We narrowed to 22,610 results for: ESC
-
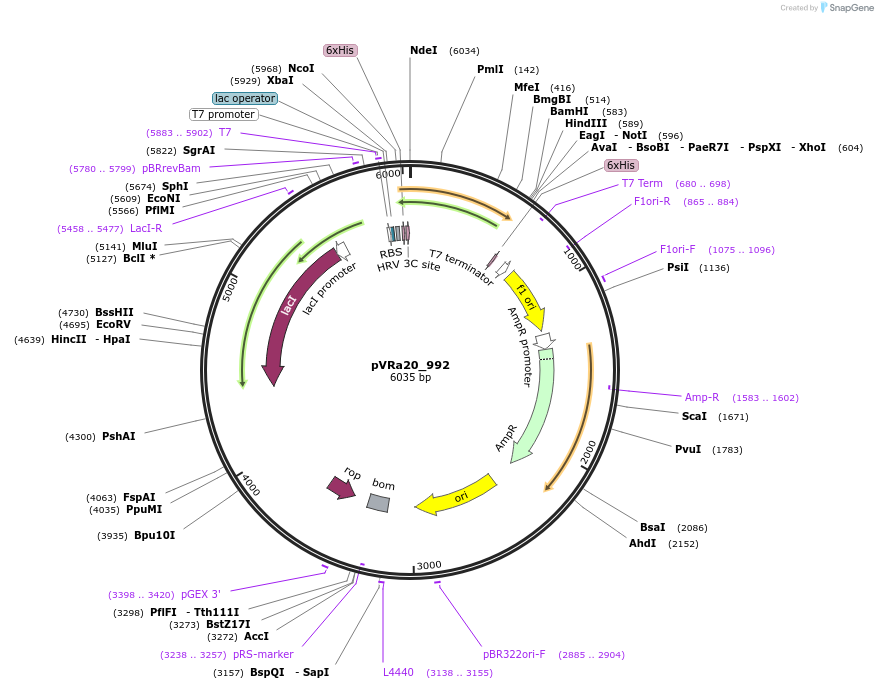
Plasmid#49673DepositorInsertECF20_992
UseSynthetic BiologyTagsHis6-PreScissionExpressionBacterialMutationCodon optimized for E.coliPromoterpT7-lacOAvailable SinceJan. 6, 2014AvailabilityAcademic Institutions and Nonprofits only -
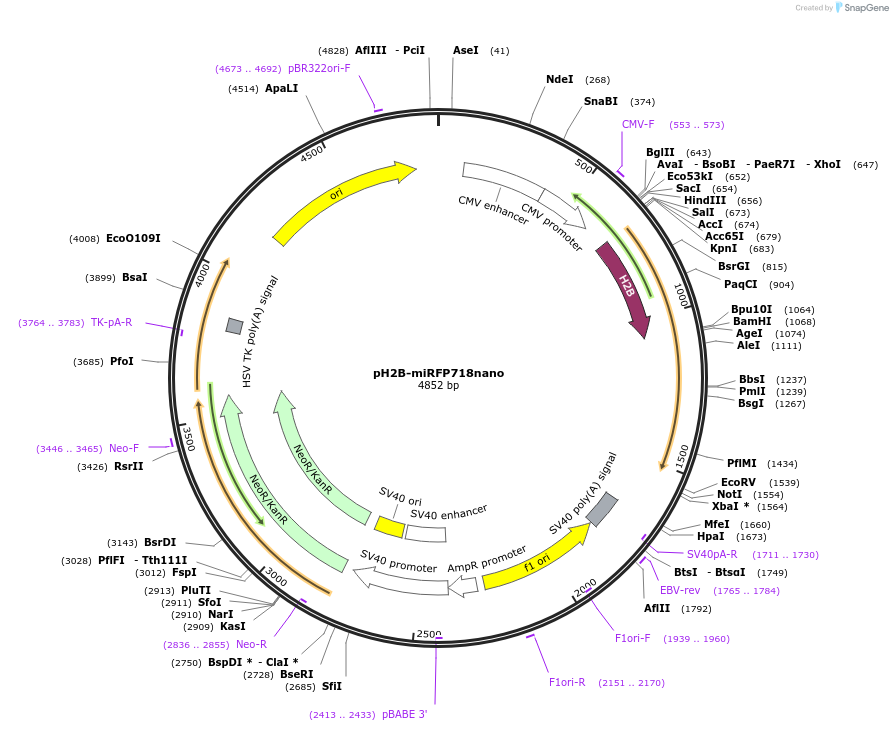
pH2B-miRFP718nano
Plasmid#195750PurposeHistone 2B labelled with near-infrared fluorescent protein miRFP718nanoDepositorInsertH2B-miRFP718nano
TagsmiRFP718nanoExpressionMammalianPromoterCMVAvailable SinceFeb. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
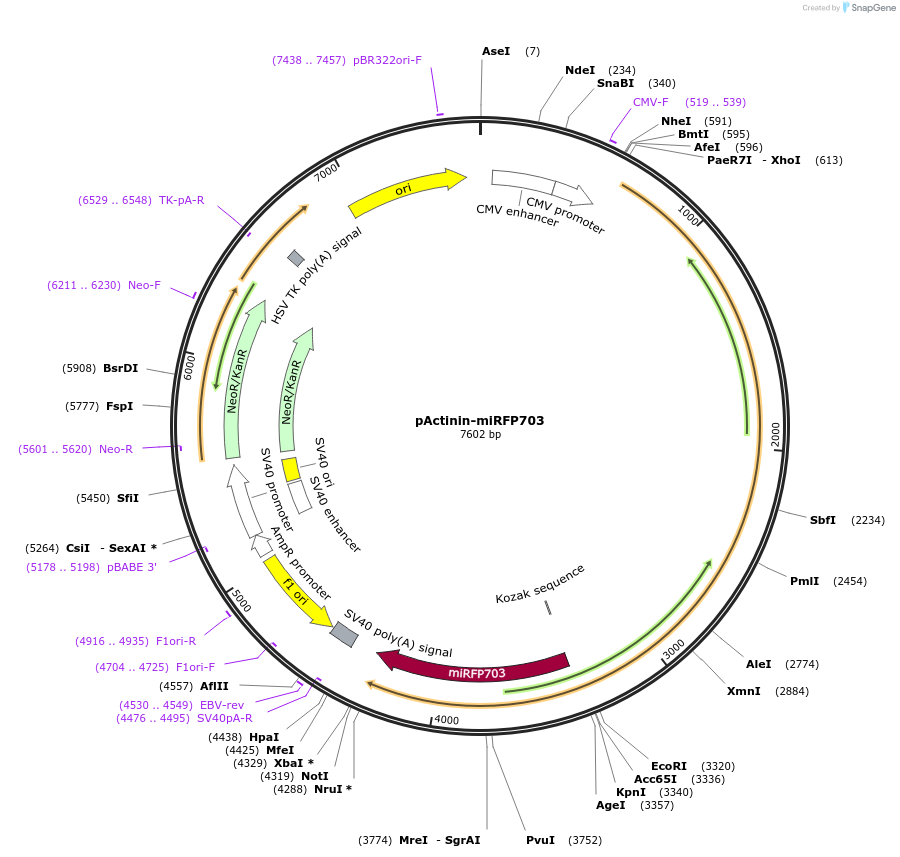
pActinin-miRFP703
Plasmid#79992PurposeAlpha actinin labeling with near-infrared fluorescent protein miRFP703DepositorAvailable SinceJuly 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
Orange Nano-lantern(Ca2+)/pcDNA3
Plasmid#71346PurposeExpress Orange Nano-lantern-based luminescent Ca2+ indicator in mammalian cellDepositorInsertOrange Nano-lantern(Ca2+)
ExpressionMammalianAvailable SinceDec. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
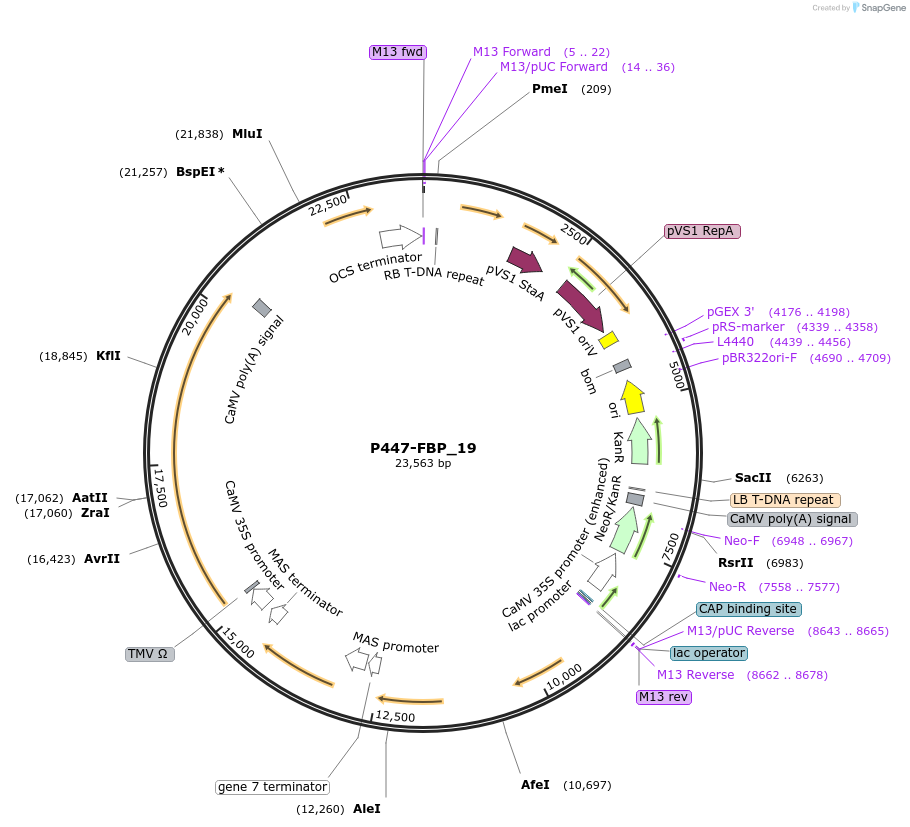
P447-FBP_19
Plasmid#139710PurposeT-DNA encoding Fungal Bioluminescent Pathway Components, complete pathway, Luz promoter is ABA responsiveDepositorInsertpSlH4:NnCPH:tSlH4, pSlRBCS2:AsNpga:tAtuG7, pAtuMas:NnH3H:tAtuMas, p35S:NnHisps:t35S, pAtRAB18:NnLuz:tAtuOcs
UseLuciferaseExpressionPlantPromotervariousAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
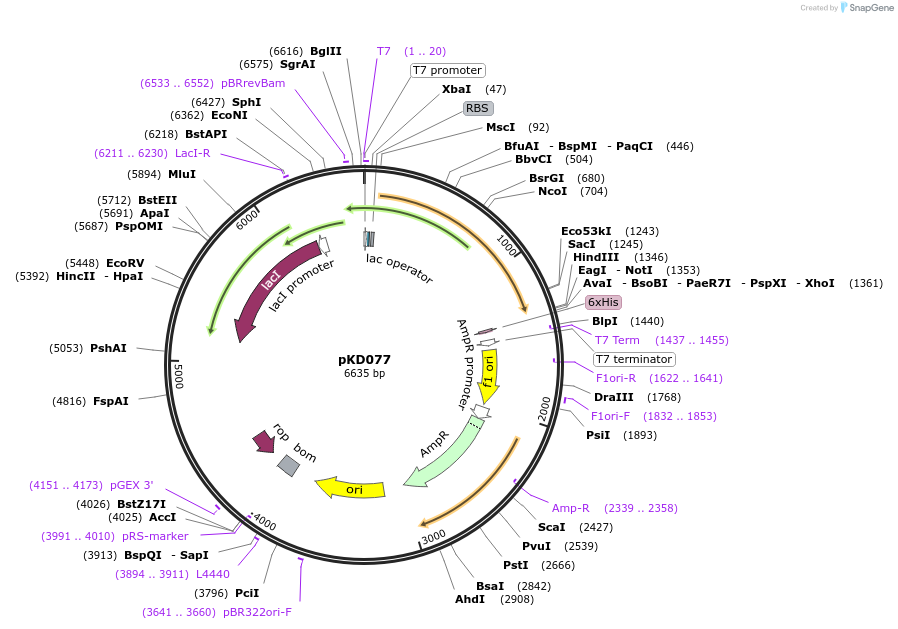
pKD077
Plasmid#136473PurposeSSB-mTur2 IDL fusion expression plasmidDepositorInsertSSB-mTur2 (ssb E. coli)
TagsmTurquoise2ExpressionBacterialMutationmTurquoise2 inserted between Phe148 and Ser149PromoterT7 promoterAvailable SinceApril 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
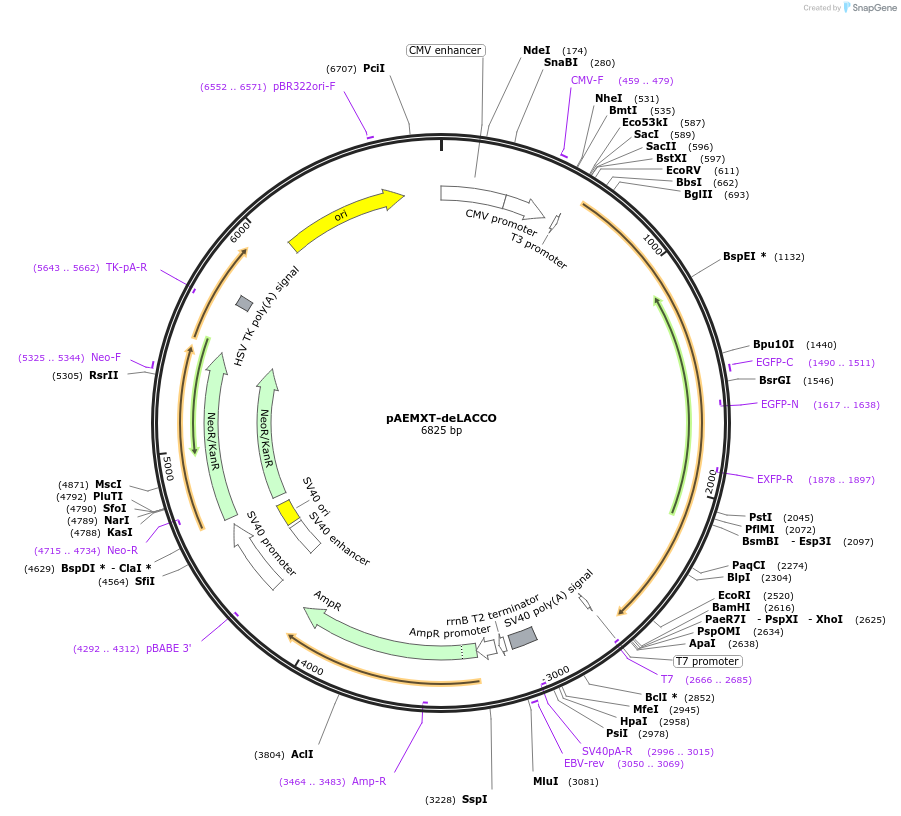
pAEMXT-deLACCO
Plasmid#167947PurposeFluorescent biosensor control for extracellular L-lactateDepositorInsertCD59-deLACCO-GPI
ExpressionMammalianPromoterCMVAvailable SinceApril 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
Orange Nano-lantern/pcDNA3
Plasmid#67661Purposeorange luminescent protein for high-speed single-cell and whole body imagingDepositorInsertOrange Nano-lantern
ExpressionMammalianAvailable SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
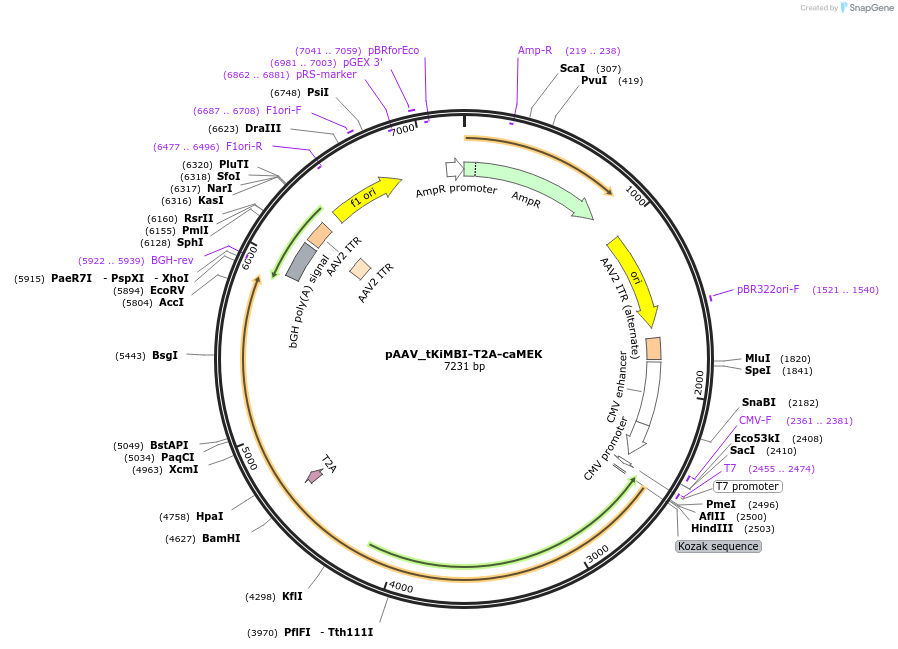
pAAV_tKiMBI-T2A-caMEK
Plasmid#199578PurposeExpresses tKiMBI and caMEK in an AAV vectorDepositorInsertsERK tdTomato-Kinase-Modulated Bioluminescent Indicator
constitutively active MEK
UseAAVExpressionMammalianPromoterCMVAvailable SinceJune 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
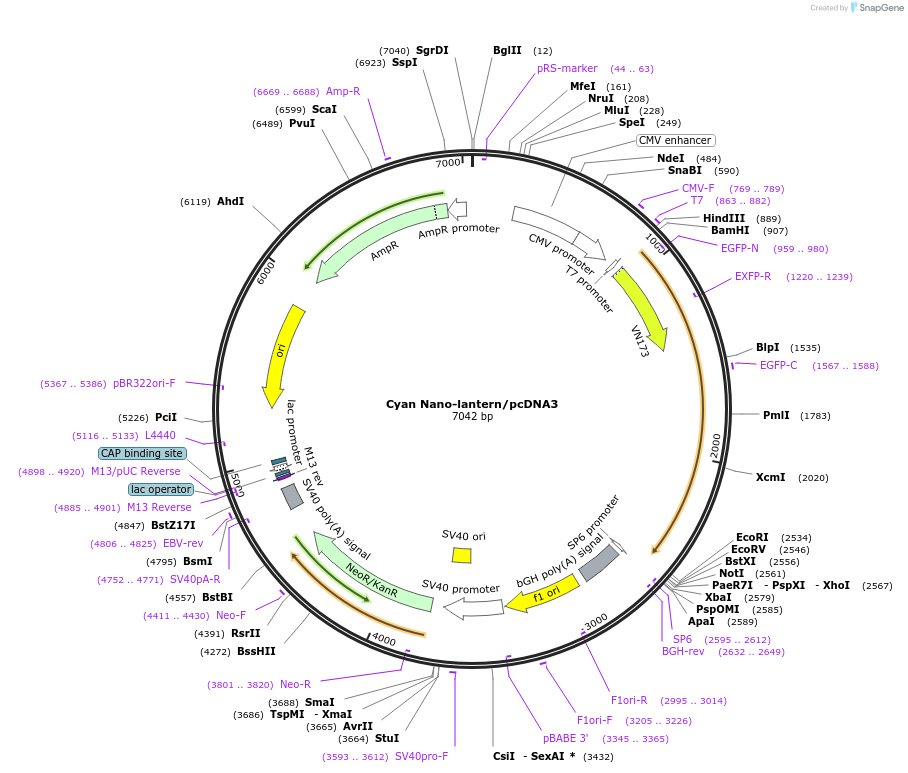
Cyan Nano-lantern/pcDNA3
Plasmid#67660Purposecyan luminescent protein for high-speed single-cell and whole body imagingDepositorInsertCyan Nano-lantern
ExpressionMammalianAvailable SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
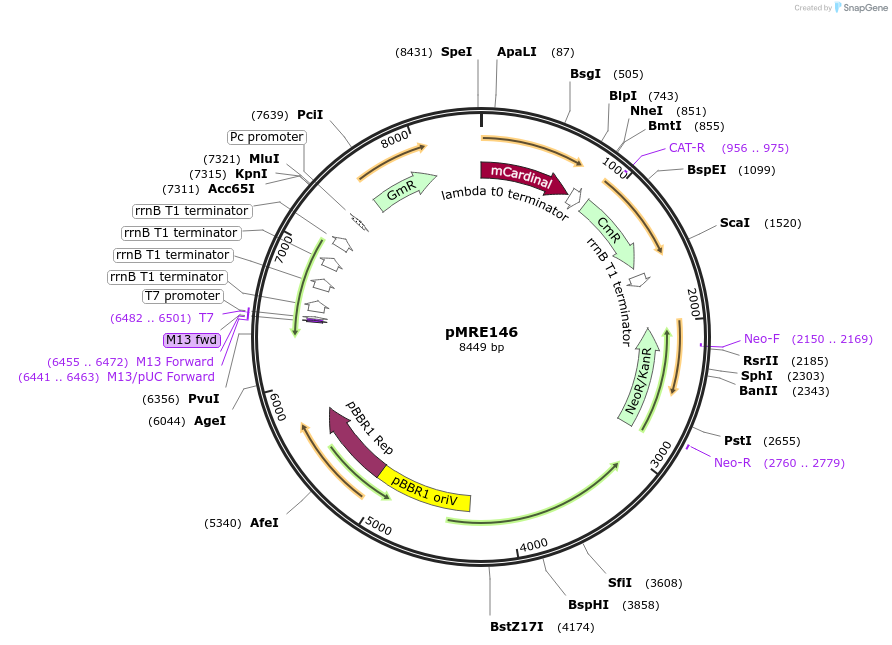
pMRE146
Plasmid#118498Purposebroad-host range plasmid to constitutively express fluorescent protein genes in bacteriaDepositorInsertmCardinal
ExpressionBacterialAvailable SinceNov. 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
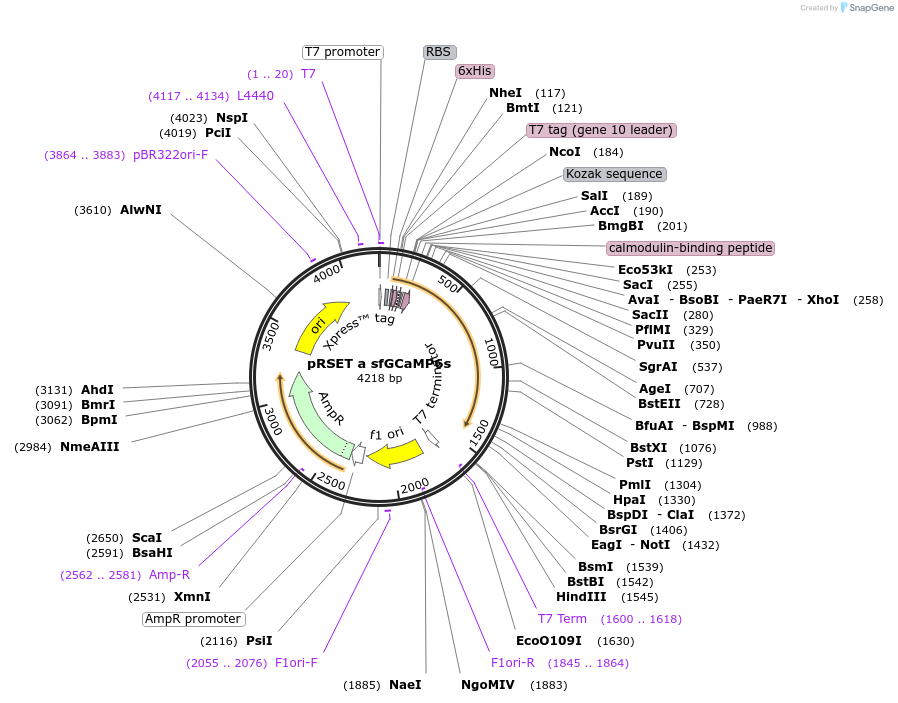
pRSET a sfGCaMP6s
Plasmid#100022PurposeBacterial expression of fluorescent reporter for calcium signaling, based off of GCaMP6s. cpEGFP replaced with a superfolder cpGFP variant.DepositorInsertsfGCaMP6s
Tags6x HIS tag and Xpress_EK tagExpressionBacterialMutationcpEGFP replaced with superfolder cpGFPPromoterT7Available SinceOct. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
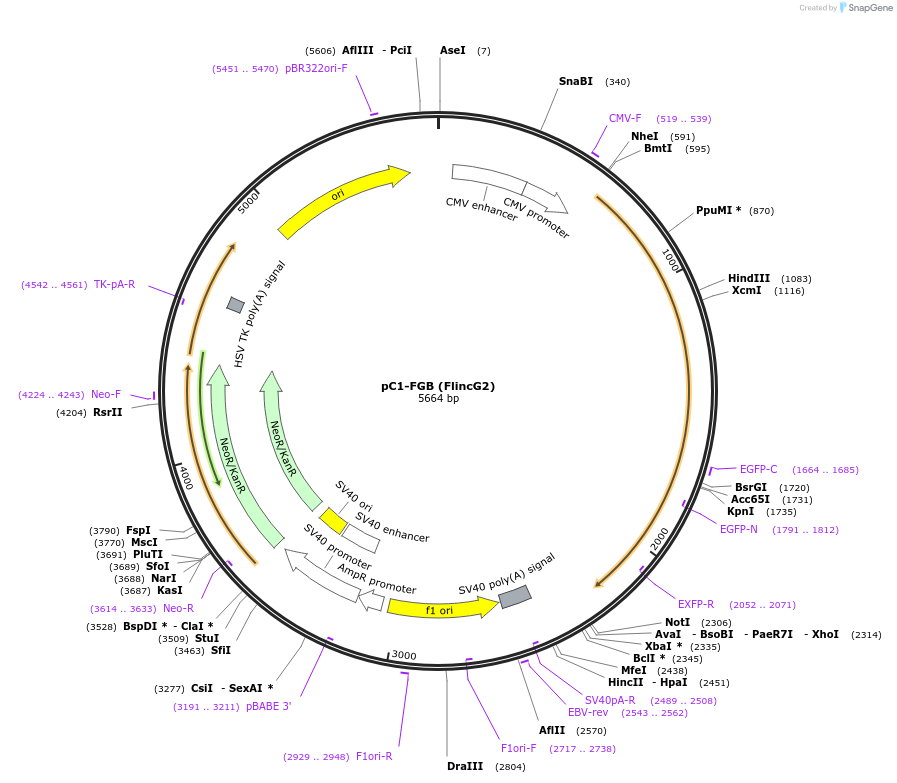
pC1-FGB (FlincG2)
Plasmid#49204PurposeFluorescent reporter for cGMPDepositorAvailable SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
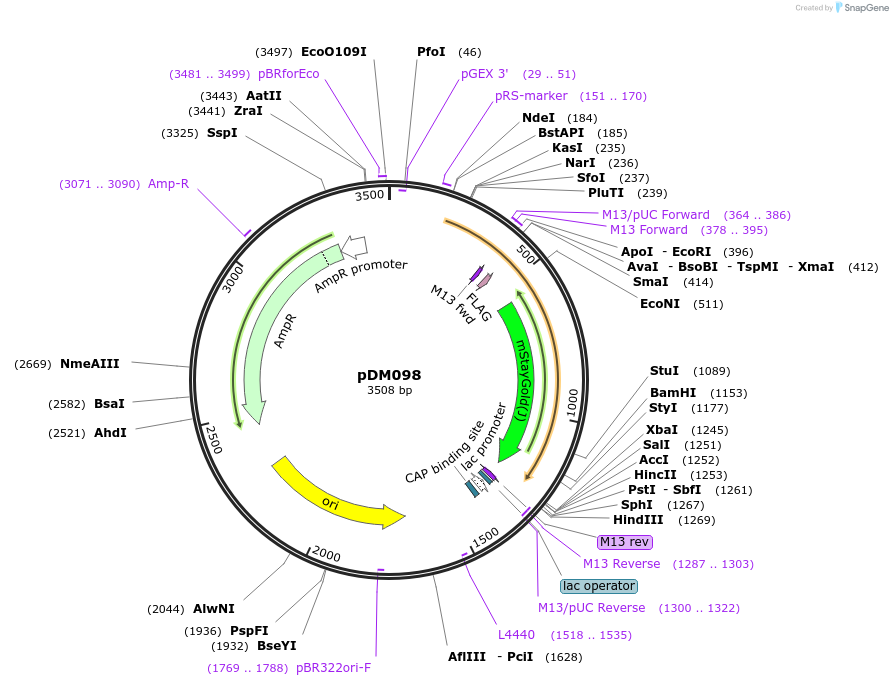
pDM098
Plasmid#216818PurposeAspergillus nidulans codon-adjusted mStayGold fluorescent protein, includes linker for C-terminal tagging.DepositorInsertmStayGold
TagsFLAG-(SGGS)x2-XTEN16-(GGGGS)x3ExpressionBacterialMutationAspergillus nidulans codon-adjustedAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
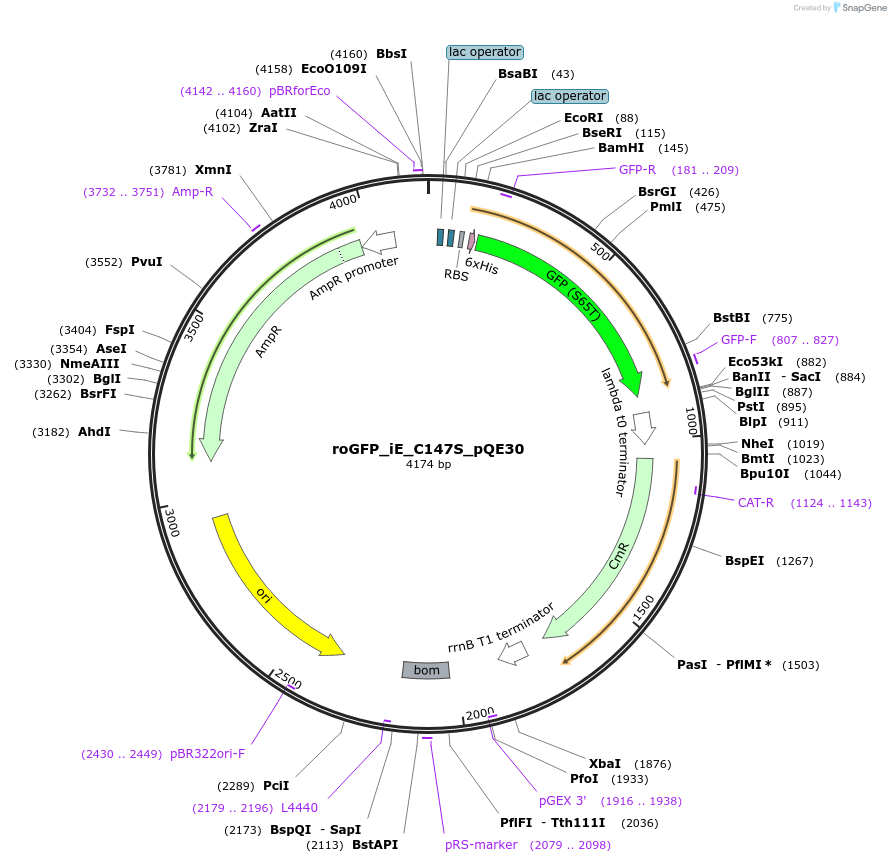
roGFP_iE_C147S_pQE30
Plasmid#48631PurposeBacterial expression vector encoding for the 6XHis tagged insensitive variant of fluorescent redox probe roGFPiE with Cysteine 147 to Serine mutation (roGFP C147S, E147b, C204)DepositorInsertroGFP1
TagsHisTagExpressionBacterialMutationinsertion E147B, C147SAvailable SinceOct. 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
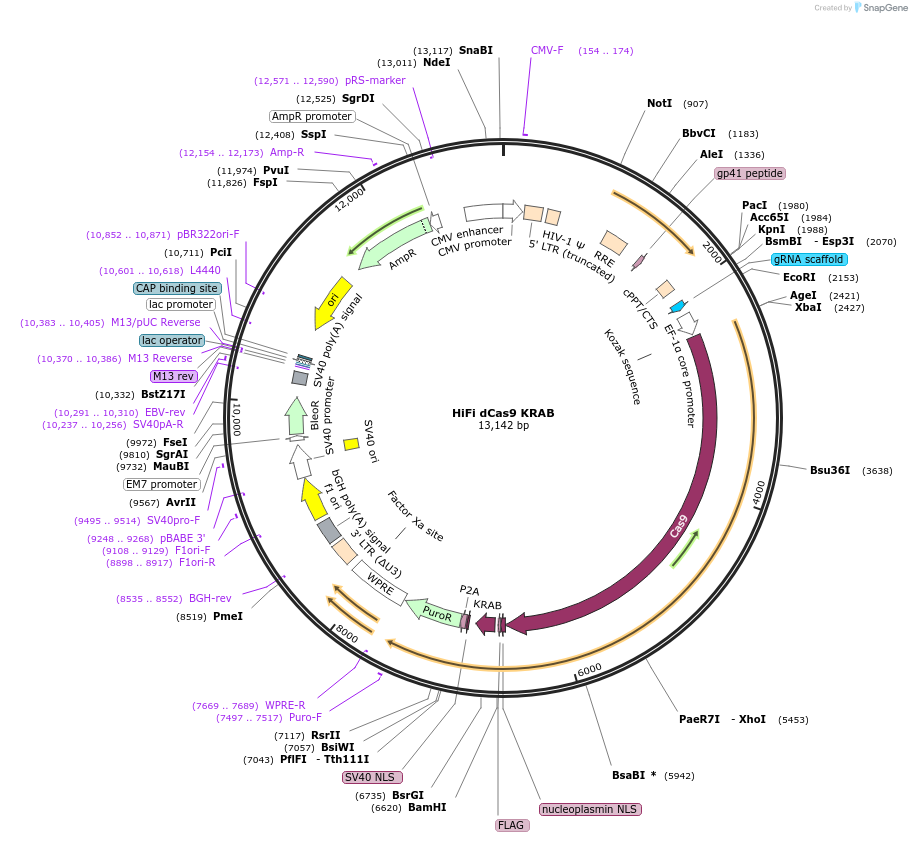
HiFi dCas9 KRAB
Plasmid#188500PurposeExpresses FLAG tagged HiFi dCas9 KRABDepositorInsertdCas9
UseCRISPR and LentiviralExpressionMammalianMutationD10A/R691A/H840APromoterEF1aAvailable SinceSept. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
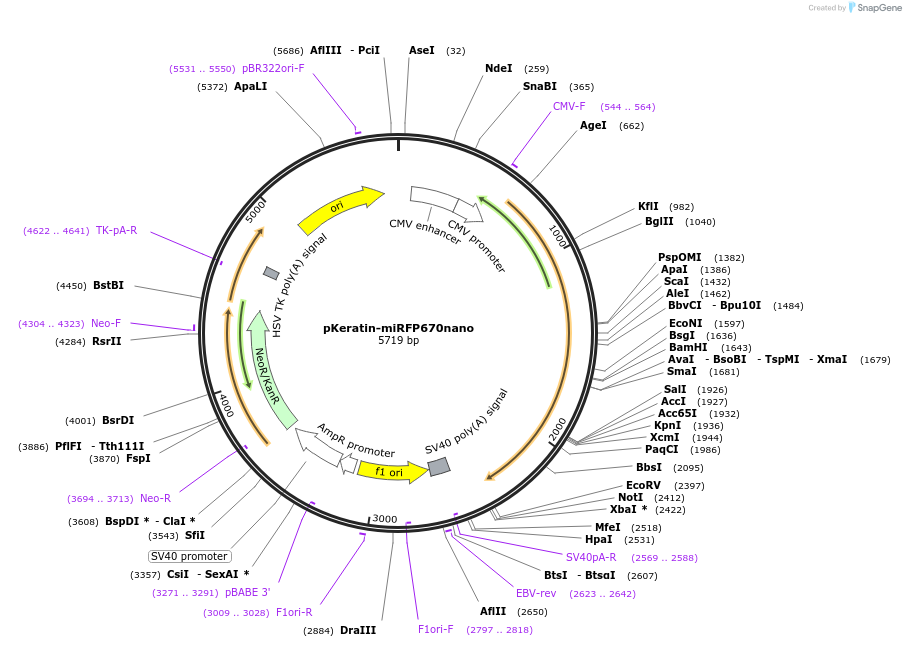
pKeratin-miRFP670nano
Plasmid#127437PurposeKeratin labeling with near-infrared fluorescent protein miRFP670nanoDepositorInsertKeratin-miRFP670nano
TagsmiRFP670nanoExpressionMammalianMutationKeratin V282L - please see depositor comments bel…PromoterCMVAvailable SinceJuly 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
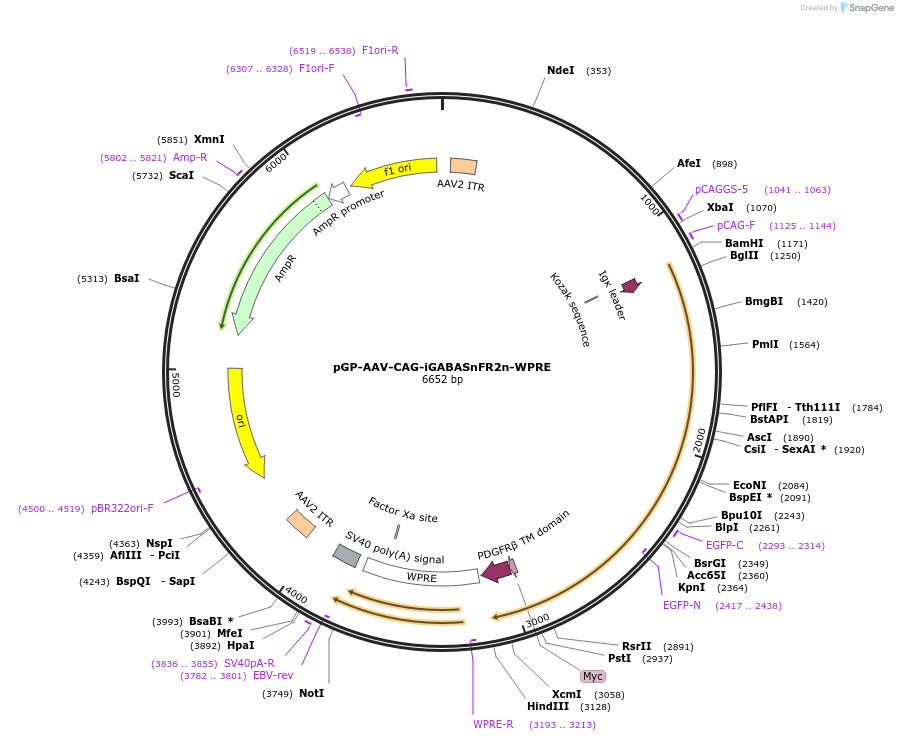
pGP-AAV-CAG-iGABASnFR2n-WPRE
Plasmid#218884PurposeAAV-mediated expression of improved GABA sensor (negative change in fluorescence)DepositorInsertiGABASnFR2n
UseAAVExpressionMammalianMutationS99A F104H R168PPromoterCAGAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
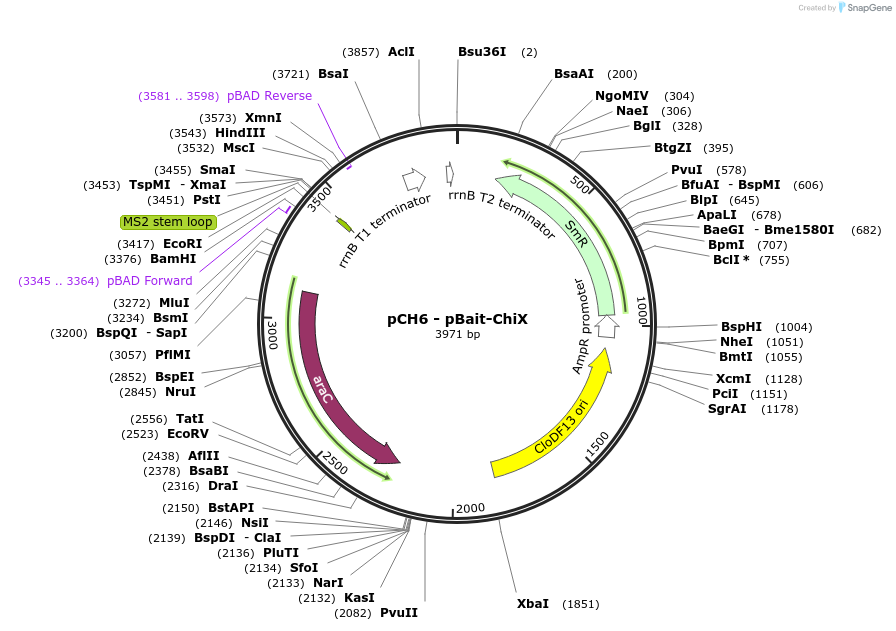
pCH6 - pBait-ChiX
Plasmid#174662Purposearabinose-induced synthesis of a hybrid RNA containing a single MS2 hairpin followed by E coli ChiX inserted between XmaI and HindIII sites. Insert can be replaced with RNA bait of interest.DepositorInsertChiX
Tags1xMS2 hairpinExpressionBacterialPromoterpBadAvailable SinceMay 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
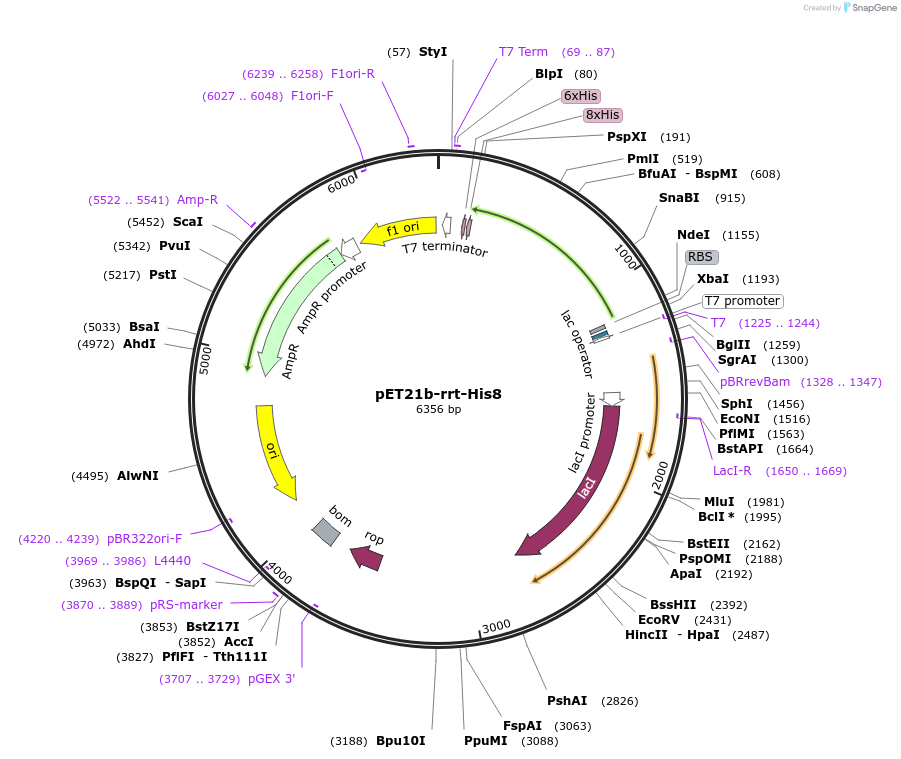
pET21b-rrt-His8
Plasmid#194088PurposeExpresses Ec86 reverse transcriptase in Escherichia coliDepositorInsertEc86 rrt
TagsHis8PromoterT7Available SinceMarch 1, 2023AvailabilityAcademic Institutions and Nonprofits only