We narrowed to 5,234 results for: LucY
-
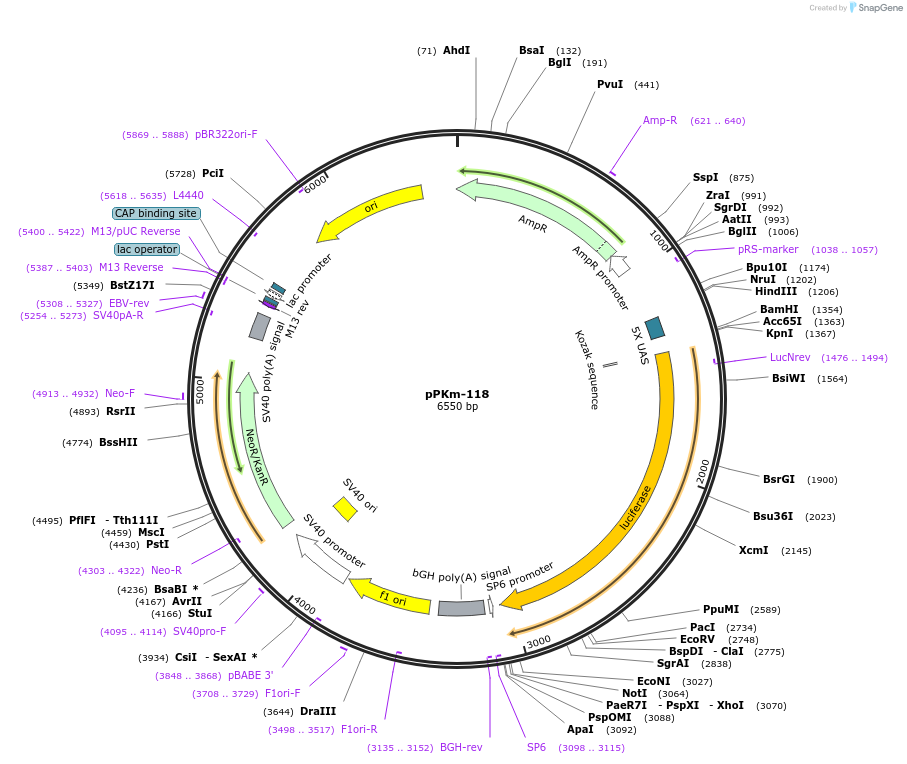
Plasmid#90491PurposeExpresses pcDNA3 - CMVmin - 5X UAS - pFR LuciferaseDepositorInsertpFR Luciferase Fluc
ExpressionMammalianAvailable SinceMarch 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
pGL4 mIL-17 1.1kb promoter
Plasmid#20125DepositorInsertmIL-17 (Il17a Mouse)
UseLuciferaseAvailable SinceJan. 16, 2009AvailabilityAcademic Institutions and Nonprofits only -
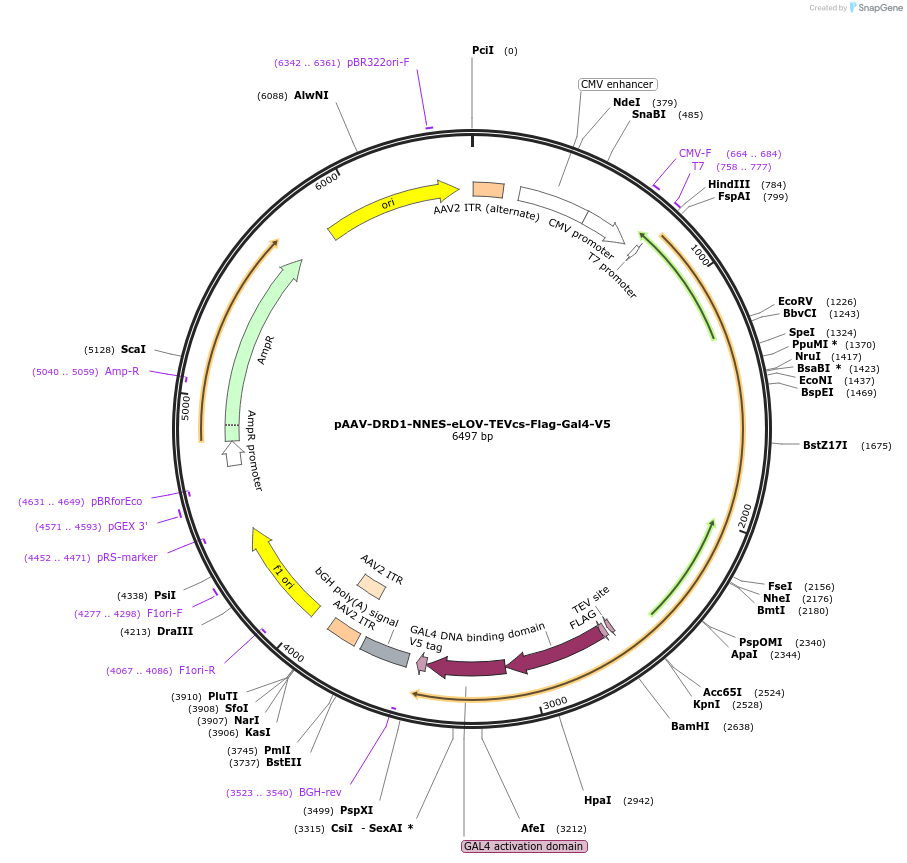
pAAV-DRD1-NNES-eLOV-TEVcs-Flag-Gal4-V5
Plasmid#125227PurposeExpresses SPARK DRD1 transmembrane component in mammalian cellsDepositorInsertDRD1-NNES-eLOV-TEVcs-Flag-Gal4-V5
UseAAVTagsFlag, V5ExpressionMammalianPromoterCMVAvailable SinceMay 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
-111 hTF m1
Plasmid#15448DepositorAvailable SinceJuly 27, 2007AvailabilityAcademic Institutions and Nonprofits only -
pCheck2 Dest (R1-R2) #BGV129
Plasmid#48955PurposeDestination dual luciferase reporter plasmid for checking effectiveness of shRNA. Compatible with standard AttL1-AttL2 Gateway Entry vectors.DepositorTypeEmpty backboneUseLuciferase; Gateway cloning vectorPromoterSV40Available SinceJune 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
-962 human cyclin D1 promoter EtsB site mutant pGL3Basic
Plasmid#32730DepositorInsertCCND1 (CCND1 Human)
UseLuciferaseMutation-778 EtsB binding site AGGAA deletionPromoter-962 CCND1Available SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
bdSUMO-R9-GFP-HiBit
Plasmid#170990PurposeFor bacterial expression of bdSUMO tagged muGFP with R9 peptide and NanoLuc complementation peptide variant #86DepositorInsert14x-His-bdSUMO-R9-muGFP-HiBiT
UseLuciferaseExpressionBacterialPromoterT7Available SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
-962 human cyclin D1 promoter EtsC site mutant pGL3Basic
Plasmid#32731DepositorInsertCCND1 promoter (CCND1 Human)
UseLuciferaseMutation-424 EtsC site TTCCT deletionPromoter-962 CCND1 promoterAvailable SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
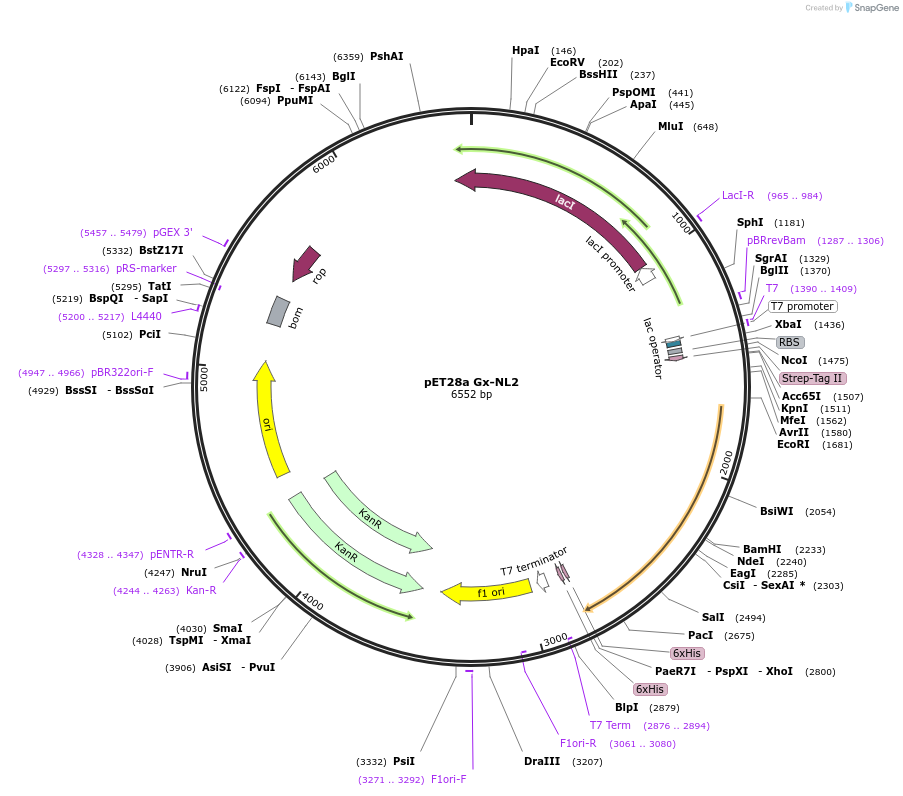
pET28a Gx-NL2
Plasmid#169107PurposeExpresses protein G fused to 2 copies of NanoLuc. The protein G domain contains an amber stop-codon at position 24 for incorporation of the unnatural amino acid para-benzoylphenylalanine.DepositorInsertprotein G - NanoLuc - NanoLuc
Tagshis-tag and strep-tagExpressionBacterialMutationAla 24 stopPromoterT7Available SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
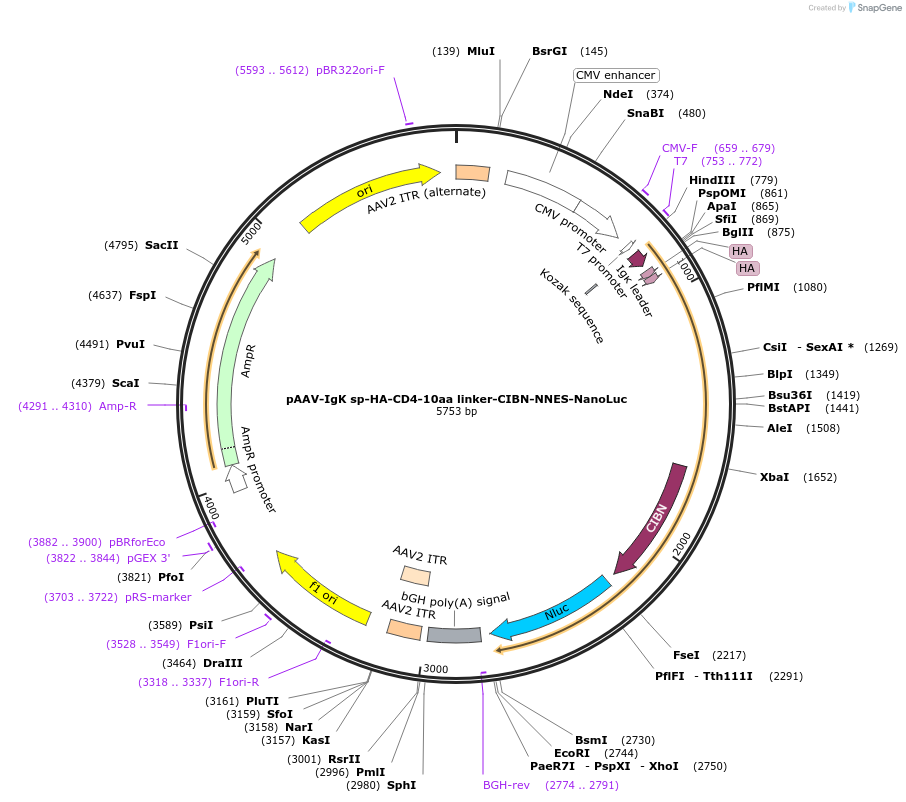
pAAV-IgK sp-HA-CD4-10aa linker-CIBN-NNES-NanoLuc
Plasmid#125229PurposeExpresses transmembrane intracellular NanoLuc in mammalian cellsDepositorInsertIgK sp-HA-CD4-10aa linker-CIBN-NNES-NanoLuc
UseAAVTagsHAExpressionMammalianPromoterCMVAvailable SinceMay 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
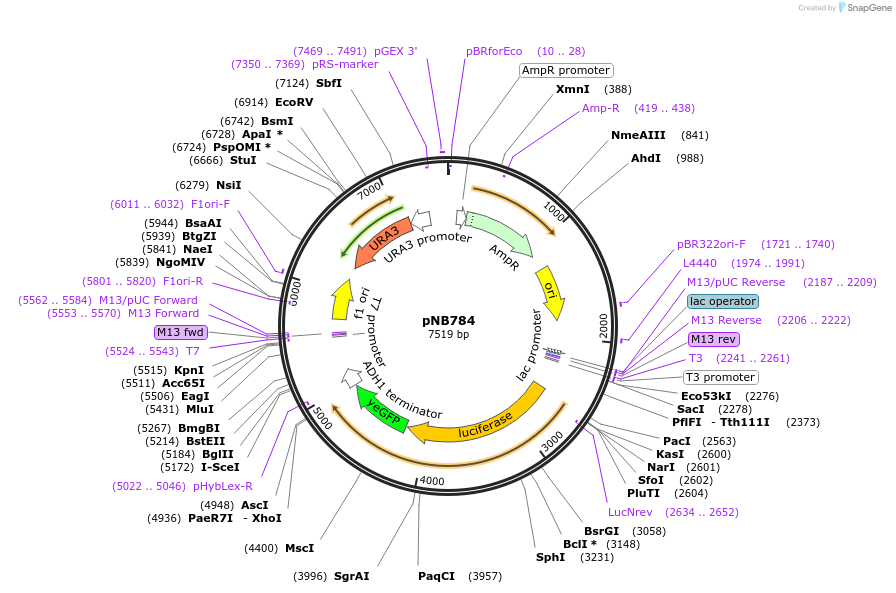
pNB784
Plasmid#60163PurposeFirefly luciferase (Promega) yeast codon-optimized yellow fluorescent protein (yEVenus) fusion driven by SIC1 promoter.DepositorInsertFLuc-yEVenus
ExpressionYeastMutationFLuc has A4V & S504G (no functional effect)PromoterSIC1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
-962 human cyclin D1 promoter TCF (1)(2) sites mutant pGL3Basic
Plasmid#32734DepositorInsertCCND1 promoter (CCND1 Human)
UseLuciferaseMutation-75 TCF(1) and -68 TCF(2) - CTTTGATCTTTGCT deleti…Promoter-962 CCND1Available SinceJan. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
pGL3-Control-E2F-3'UTR-Mut
Plasmid#21171DepositorInsertE2F 3'UTR Mutant (E2F1 Human)
UseLuciferaseExpressionMammalianMutation2 miR binding sites mutated. Changed from GCACTTT…Available SinceJune 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
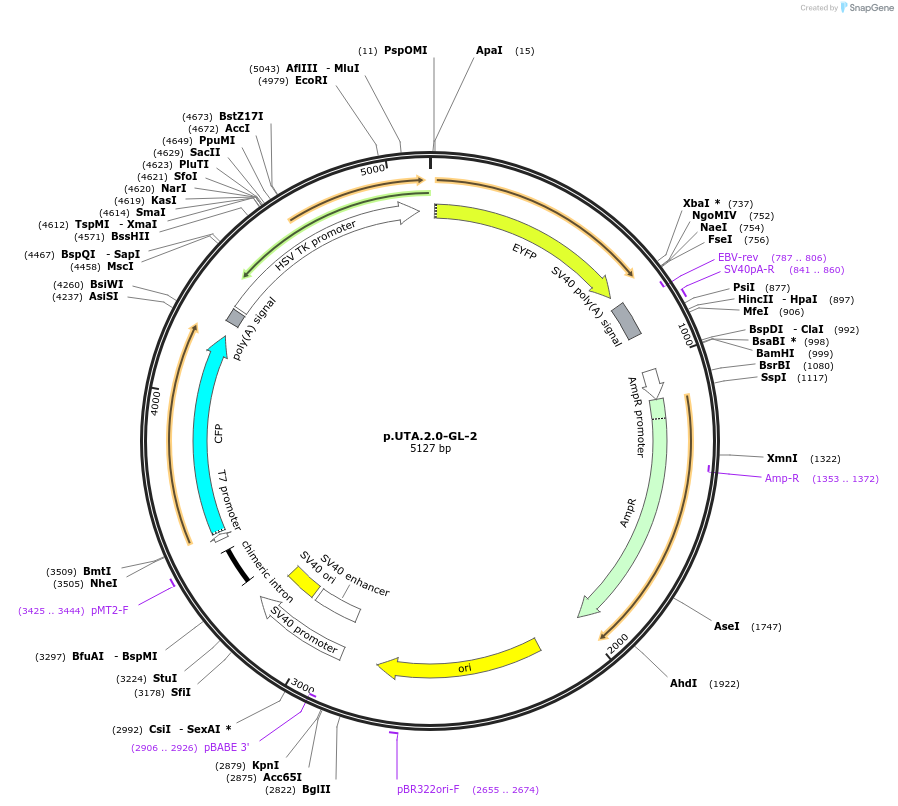
p.UTA.2.0-GL-2
Plasmid#82440PurposeDual Fluorescent endogenous miRNA sensor. Perfect complementary region of an siRNA control targeting firefly luciferase encoded by the pGL2DepositorInsertLuciferase
MutationThe complementary region for GL2 siRNA control wa…Available SinceMarch 24, 2017AvailabilityAcademic Institutions and Nonprofits only -
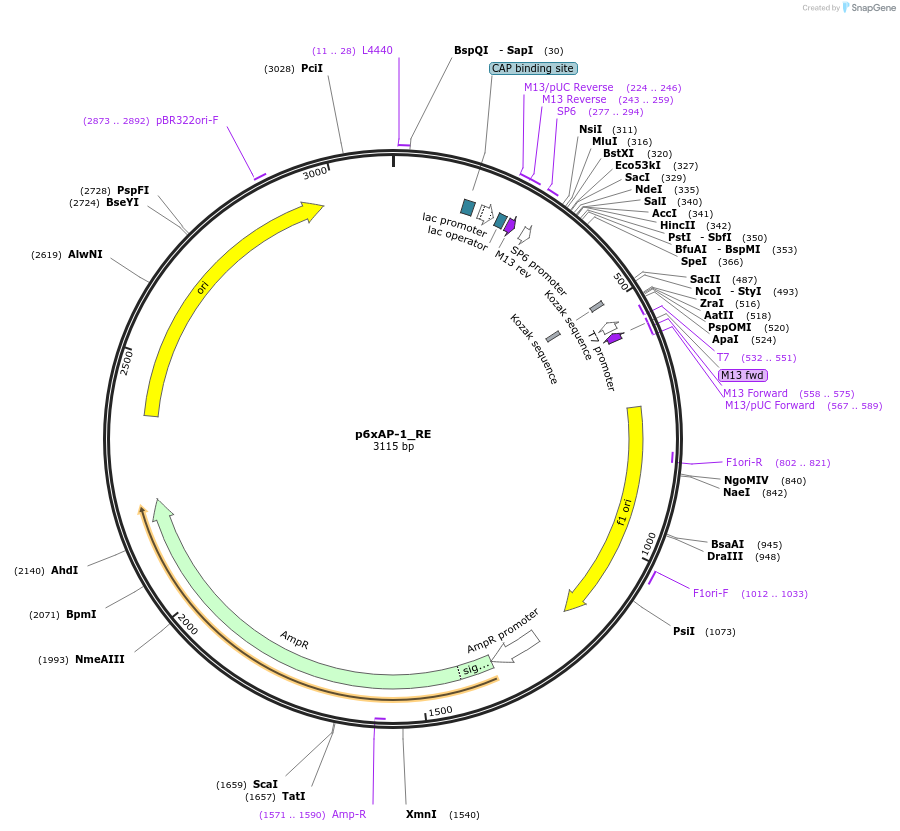
p6xAP-1_RE
Plasmid#118054PurposeGBPart - Operator/DNA Response element - 6 copies of the AP-1 binding motifDepositorInsert6 copies of the AP-1 binding motif
UseSynthetic BiologyExpressionMammalianAvailable SinceDec. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
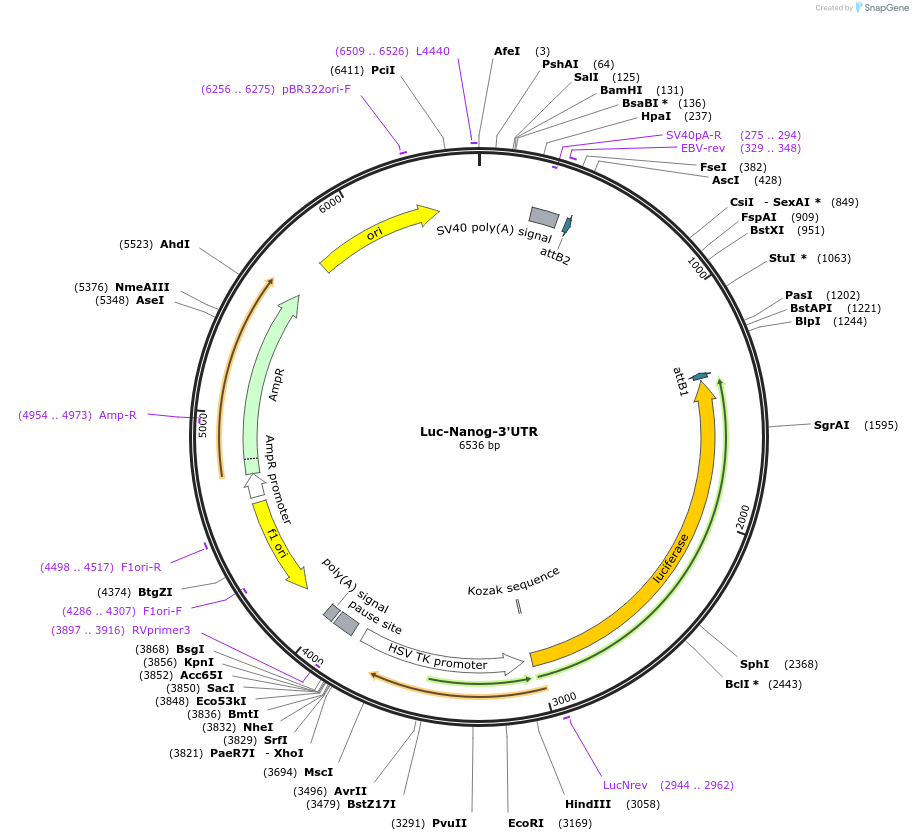
Luc-Nanog-3'UTR
Plasmid#63893PurposeLuciferase reporter of mouse wildtype Nanog 3'UTRDepositorInsertNanog homeobox (Nanog Mouse)
ExpressionMammalianAvailable SinceJune 24, 2015AvailabilityAcademic Institutions and Nonprofits only -
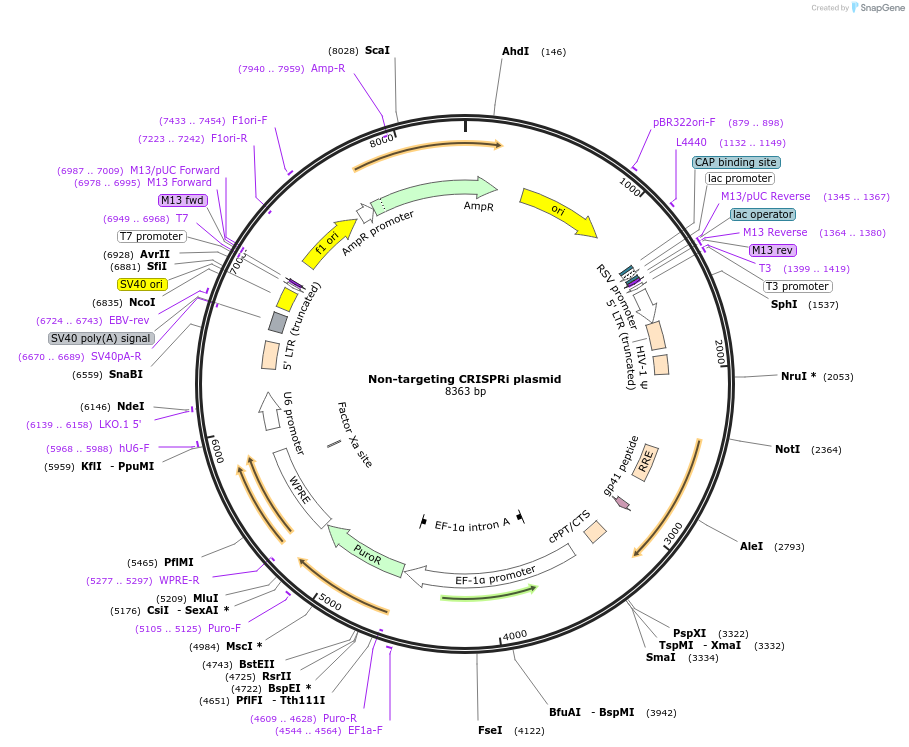
Non-targeting CRISPRi plasmid
Plasmid#154891PurposeHuman non-targeting CRISPRi gRNADepositorInsertNontargeting gRNA
UseCRISPR and LentiviralAvailable SinceNov. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
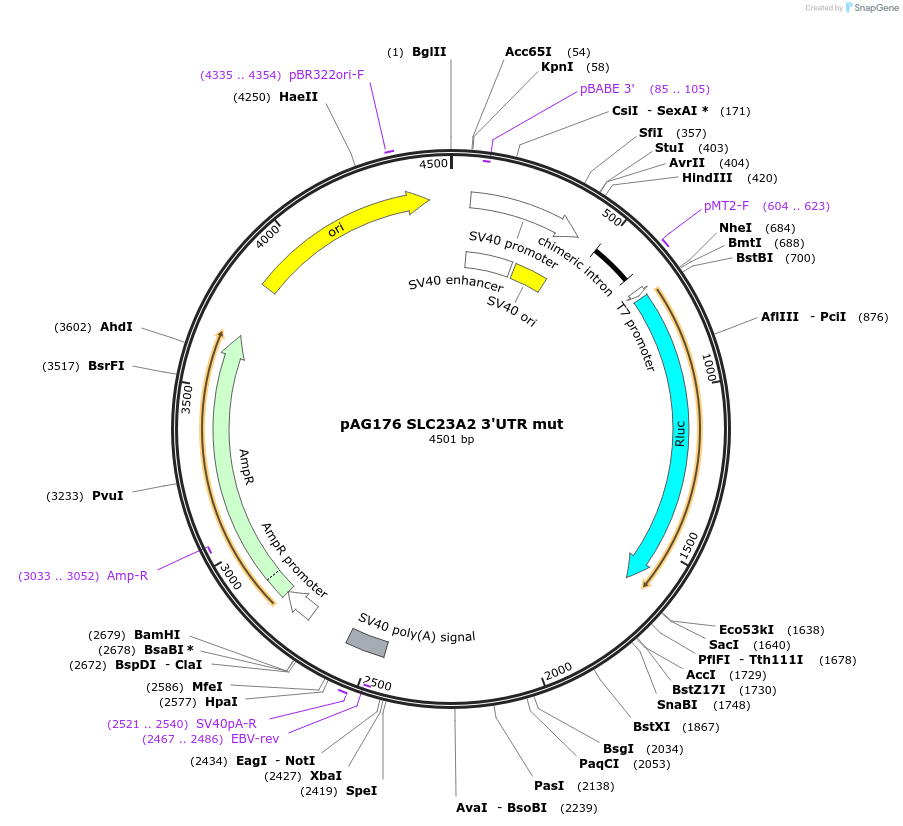
pAG176 SLC23A2 3'UTR mut
Plasmid#12034DepositorInsertN9 3'UTR mut (SLC23A2 Human)
UseLuciferaseExpressionMammalianMutationMutations in microRNA recognition sites (seed mat…Available SinceJune 8, 2006AvailabilityAcademic Institutions and Nonprofits only -
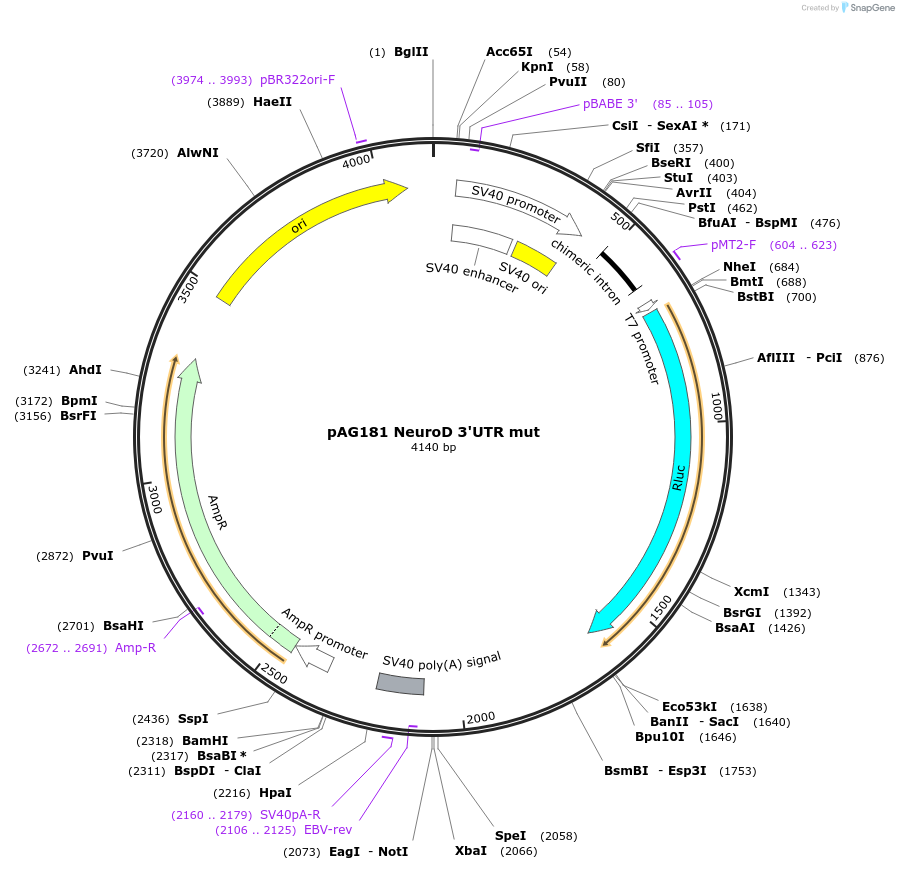
pAG181 NeuroD 3'UTR mut
Plasmid#12039DepositorInsertN14 3'UTR mut (NEUROD1 Human)
UseLuciferaseExpressionMammalianMutationMutations in microRNA recognition sites (seed mat…Available SinceJune 8, 2006AvailabilityAcademic Institutions and Nonprofits only -
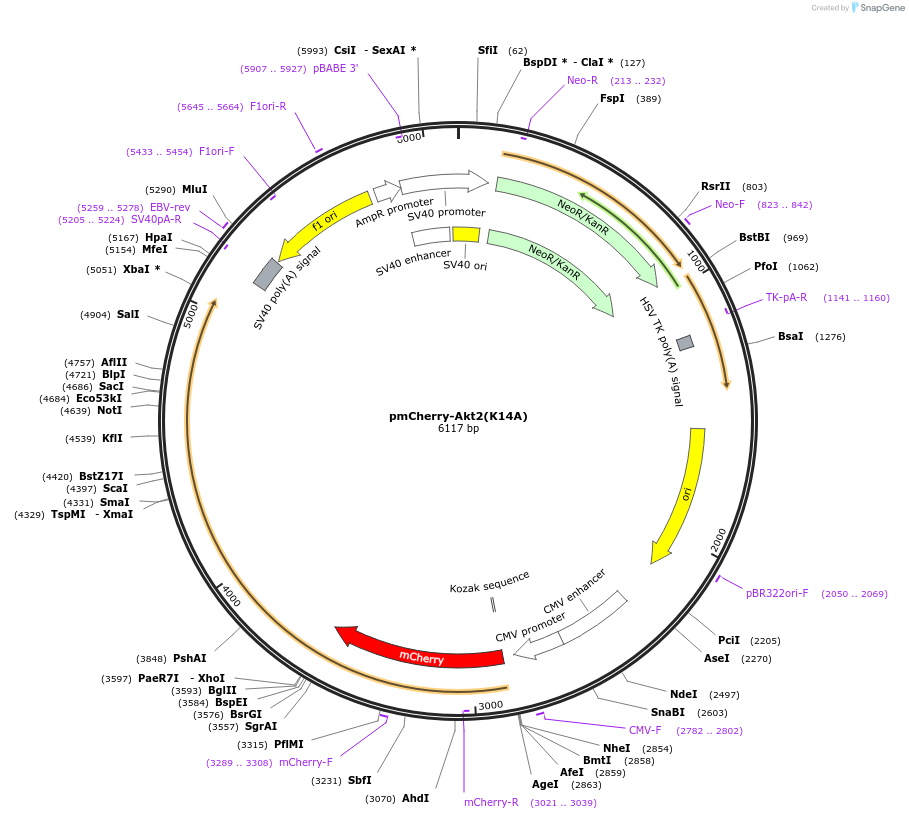
pmCherry-Akt2(K14A)
Plasmid#86624PurposeExpresses mCherry-tagged human Akt2(K14A) membrane binding mutantDepositorInsertAkt2(K14A) (AKT2 )
ExpressionMammalianAvailable SinceMarch 7, 2017AvailabilityAcademic Institutions and Nonprofits only