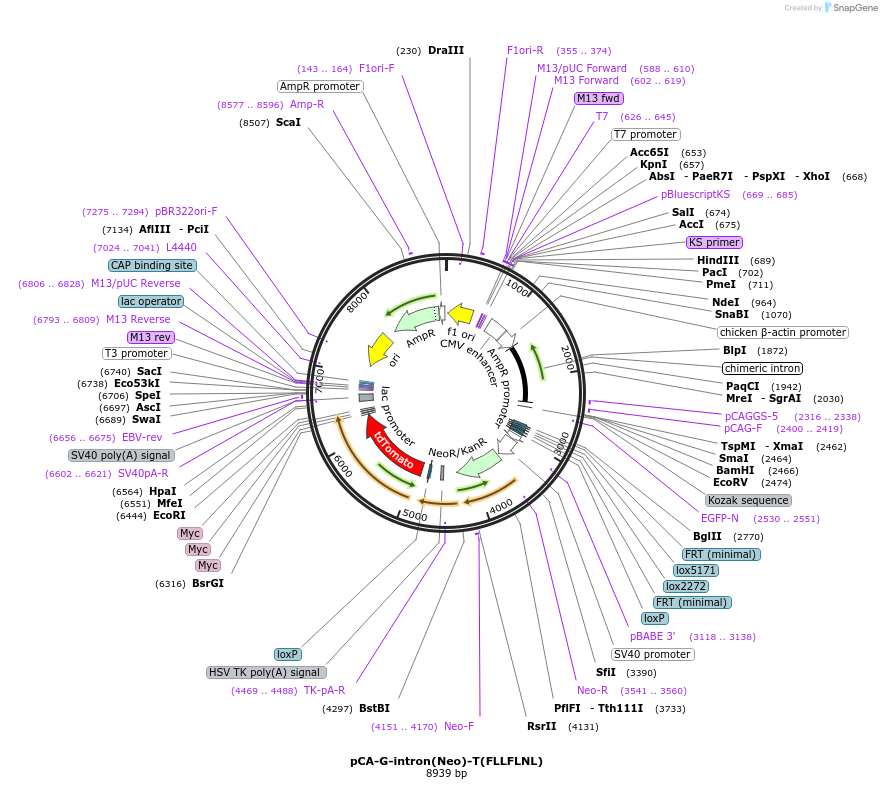
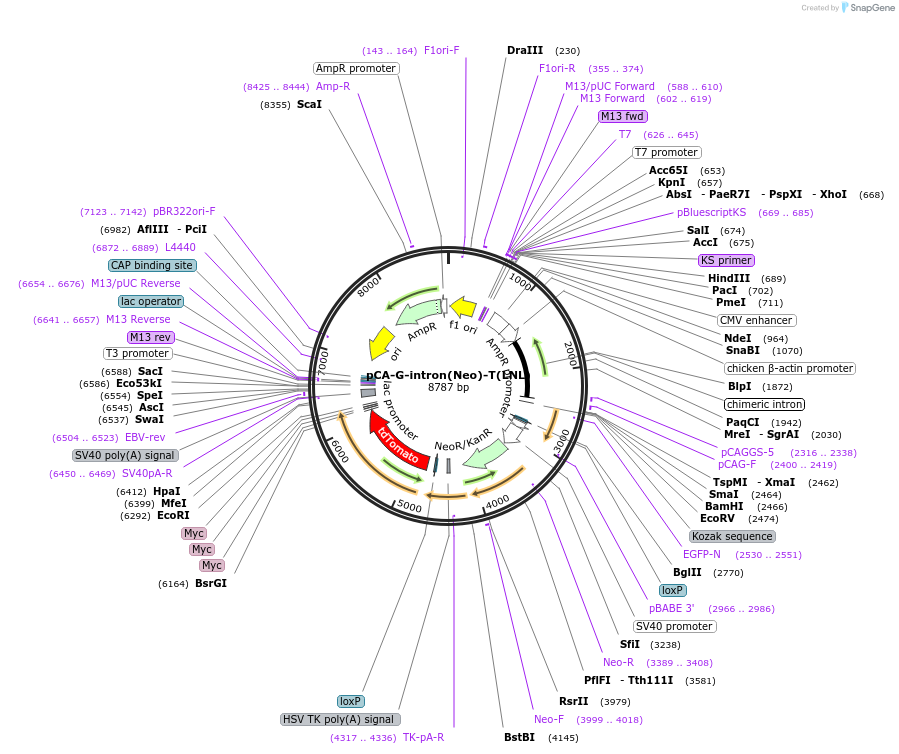
pCA-G-intron(Neo)-T(FLLFLNL)
Plasmid#40020DepositorInsertsGFP
tdT-3Myc
beta-globin intron
Neo
Tags3 Myc tagsExpressionMammalianMutationdeleted nucleotides after nucleotide 274, inserti…PromoterCAG (chicken beta globin promoter and CMV enhance…Available SinceOct. 12, 2012AvailabilityAcademic Institutions and Nonprofits only -
psiCheck-DNAJA4CDS
Plasmid#41849DepositorInsertDNAJA4 CDS (DNAJA4 Human)
UseLuciferaseTagsrenilla luciferaseMutationStart codon of DNAJA4 removedAvailable SinceFeb. 6, 2013AvailabilityAcademic Institutions and Nonprofits only -
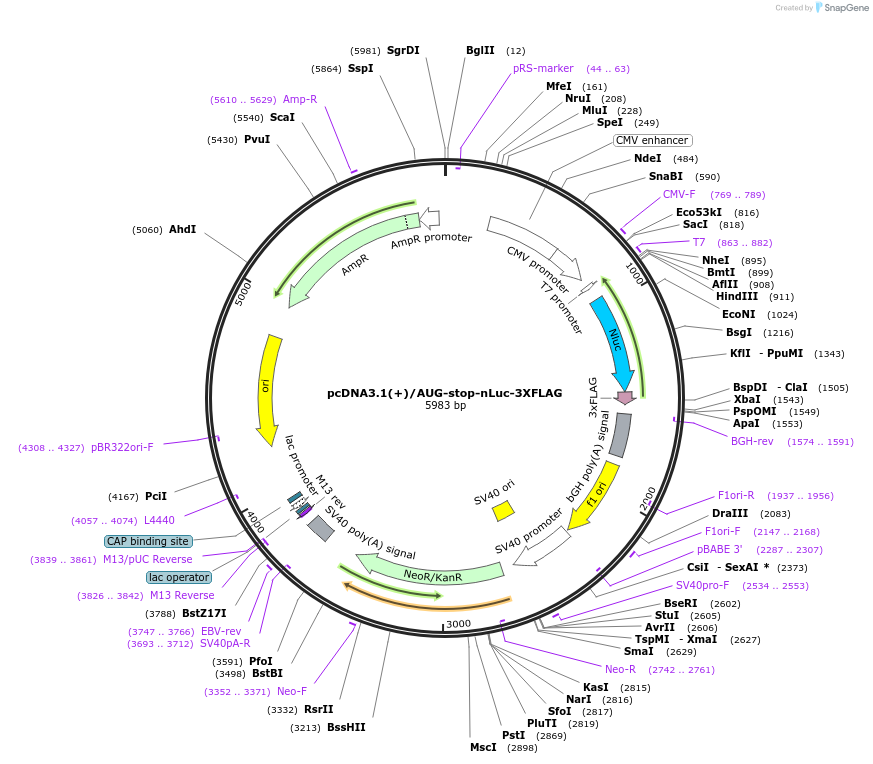
pcDNA3.1(+)/AUG-stop-nLuc-3XFLAG
Plasmid#127308PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from AUG start codon, but translation terminated immediately by stop codonDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
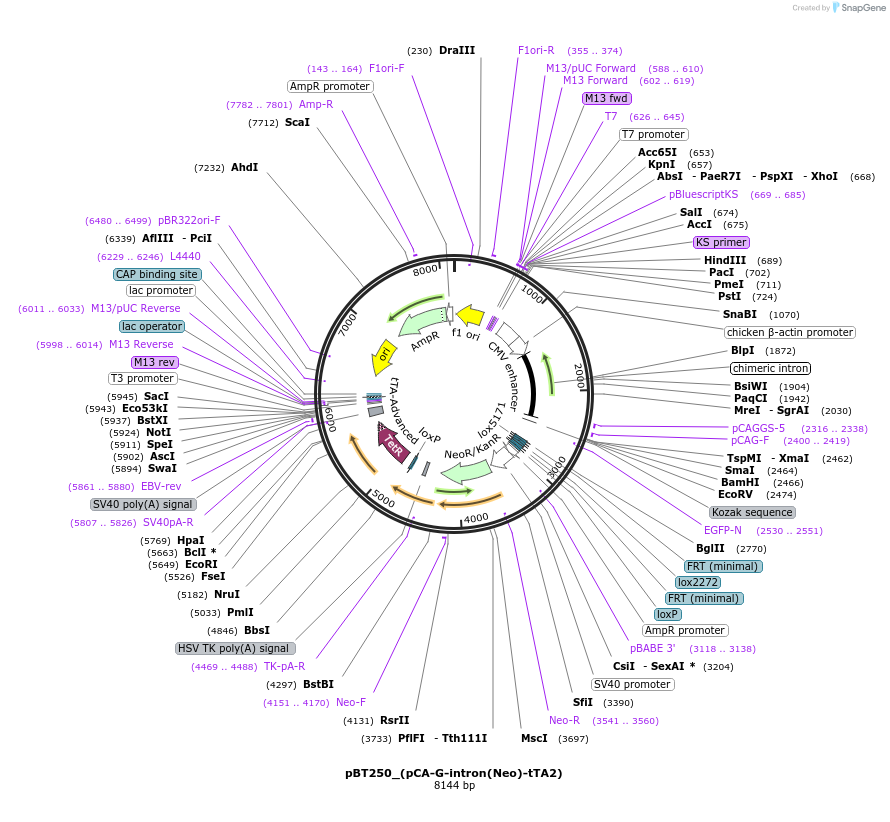
pBT250_(pCA-G-intron(Neo)-tTA2)
Plasmid#36876DepositorInsertsGFP
tTA2
beta-globin intron
Neo
ExpressionMammalianMutationdeleted nucleotides after nucleotide 274, inserti…PromoterCAG (chicken beta actin promoter and CMV enhancer…Available SinceAug. 8, 2012AvailabilityAcademic Institutions and Nonprofits only -
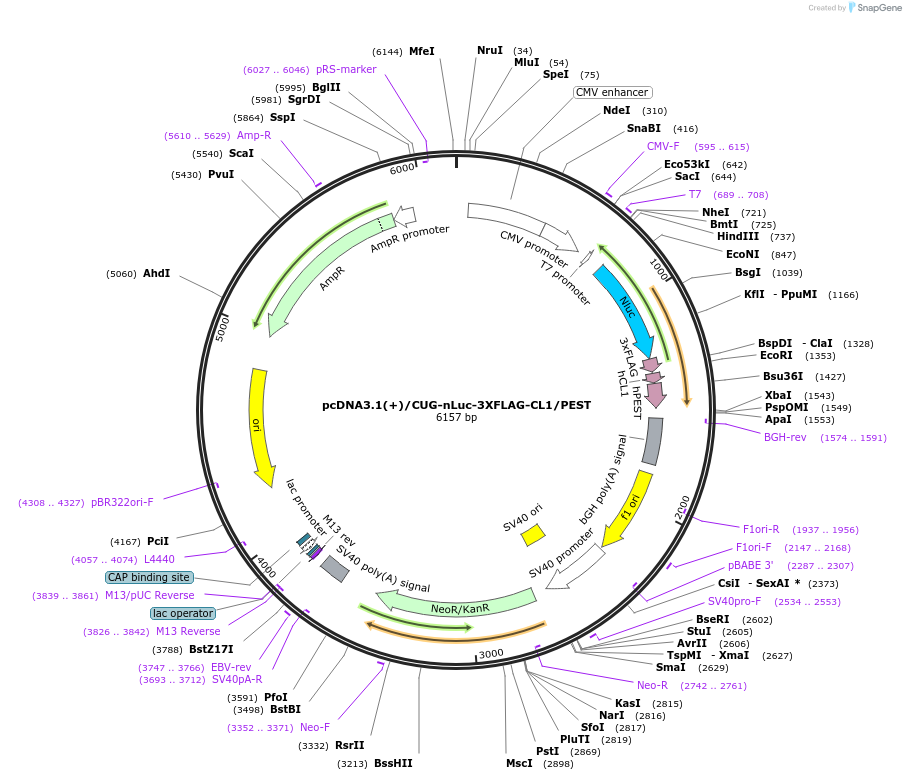
pcDNA3.1(+)/CUG-nLuc-3XFLAG-CL1/PEST
Plasmid#127316PurposeExpress 3xFLAG tagged highly destabilized nanoLuciferase (nLuc) protein from CUG start codonDepositorInsertnanoLuciferase
Tags3xFLAG and CL1/PESTExpressionMammalianPromoterCMVAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
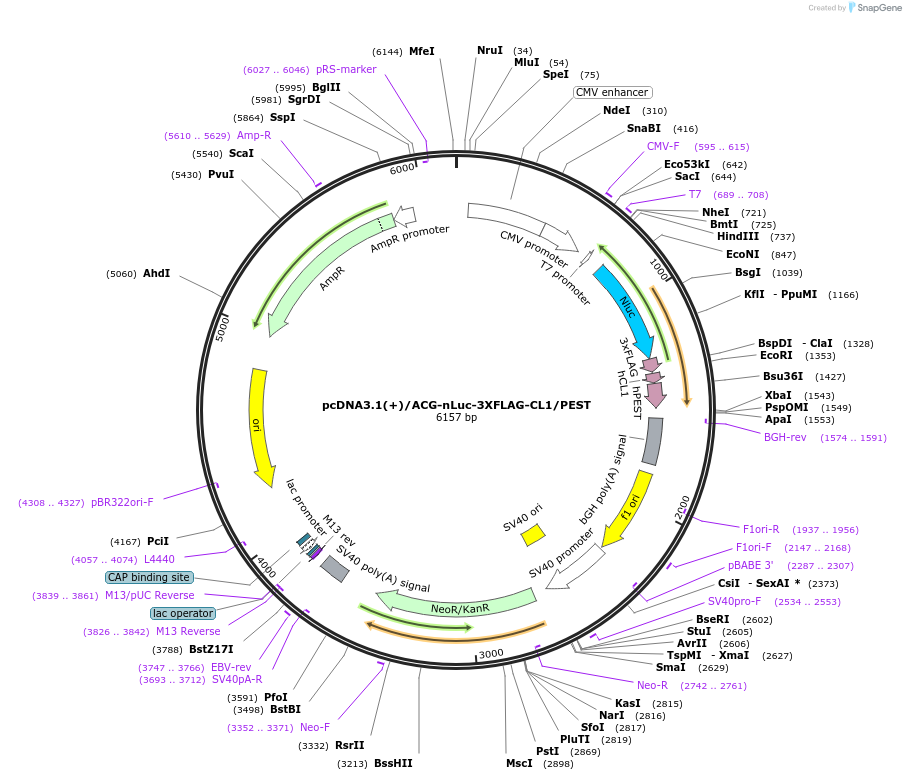
pcDNA3.1(+)/ACG-nLuc-3XFLAG-CL1/PEST
Plasmid#127318PurposeExpress 3xFLAG tagged highly destabilized nanoLuciferase (nLuc) protein from ACG start codonDepositorInsertnanoLuciferase
Tags3xFLAG and CL1/PESTExpressionMammalianPromoterCMVAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
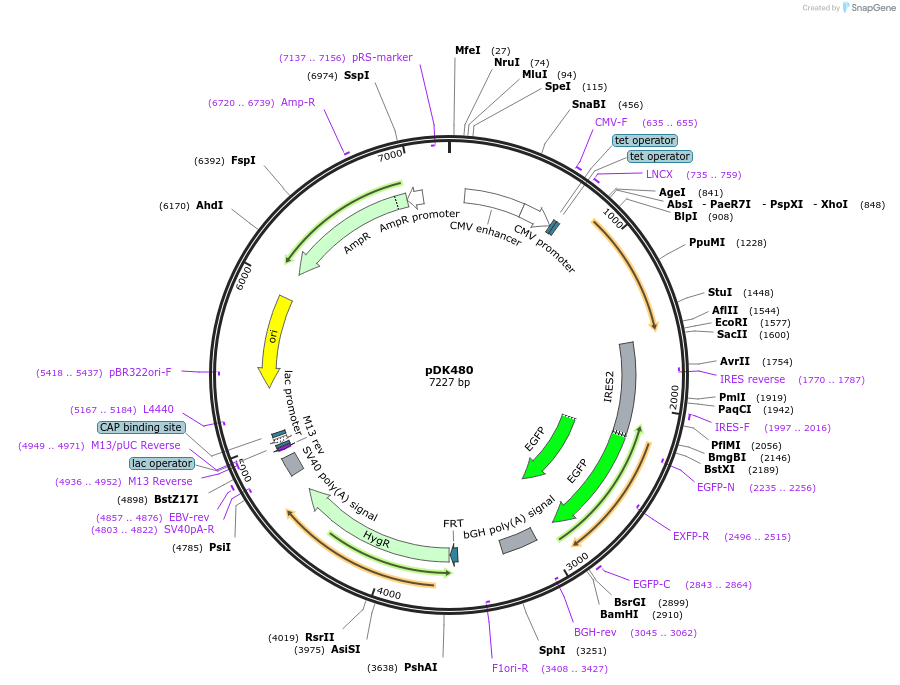
pDK480
Plasmid#134197PurposeMammalian expression of Flip in SKAP and IRES GFPDepositorInsertuntagged AltSKAP (hardened to G5siRNA) (KNSTRN Human)
TagsIRES-GFPExpressionMammalianMutationAlternate Start, G5 siRNA hardenedAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
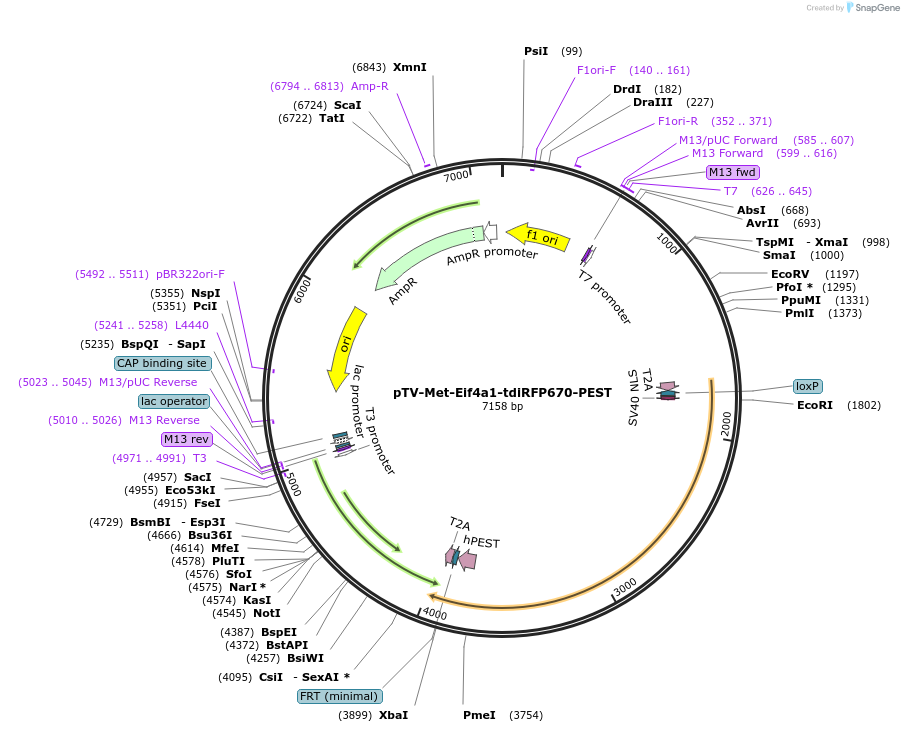
pTV-Met-Eif4a1-tdiRFP670-PEST
Plasmid#122371PurposeTargeting vector for mouse Eif4a1 to insert 2A-tdiRFP670-PEST-2A into immediately downstream of start codon.DepositorInsert2A-tdiRFP-PEST-2A with homology arms for Eif4a1
UseMouse TargetingAvailable SinceJune 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
pCA-G-intron(Neo)-T(LNL)
Plasmid#36907DepositorInsertsGFP
tdTomato-3Myc
beta-globin intron
Neo
Tags3 Myc tagsExpressionMammalianMutationdeleted nucleotides after nucleotide 274, inserti…PromoterCAG (chicken beta actin promoter and CMV enhancer…Available SinceAug. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
pETDUET1-hUCH37 (ISF1)-NFRKB(39-156)
Plasmid#61943PurposeCo-expression of hUCH37 (ISF1) and hNFRKB (39-156) in E.coliDepositorInsertsTags6x HISExpressionBacterialMutationResidues 2-39 deleted resulting in construct star…PromoterT7Available SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
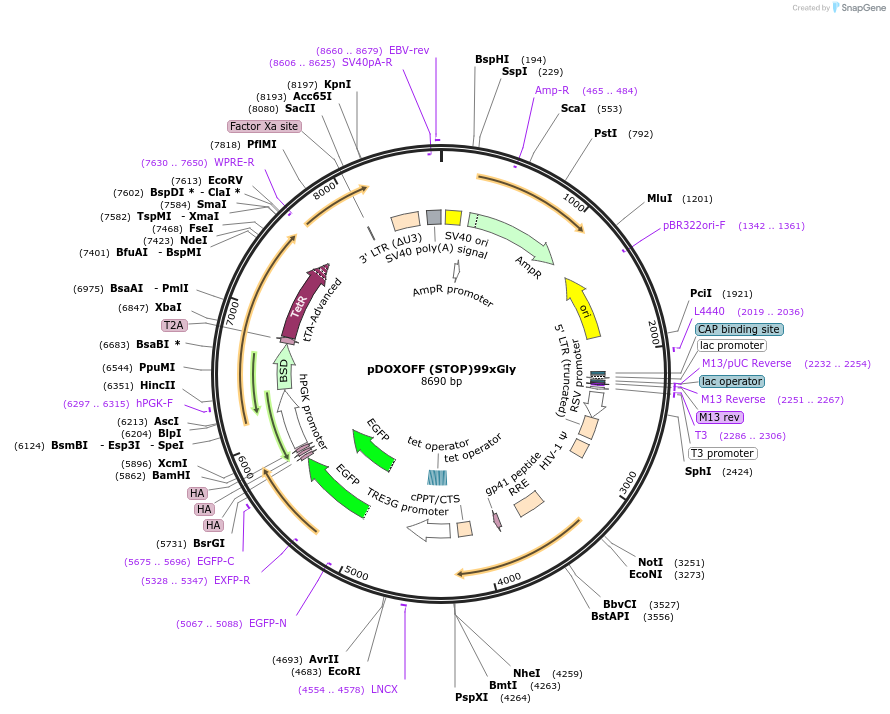
pDOXOFF (STOP)99xGly
Plasmid#211349PurposeDOX-regulated lentiviral expression of 99xGly-EGFP-3xHA lacking initiation codonDepositorInsert(STOP)99xGly-EGFP-3xHA
UseLentiviralTagsEGFP-3xHAMutationStart (ATG) codon replaced with stop (TAG) codonAvailable SinceOct. 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
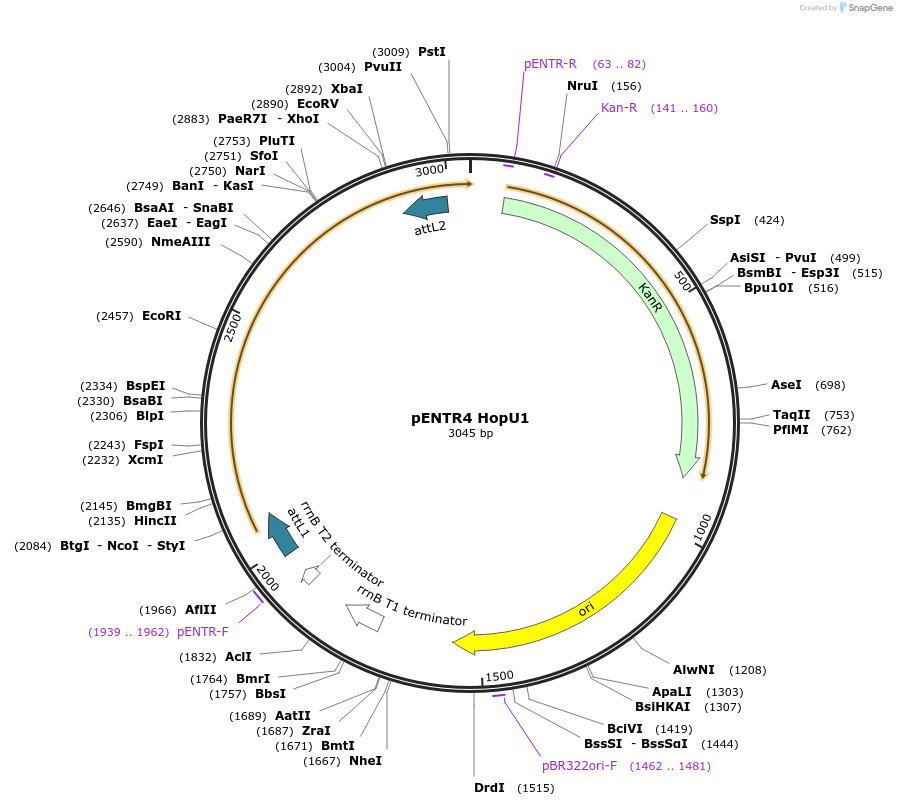
pENTR4 HopU1
Plasmid#228395PurposepENTR4 plasmid with CDS of Pseudomonas syringae pv tomato type III effector HopU1 without stop codon for C-terminal epitope tagsDepositorInsertHopU1
UseGateway-compatible entry vectorMutationAlanine inserted after start Methionine due to Nc…Available SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
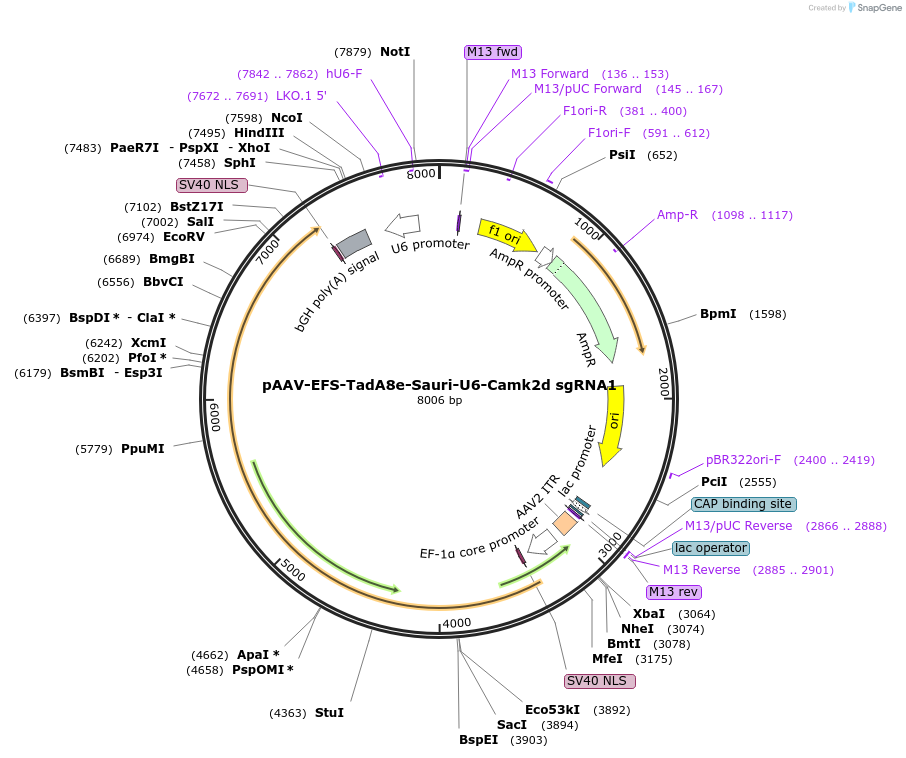
pAAV-EFS-TadA8e-Sauri-U6-Camk2d sgRNA1
Plasmid#220117PurposeExpresses SauriABE8e by EFS promoter and sgRNA targeting murine Camk2d start codon site by U6 promoterDepositorInsertTadA8e, nSauriCas9
UseAAVPromoterEFSAvailable SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
pAAV-MYL2-TadA8e-Sauri-U6-Camk2d sgRNA1
Plasmid#220120PurposeExpresses SauriABE8e by MYL2 promoter and sgRNA targeting murine Camk2d start codon site by U6 promoterDepositorInsertMYL2, TadA, nSauriCas9
UseAAVPromoterMYL2Available SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
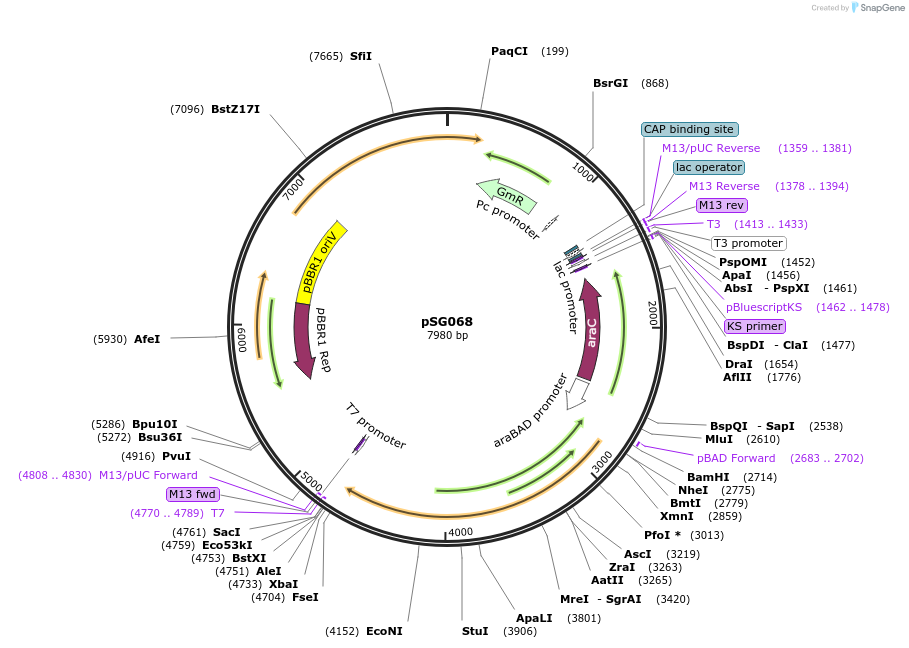
pSG068
Plasmid#214259PurposepJN105 with PaftsH2SD and PaFtsH1 ORF cloned between NheI/XbaIDepositorInsertCore genome PaFtsH1 proteinase from Pseudomonas aeruginosa SG17M
ExpressionBacterialMutation18 nucleotide 5' of the start codon (Shine-D…PromoteraraBAD promoterAvailable SinceOct. 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
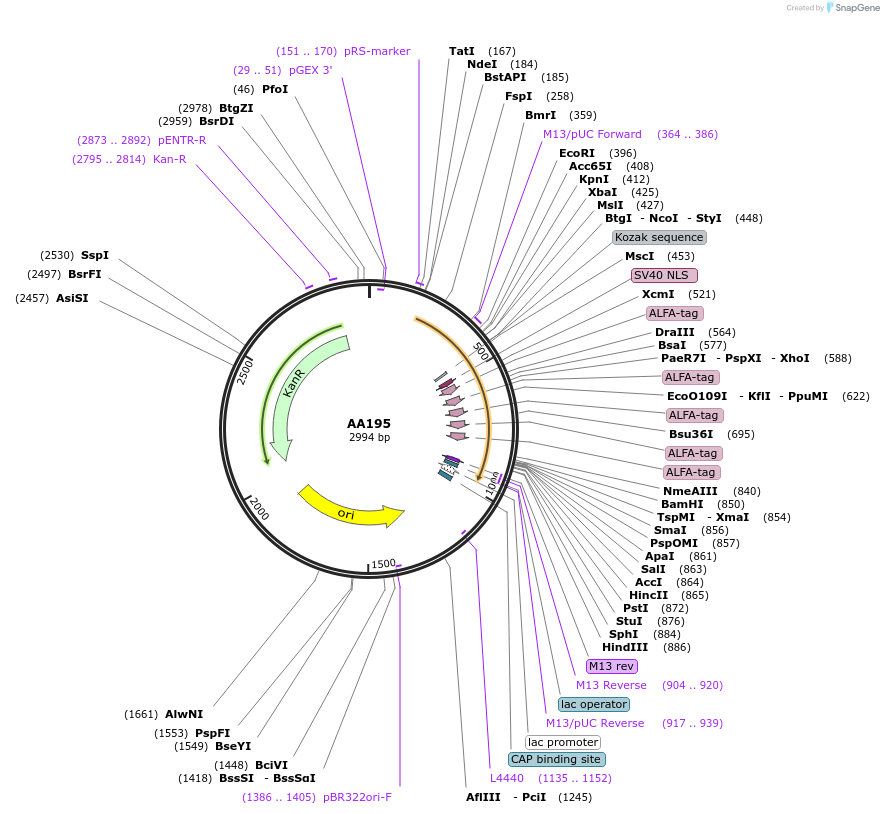
AA195
Plasmid#215984PurposeFragmid fragment: (N' terminus) ALFA tag for recruiting via NbALFADepositorHas ServiceCloning Grade DNAInsertStart; BPNLS_v1.1; ALFA_v1.3; ALFA_v1.2; ALFA_v1.1; ALFA_v1.4; ALFA_v1.5
UseFragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
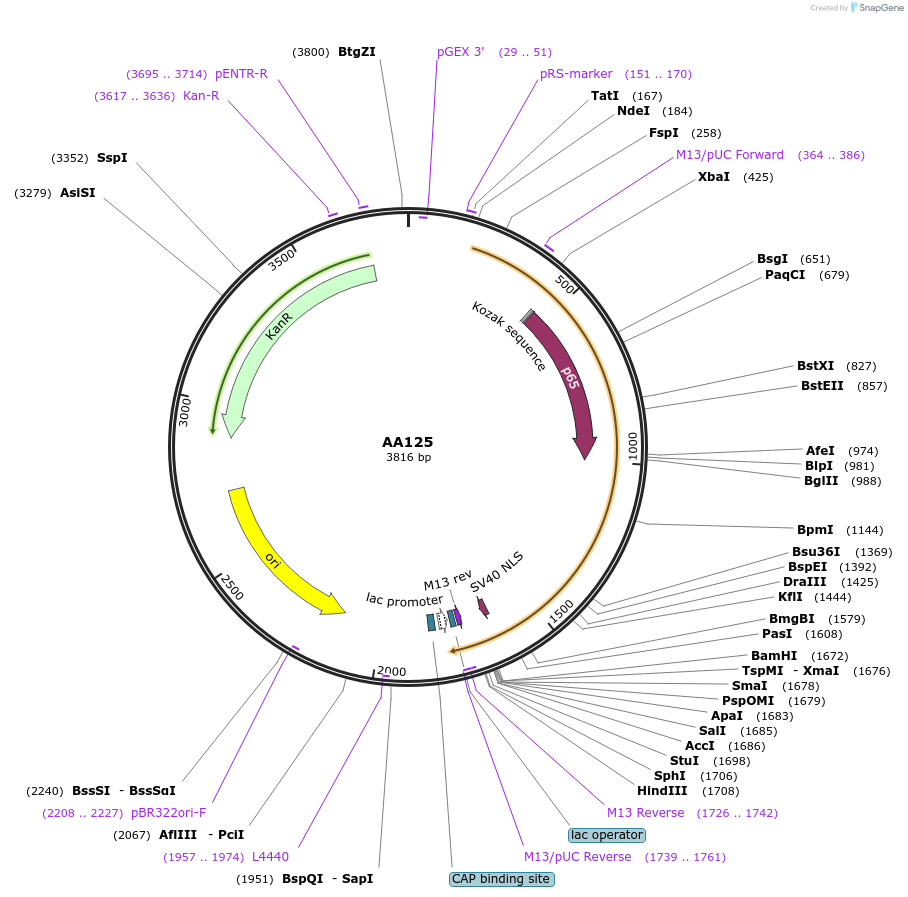
AA125
Plasmid#215982PurposeFragmid fragment: (N' terminus) activator from p65 subunit of mouse NfkbDepositorHas ServiceCloning Grade DNAInsertStart; p65_v1.3; HSF1_v1.2; Linker_v3.1; SV40NLS_v1.6
UseFragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
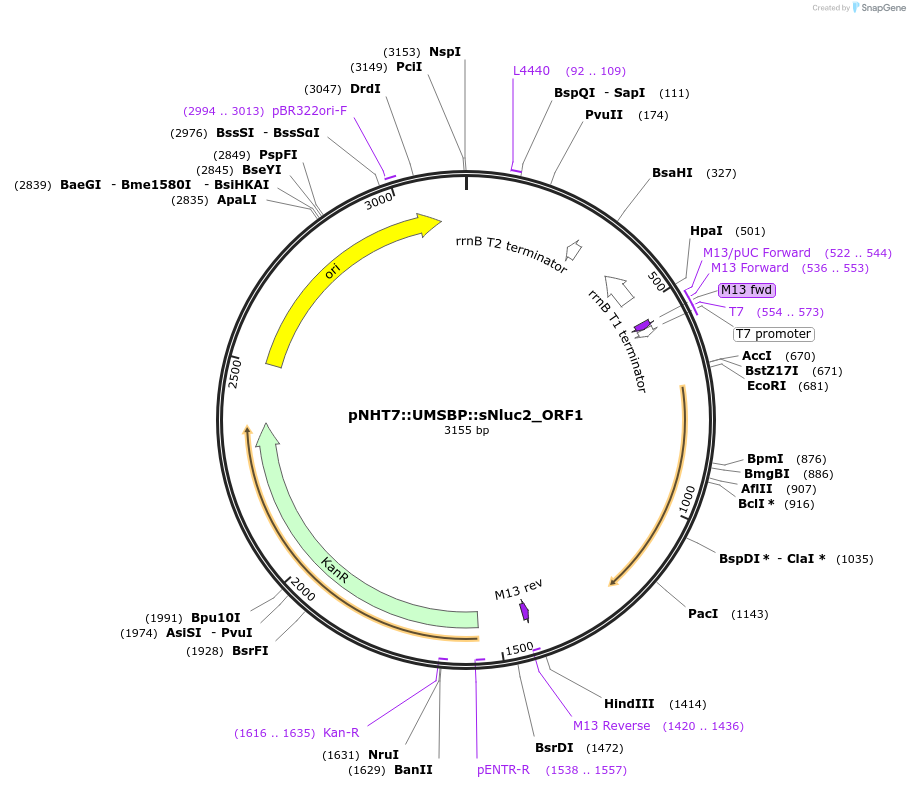
pNHT7::UMSBP::sNluc2_ORF1
Plasmid#186765PurposeAn mRNA production vector encoding planarian codon-optimized sNluc2 with UMSBP 5' and 3' UTRs beginning at the endogenous gene's start codon.DepositorInsertNanoluciferase
PromoterT7Available SinceOct. 27, 2022AvailabilityAcademic Institutions and Nonprofits only