We narrowed to 14,145 results for: TIM
-
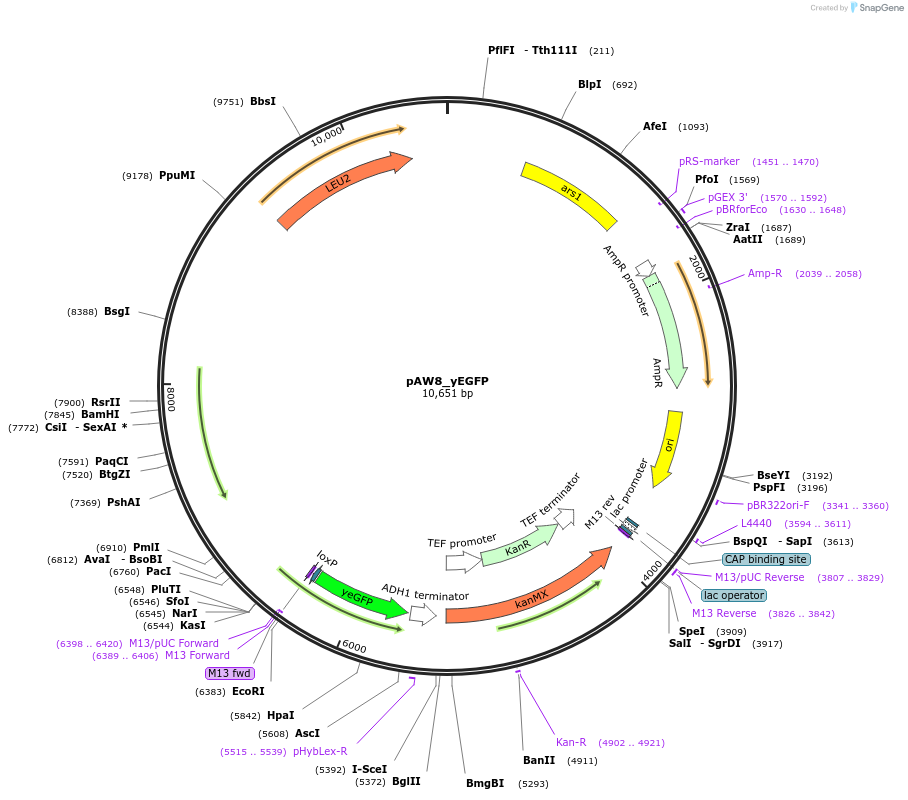
Plasmid#110229PurposeCre/Lox C-terminal tagging construct encoding codon-optimized green fluorescent proteinDepositorTypeEmpty backboneUseCre/LoxAvailable SinceOct. 15, 2018AvailabilityAcademic Institutions and Nonprofits only
-
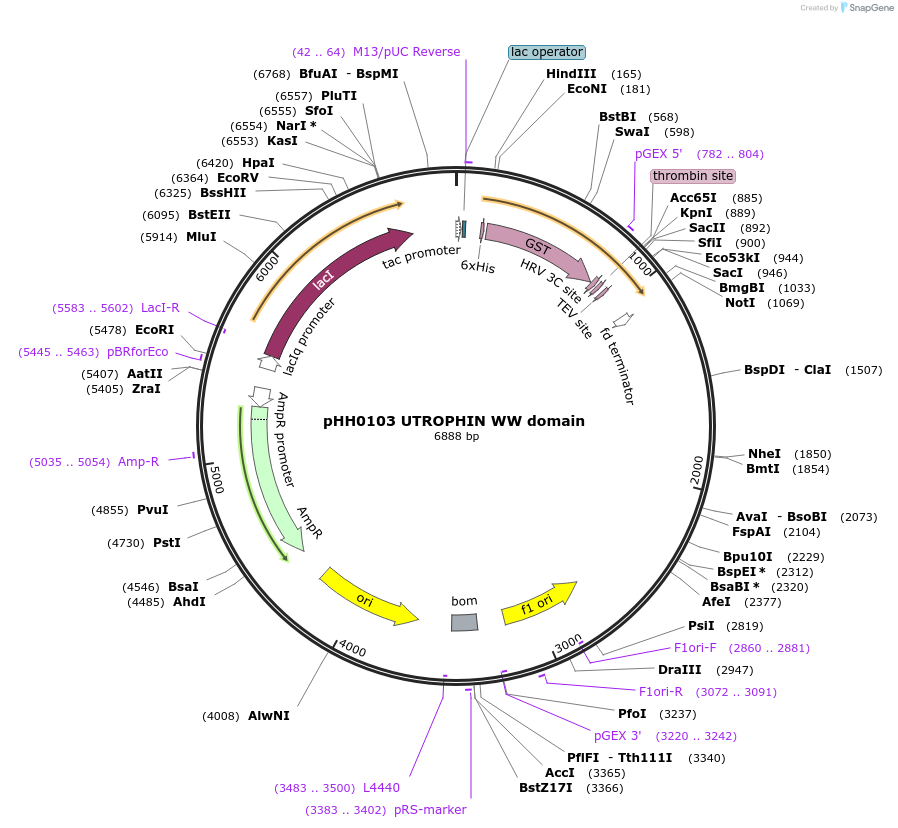
pHH0103 UTROPHIN WW domain
Plasmid#104275PurposeBacterial expression of WW domain from UTROPHINDepositorInsertUTROPHIN WW domain (UTRN Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
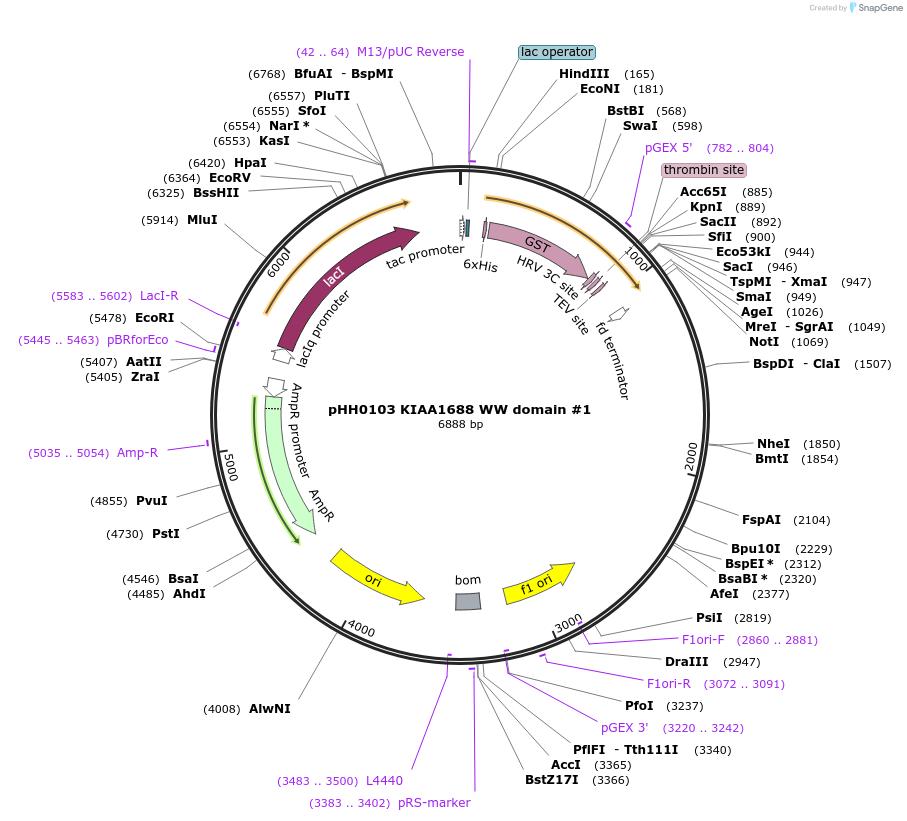
pHH0103 KIAA1688 WW domain #1
Plasmid#104277PurposeBacterial expression of WW domain #1 from KIAA1688DepositorInsertKIAA1688 WW domain #1 (ARHGAP39 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
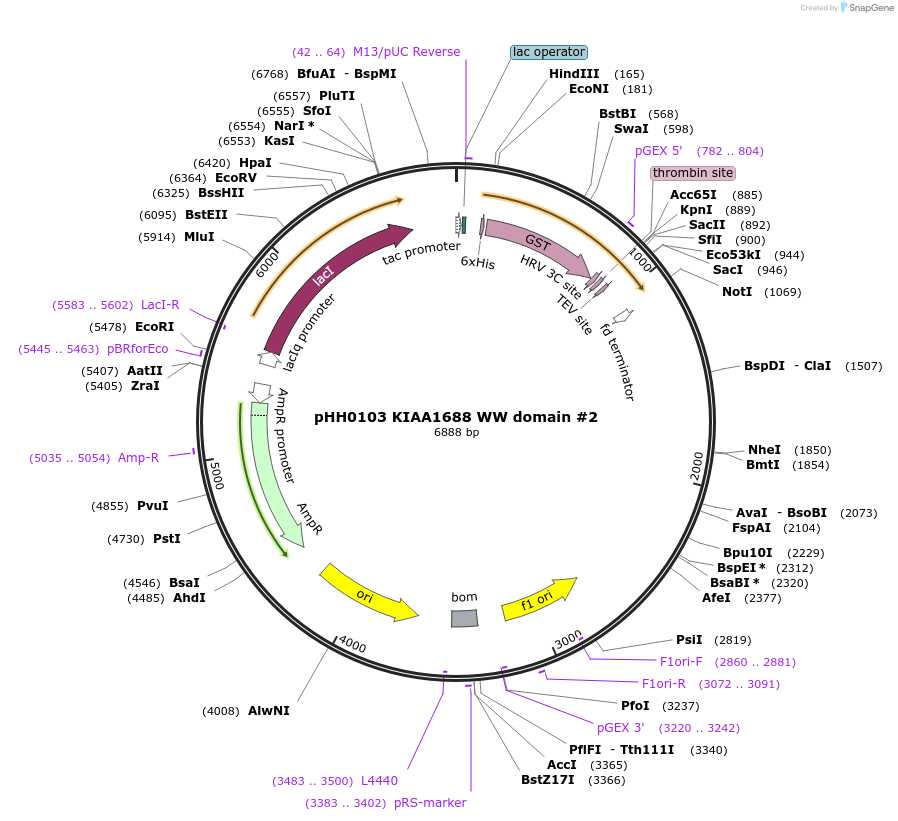
pHH0103 KIAA1688 WW domain #2
Plasmid#104278PurposeBacterial expression of WW domain #2 from KIAA1688DepositorInsertKIAA1688 WW domain #2 (ARHGAP39 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
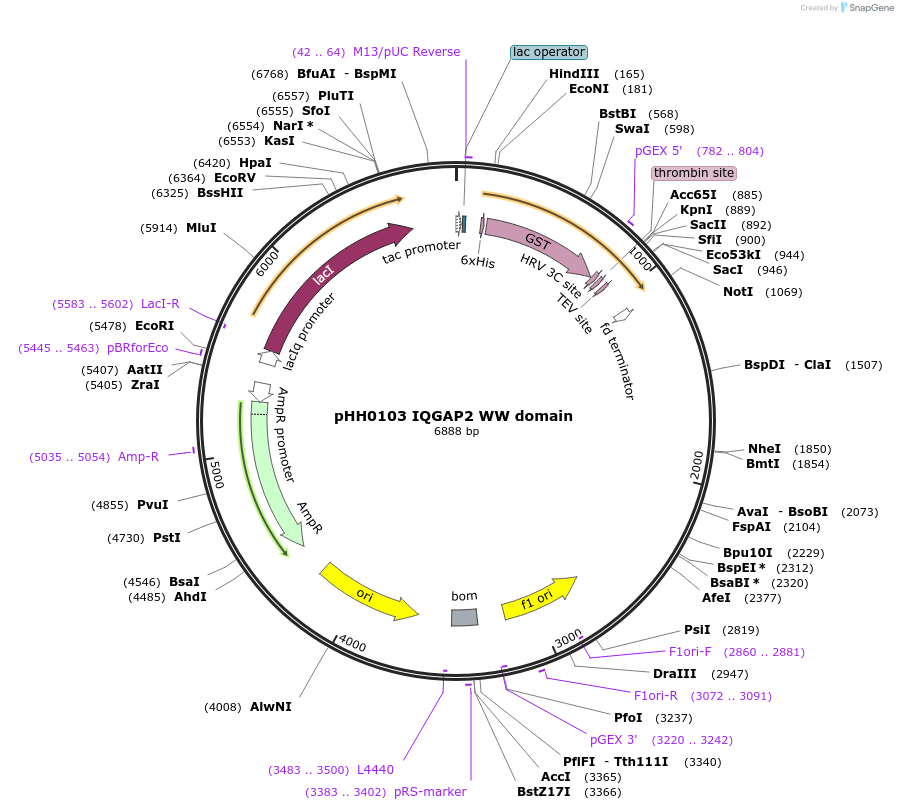
pHH0103 IQGAP2 WW domain
Plasmid#104280PurposeBacterial expression of WW domain from IQGAP2DepositorInsertIQGAP2 WW domain (IQGAP2 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
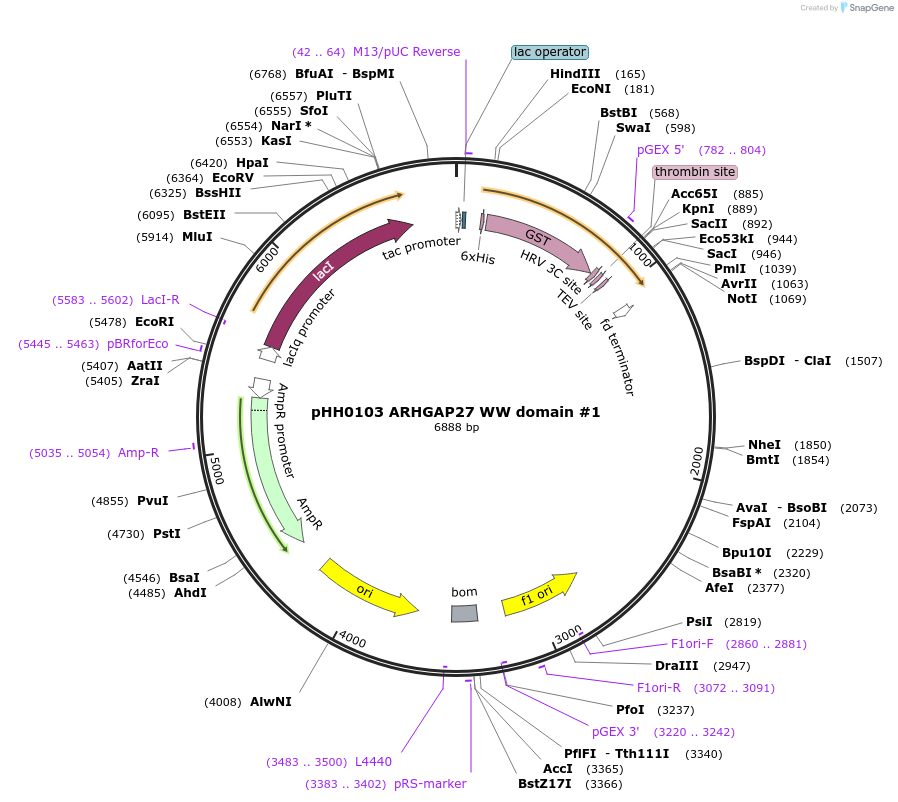
pHH0103 ARHGAP27 WW domain #1
Plasmid#104282PurposeBacterial expression of WW domain #1 from ARHGAP27DepositorInsertARHGAP27 WW domain #1 (ARHGAP27 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
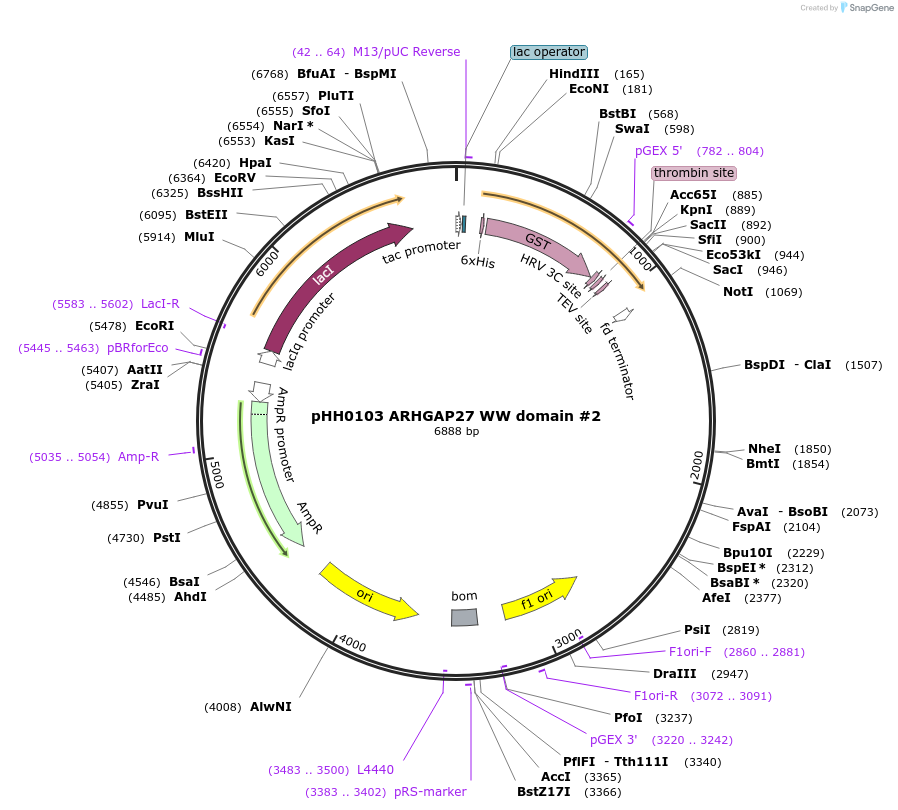
pHH0103 ARHGAP27 WW domain #2
Plasmid#104283PurposeBacterial expression of WW domain #2 from ARHGAP27DepositorInsertARHGAP27 WW domain #2 (ARHGAP27 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
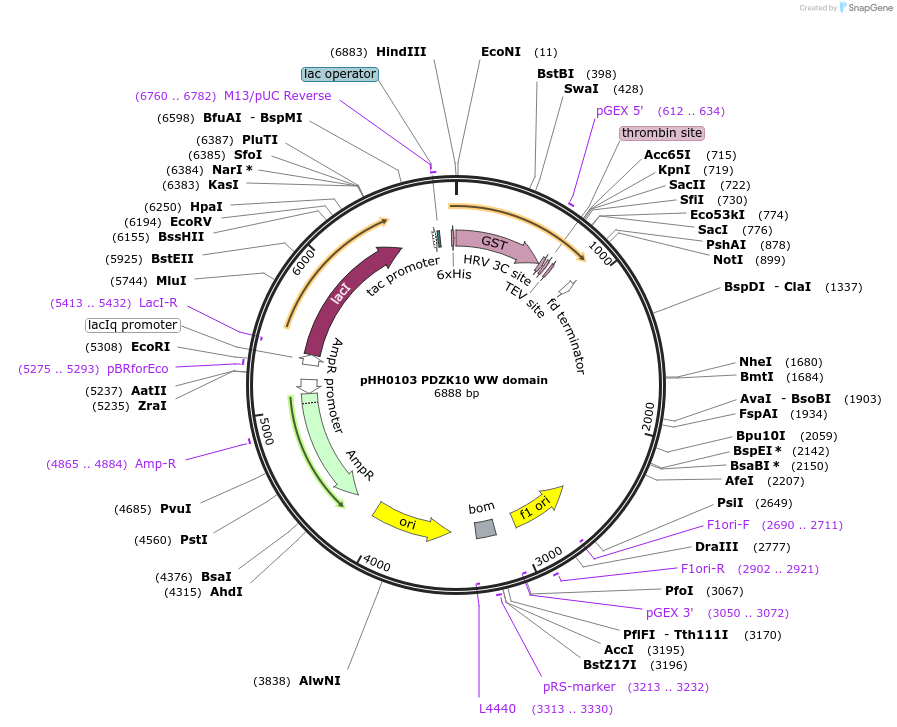
pHH0103 PDZK10 WW domain
Plasmid#104285PurposeBacterial expression of WW domain from PDZK10DepositorInsertPDZK10 WW domain (FRMPD4 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
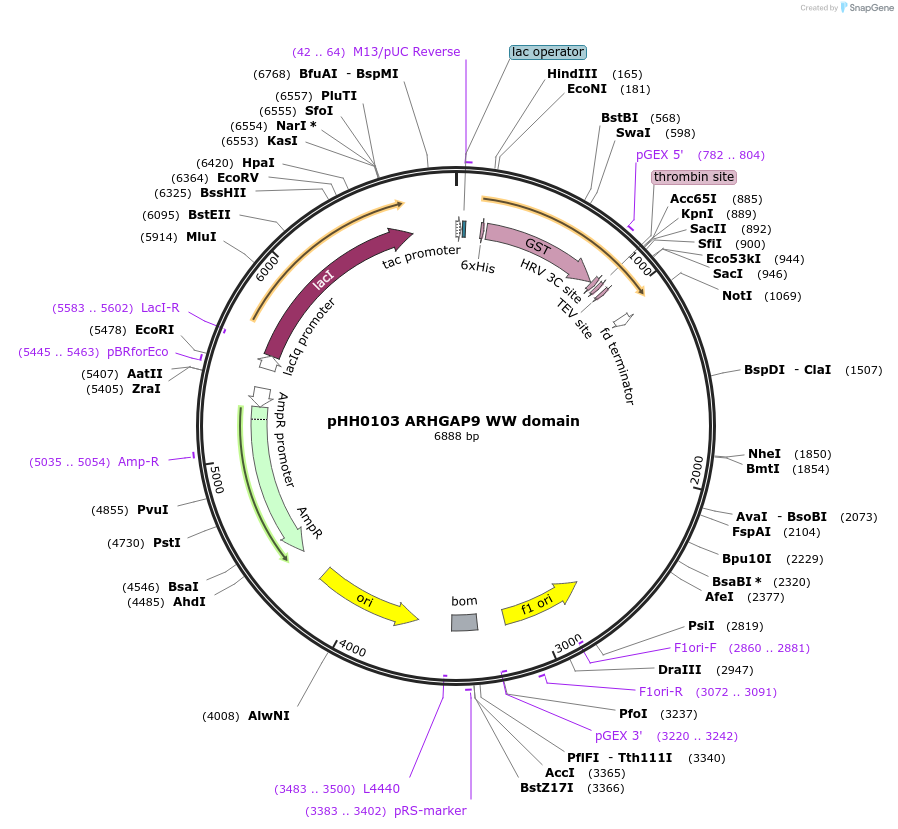
pHH0103 ARHGAP9 WW domain
Plasmid#104286PurposeBacterial expression of WW domain from ARHGAP9DepositorInsertARHGAP9 WW domain (ARHGAP9 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
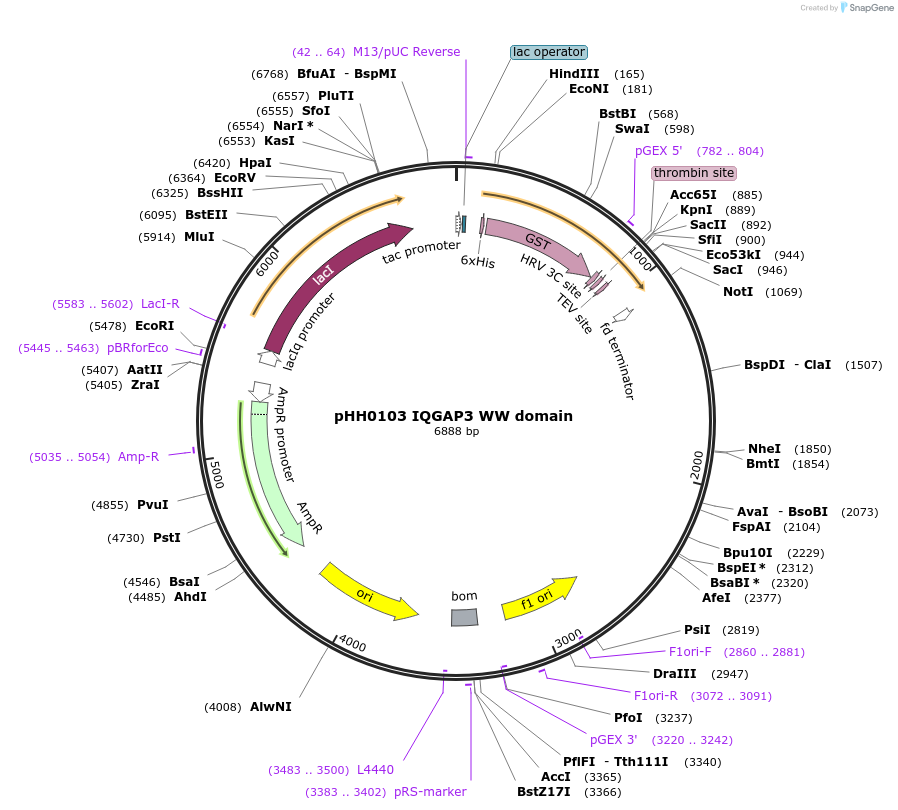
pHH0103 IQGAP3 WW domain
Plasmid#104289PurposeBacterial expression of WW domain from IQGAP3DepositorInsertIQGAP3 WW domain (IQGAP3 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
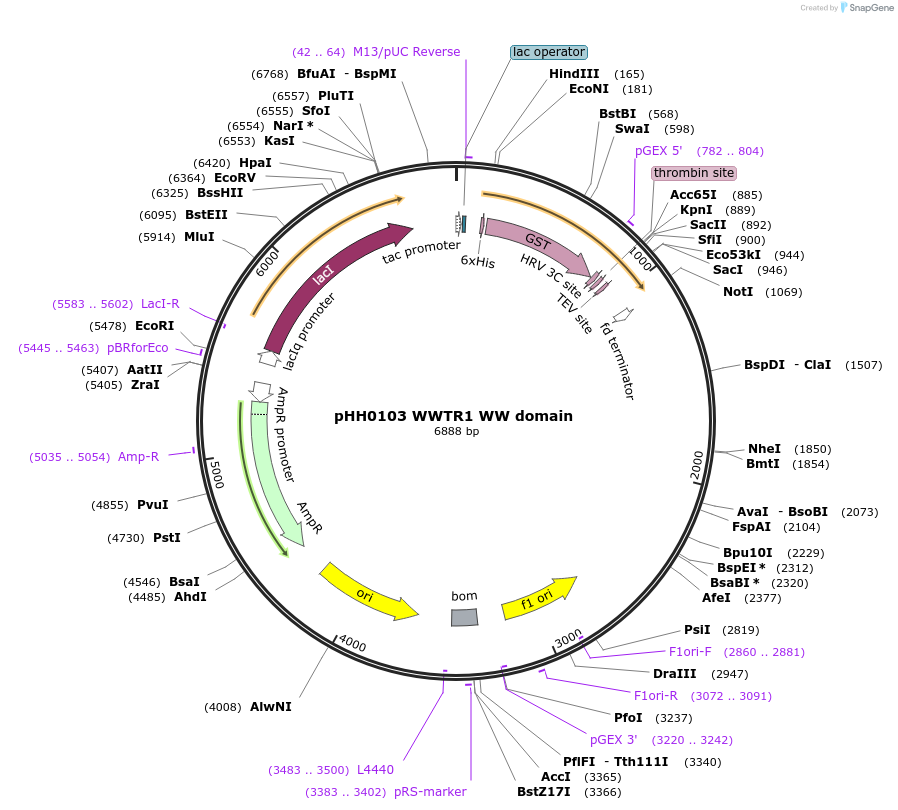
pHH0103 WWTR1 WW domain
Plasmid#104261PurposeBacterial expression of WW domain from WWTR1DepositorInsertWWTR1 WW domain (WWTR1 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
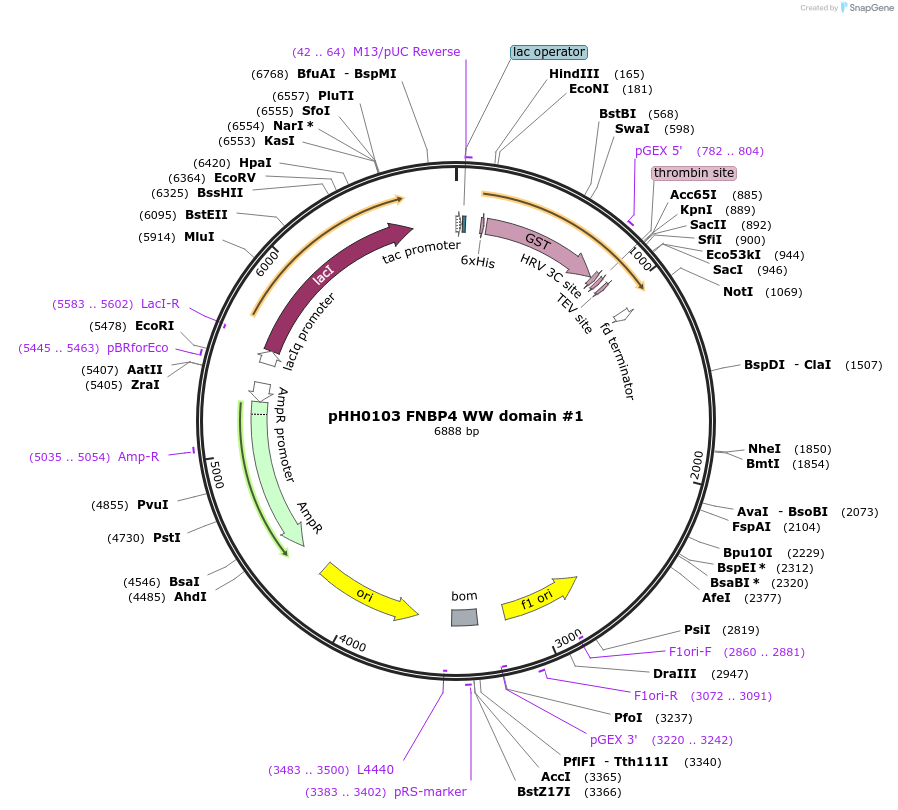
pHH0103 FNBP4 WW domain #1
Plasmid#104263PurposeBacterial expression of WW domain #1 from FNBP4DepositorInsertFNBP4 WW domain #1 (FNBP4 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
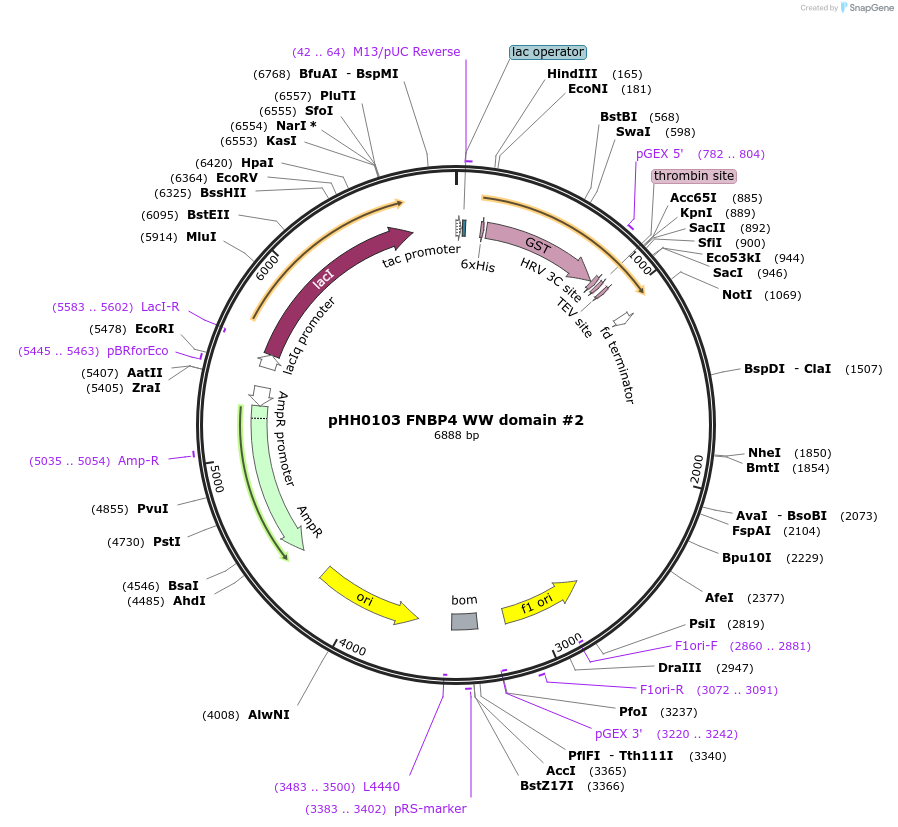
pHH0103 FNBP4 WW domain #2
Plasmid#104264PurposeBacterial expression of WW domain #2 from FNBP4DepositorInsertFNBP4 WW domain #2 (FNBP4 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
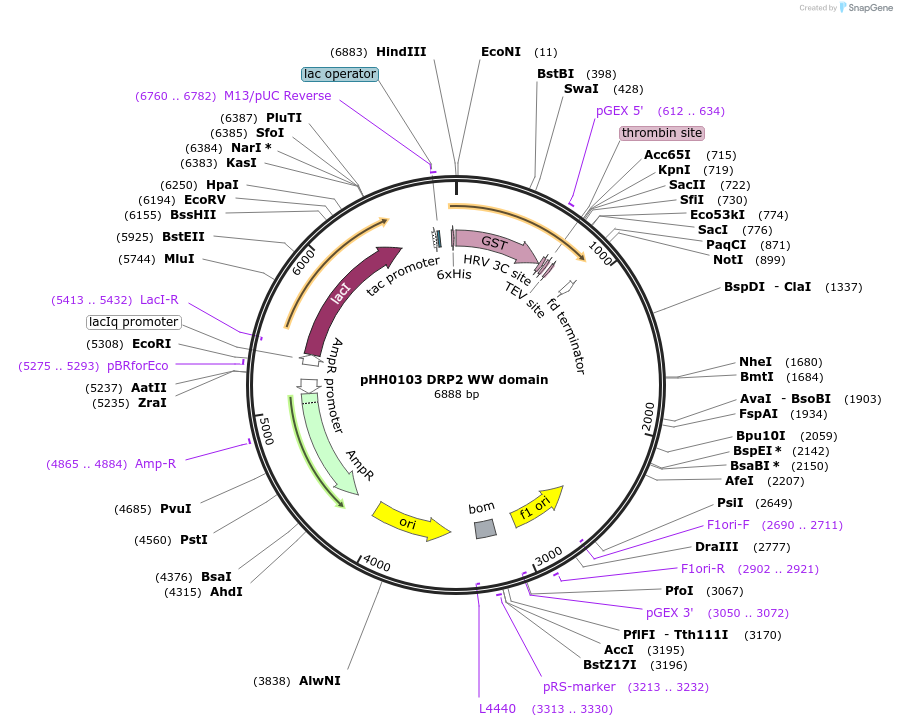
pHH0103 DRP2 WW domain
Plasmid#104270PurposeBacterial expression of WW domain from DRP2DepositorInsertDRP2 WW domain (DRP2 Human)
TagsGSTExpressionBacterialMutationcodon optimized for expression in bacteriaPromotertacAvailable SinceAug. 28, 2018AvailabilityAcademic Institutions and Nonprofits only -
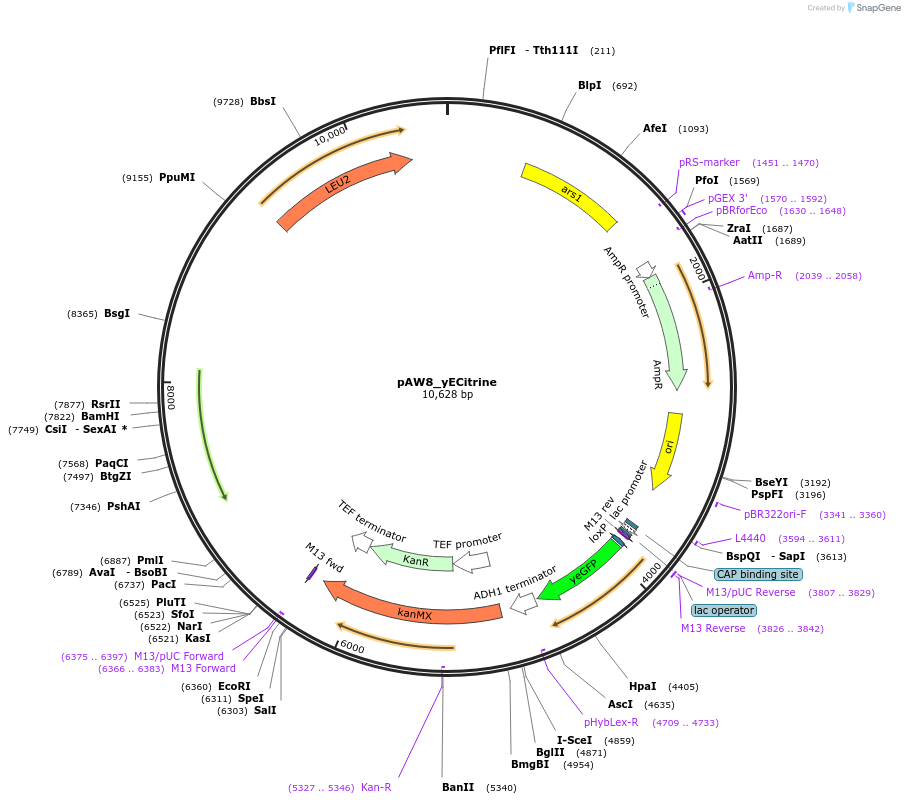
pAW8_yECitrine
Plasmid#110231PurposeCre/Lox C-terminal tagging construct encoding codon-optimized yellow fluorescent proteinDepositorTypeEmpty backboneUseCre/LoxAvailable SinceJuly 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
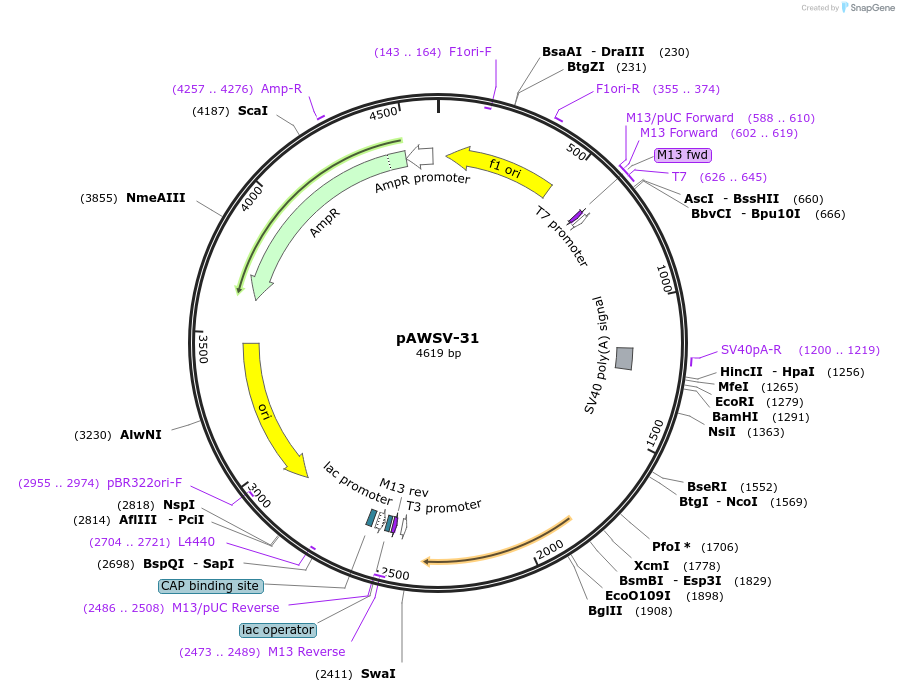
pAWSV-31
Plasmid#72784PurposepBT264-[miR-376a]DepositorInsert[miR-376a]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
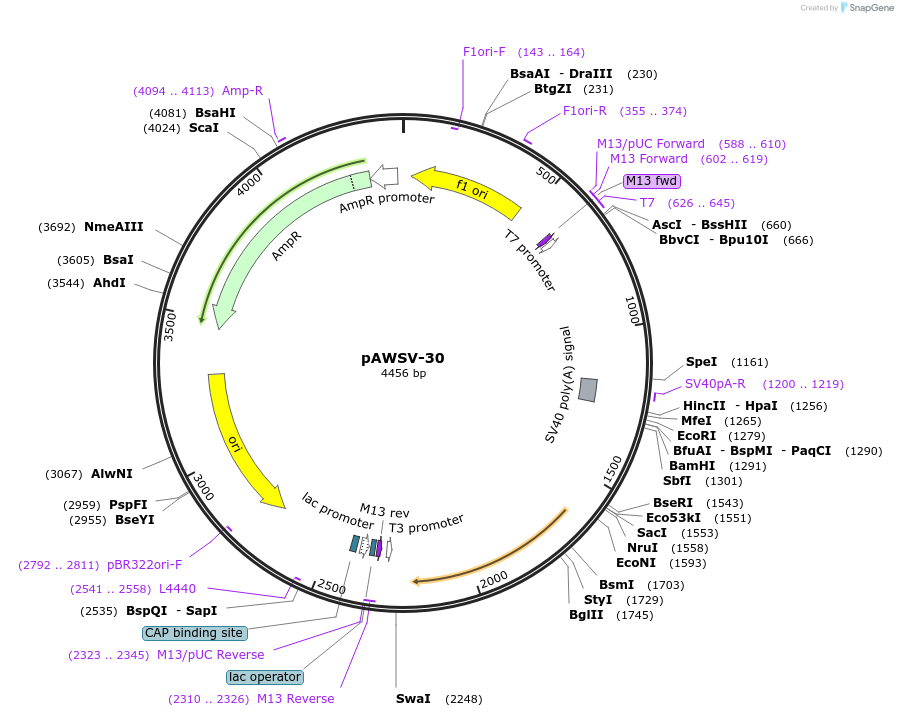
pAWSV-30
Plasmid#72783PurposepBT264-[miR-373]DepositorInsert[miR-373]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
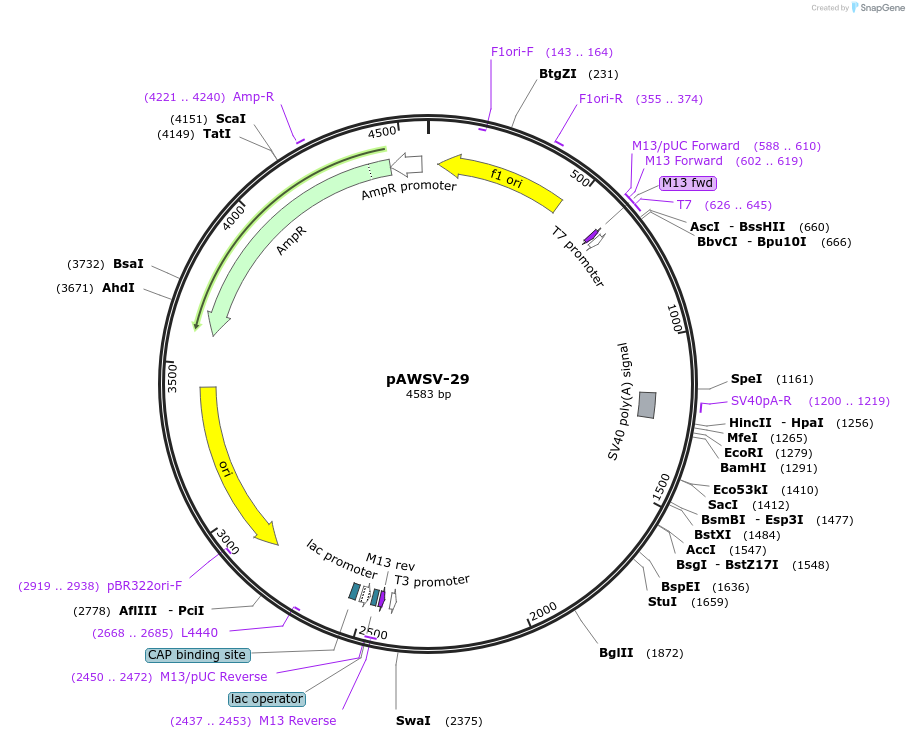
pAWSV-29
Plasmid#72782PurposepBT264-[miR-34a]DepositorInsert[miR-34a]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
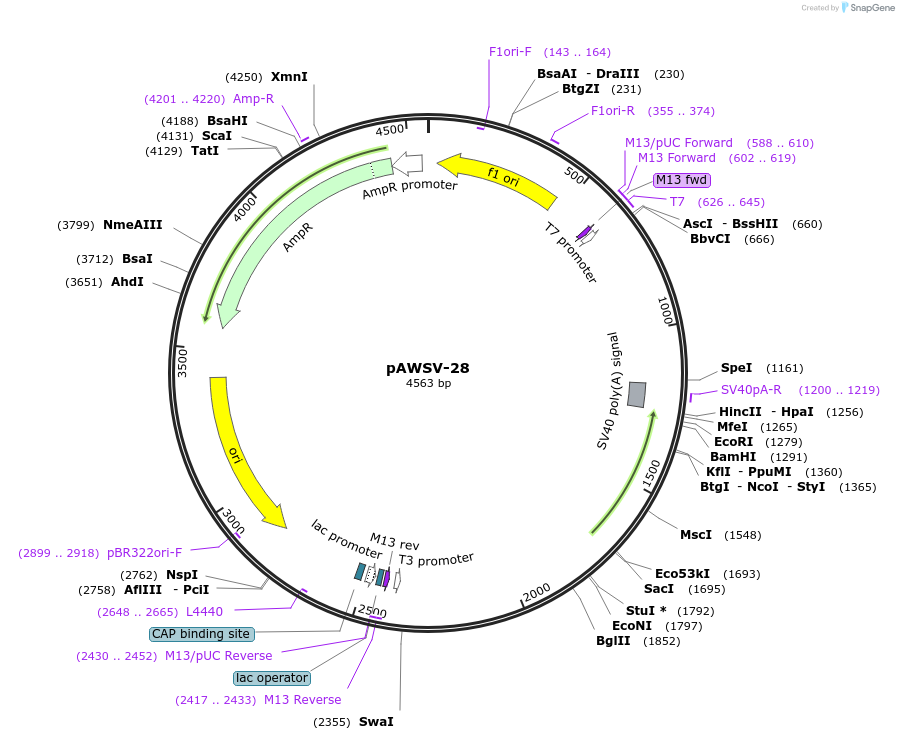
pAWSV-28
Plasmid#72781PurposepBT264-[miR-328]DepositorInsert[miR-328]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
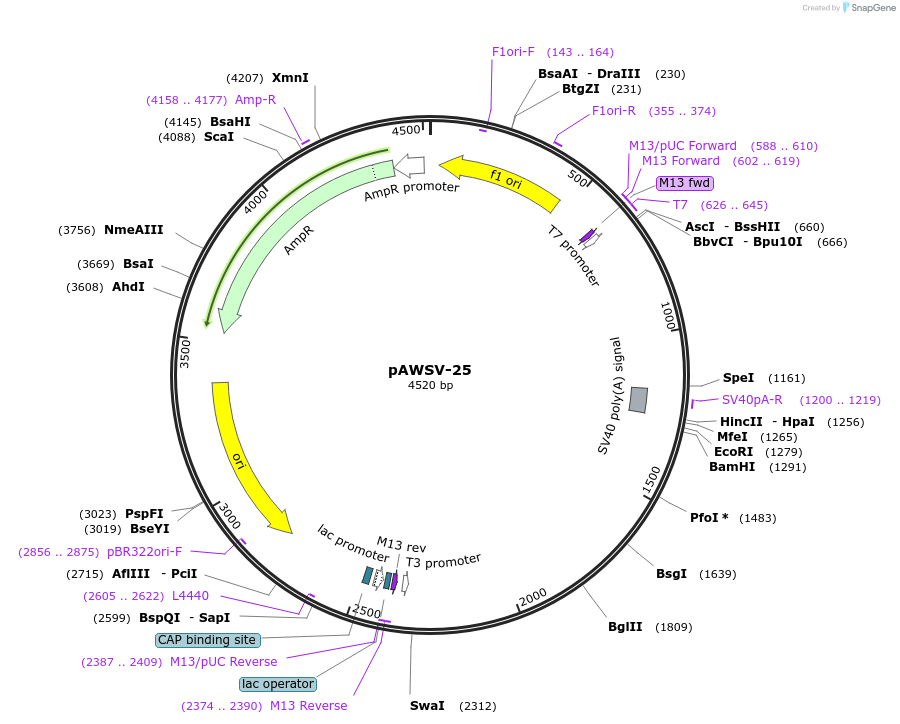
pAWSV-25
Plasmid#72778PurposepBT264-[miR-29c]DepositorInsert[miR-29c]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
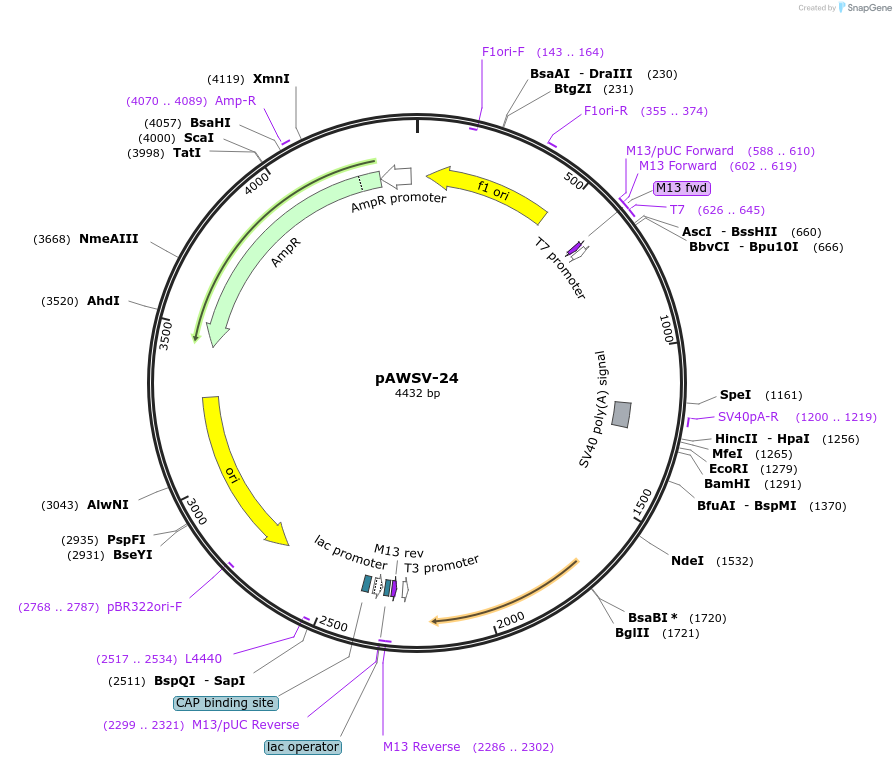
pAWSV-24
Plasmid#72777PurposepBT264-[miR-29b]DepositorInsert[miR-29b]
Available SinceFeb. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
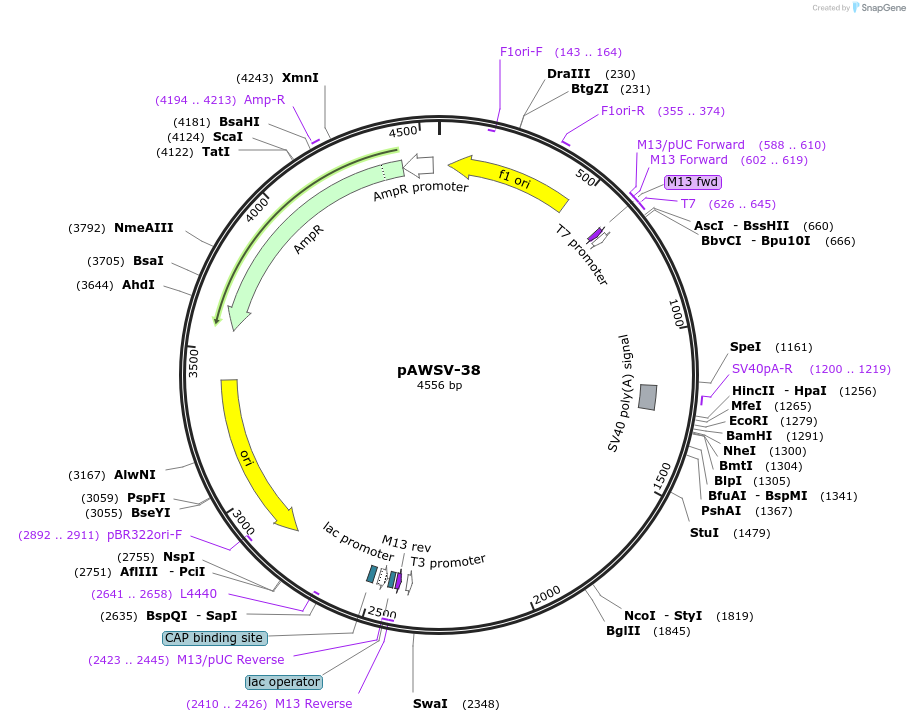
pAWSV-38
Plasmid#72791PurposepBT264-[miR-93/106b cluster]DepositorInsert[miR-93/106b cluster]
Available SinceFeb. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
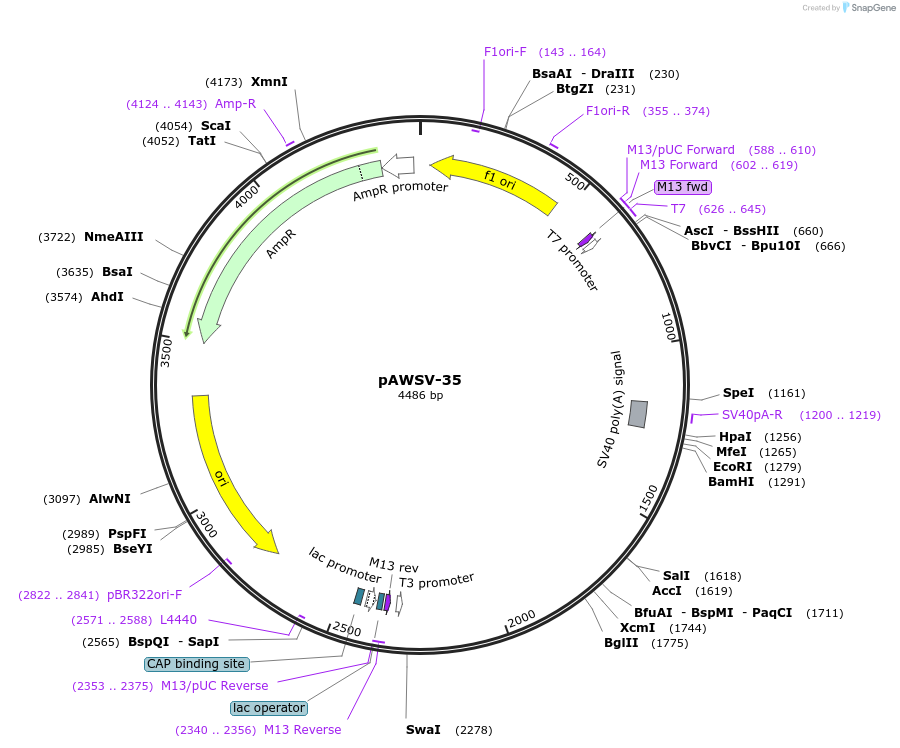
pAWSV-35
Plasmid#72788PurposepBT264-[miR-489]DepositorInsert[miR-489]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
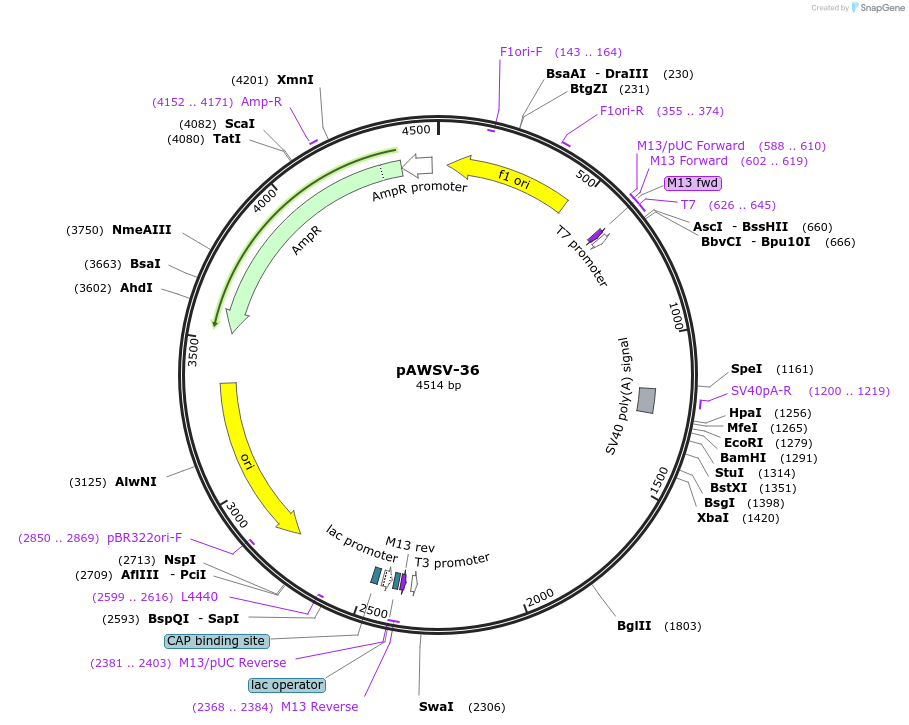
pAWSV-36
Plasmid#72789PurposepBT264-[miR-708]DepositorInsert[miR-708]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
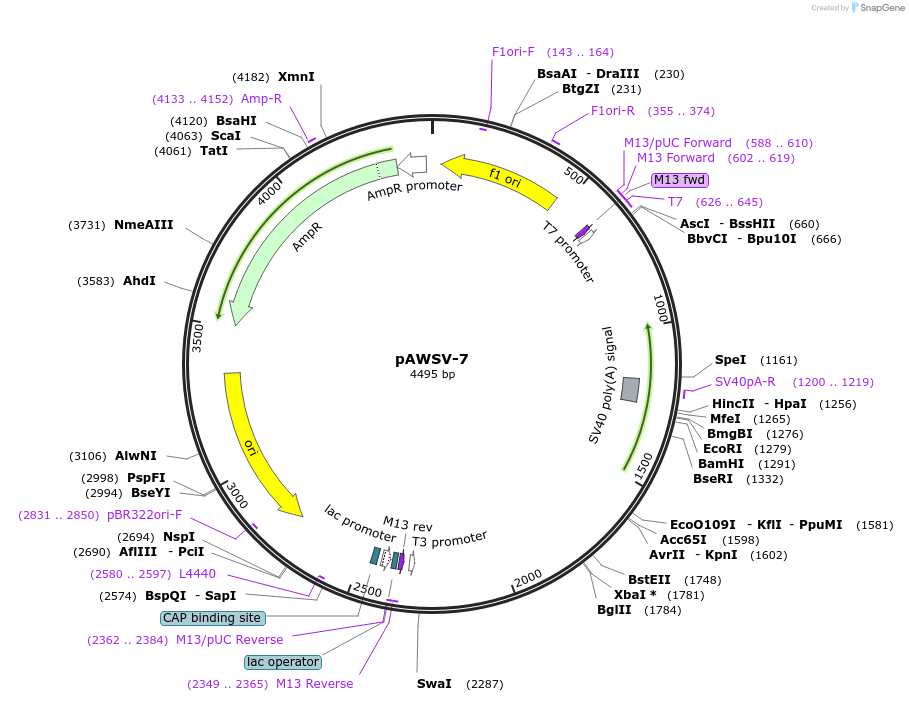
pAWSV-7
Plasmid#72760PurposepBT264-[miR-10b]DepositorInsert[miR-10b]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
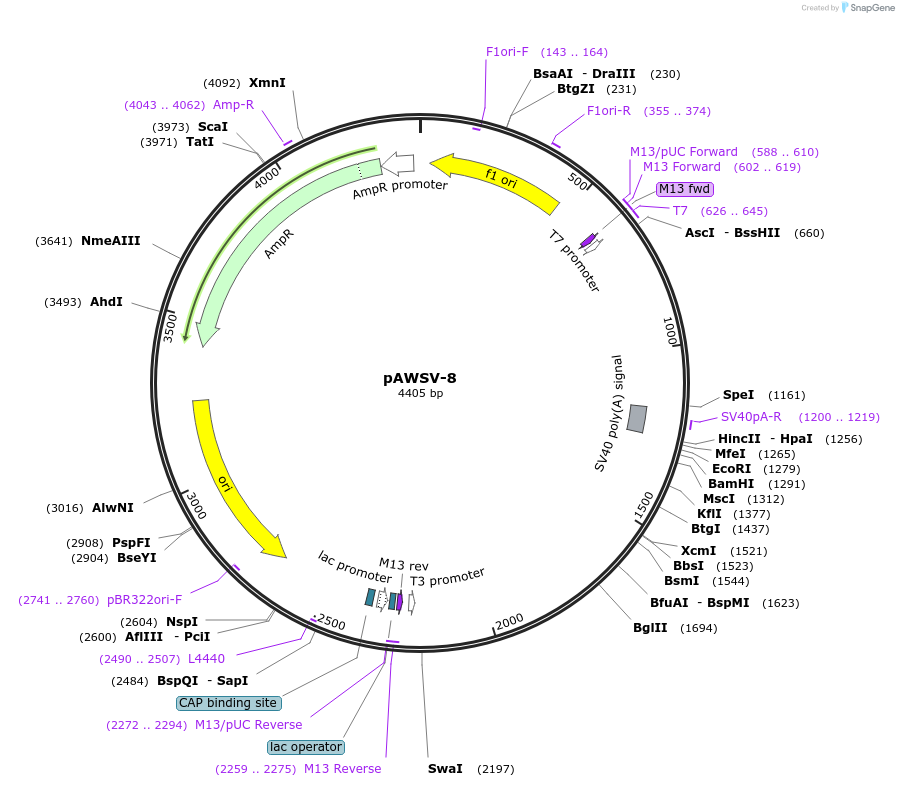
pAWSV-8
Plasmid#72761PurposepBT264-[miR-126]DepositorInsert[miR-126]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
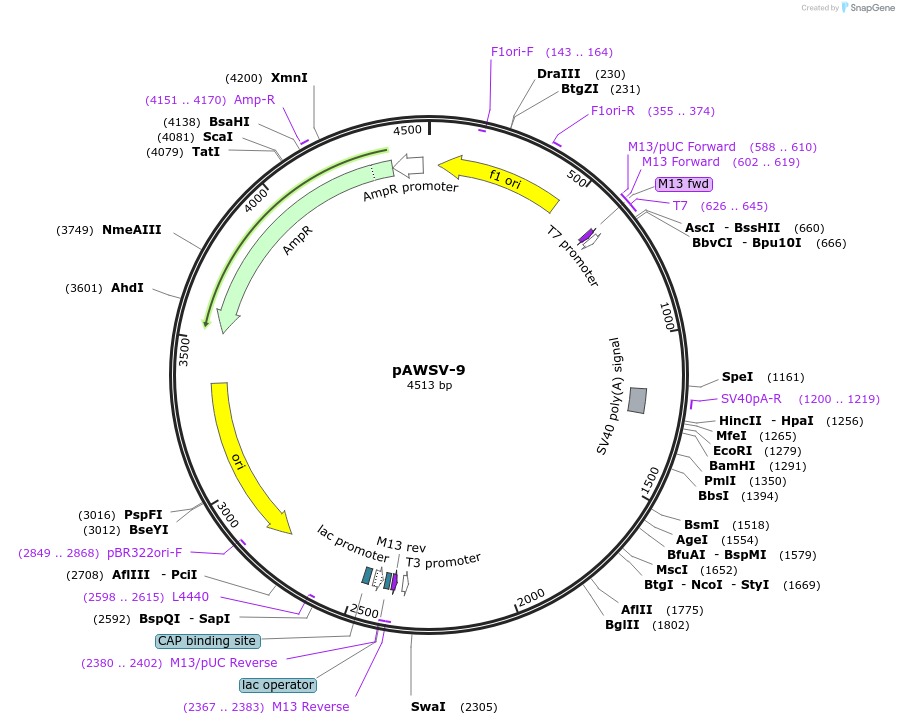
pAWSV-9
Plasmid#72762PurposepBT264-[miR-128b]DepositorInsert[miR-128b]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
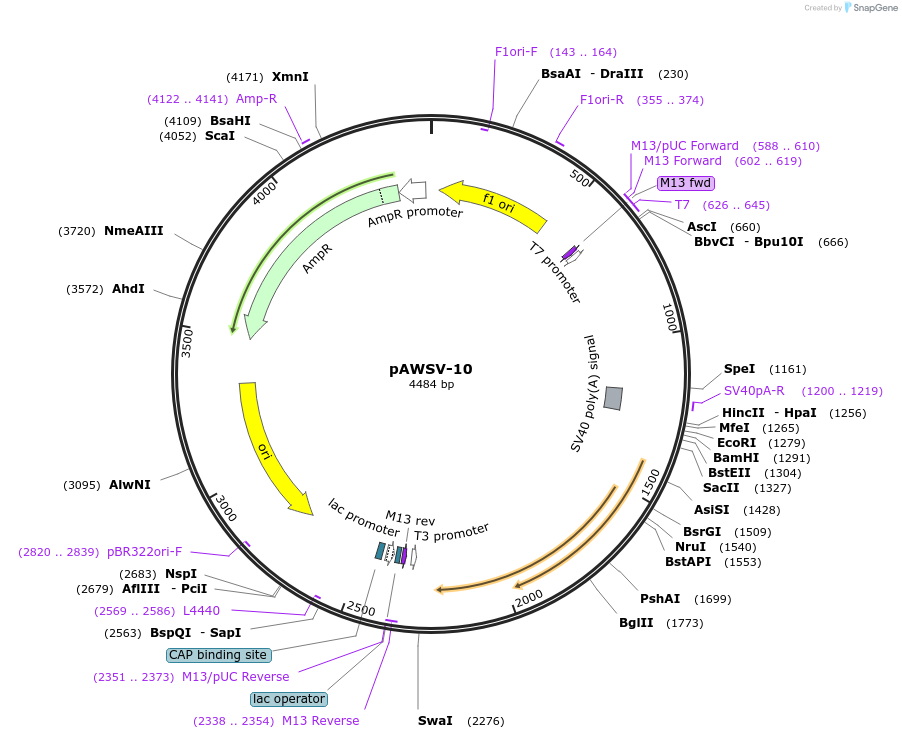
pAWSV-10
Plasmid#72763PurposepBT264-[miR-129-2]DepositorInsert[miR-129-2]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
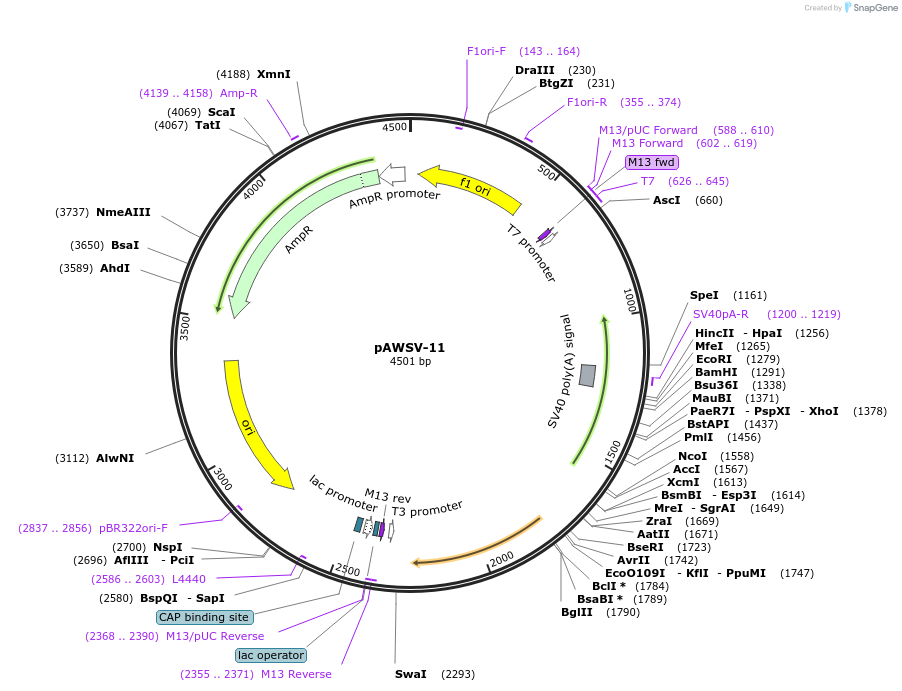
pAWSV-11
Plasmid#72764PurposepBT264-[miR-132]DepositorInsert[miR-132]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
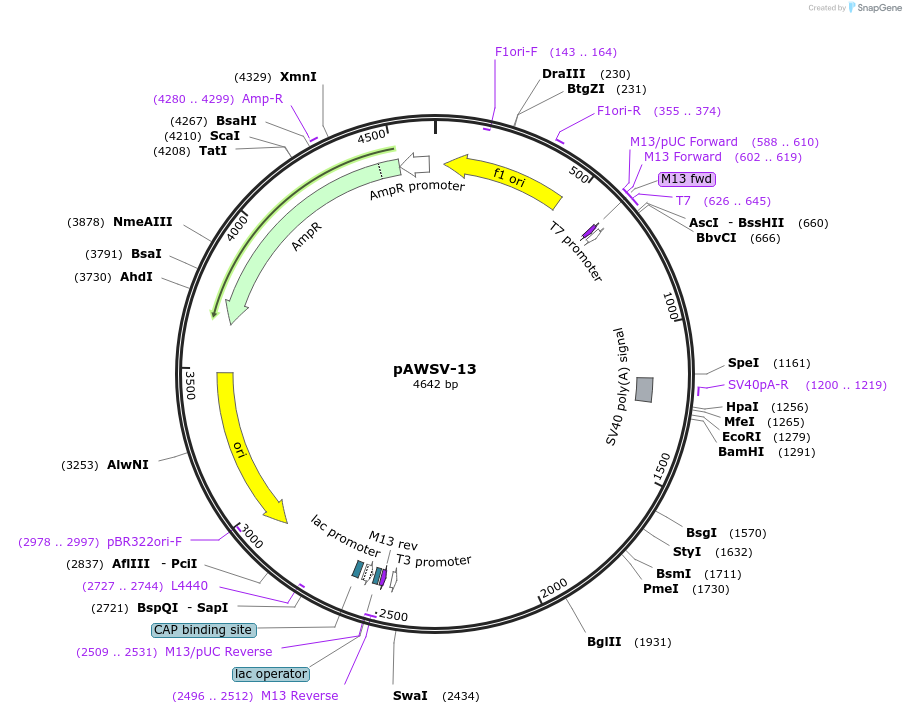
pAWSV-13
Plasmid#72766PurposepBT264-[miR-16-1/15a cluster]DepositorInsert[miR-16-1/15a cluster]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
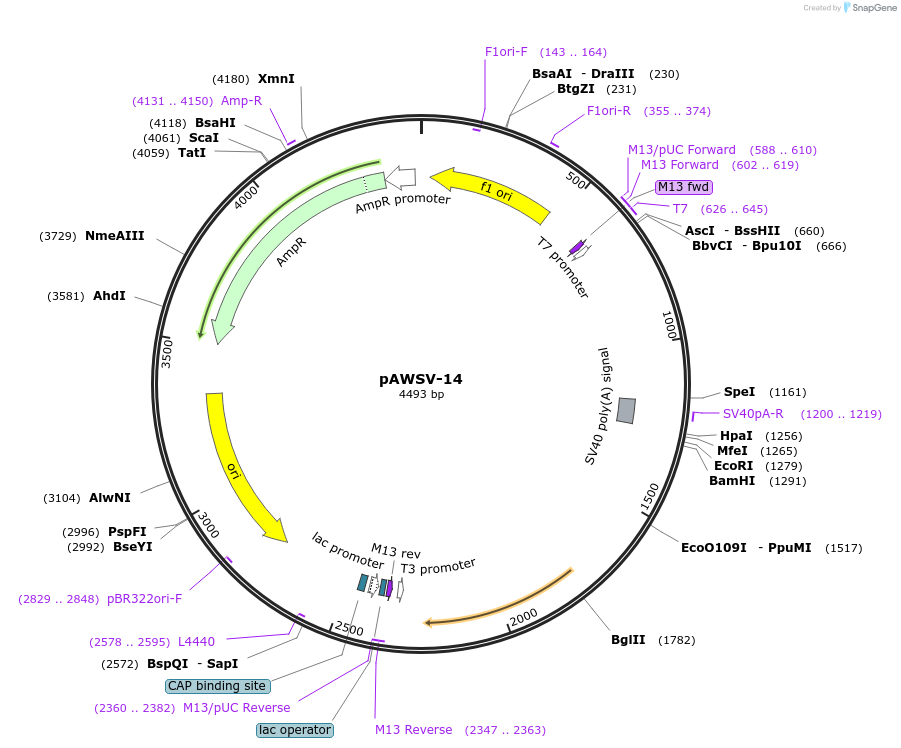
pAWSV-14
Plasmid#72767PurposepBT264-[miR-181a]DepositorInsert[miR-181a]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
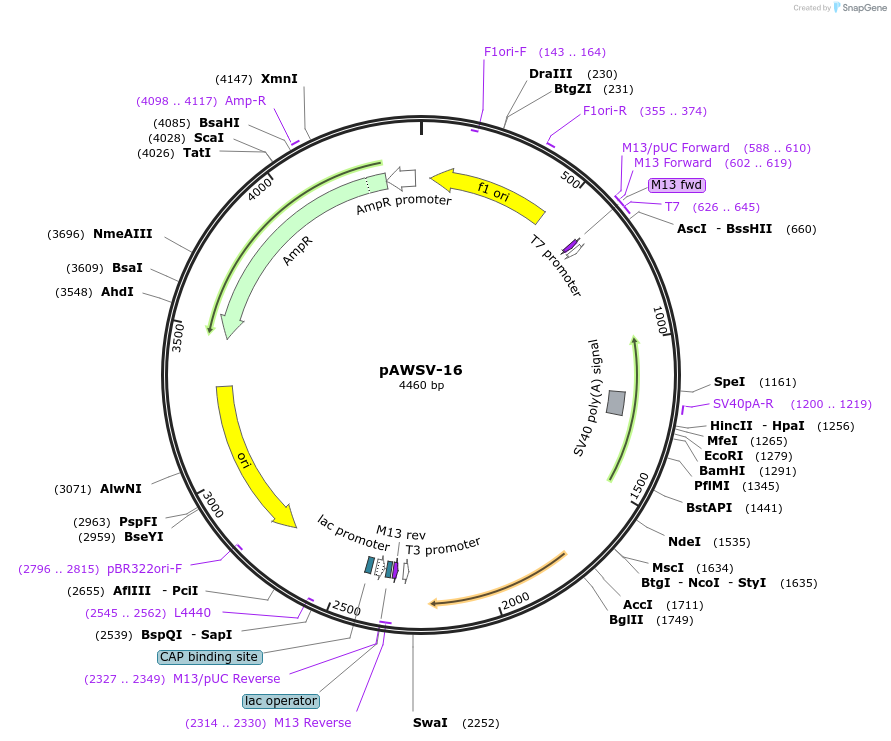
pAWSV-16
Plasmid#72769PurposepBT264-[miR-188]DepositorInsert[miR-188]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
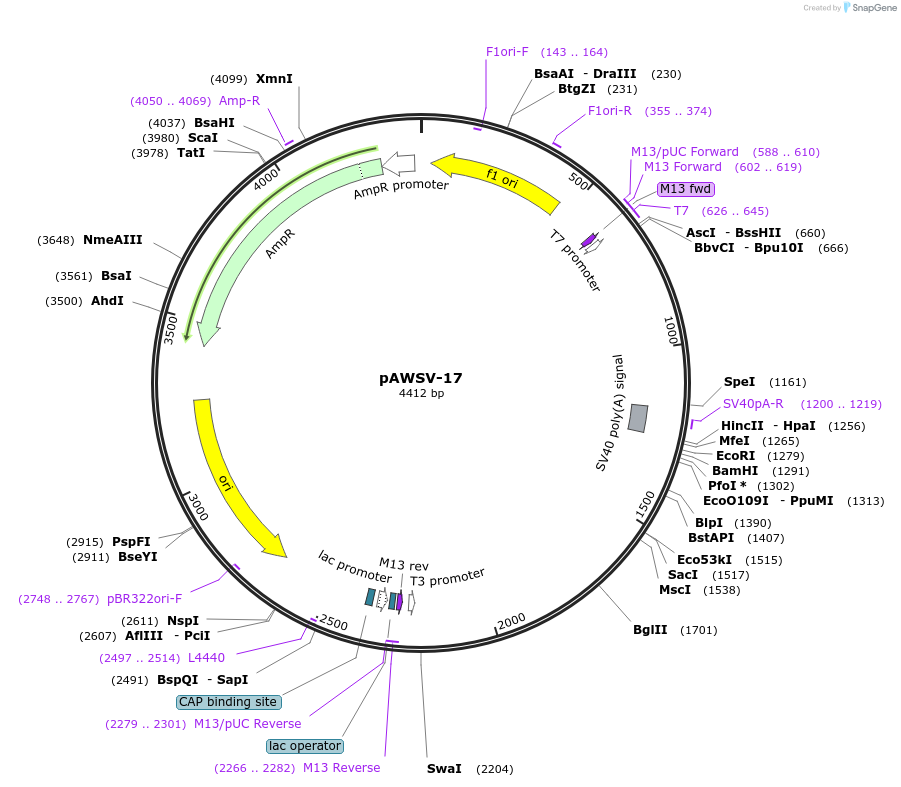
pAWSV-17
Plasmid#72770PurposepBT264-[miR-196]DepositorInsert[miR-196]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
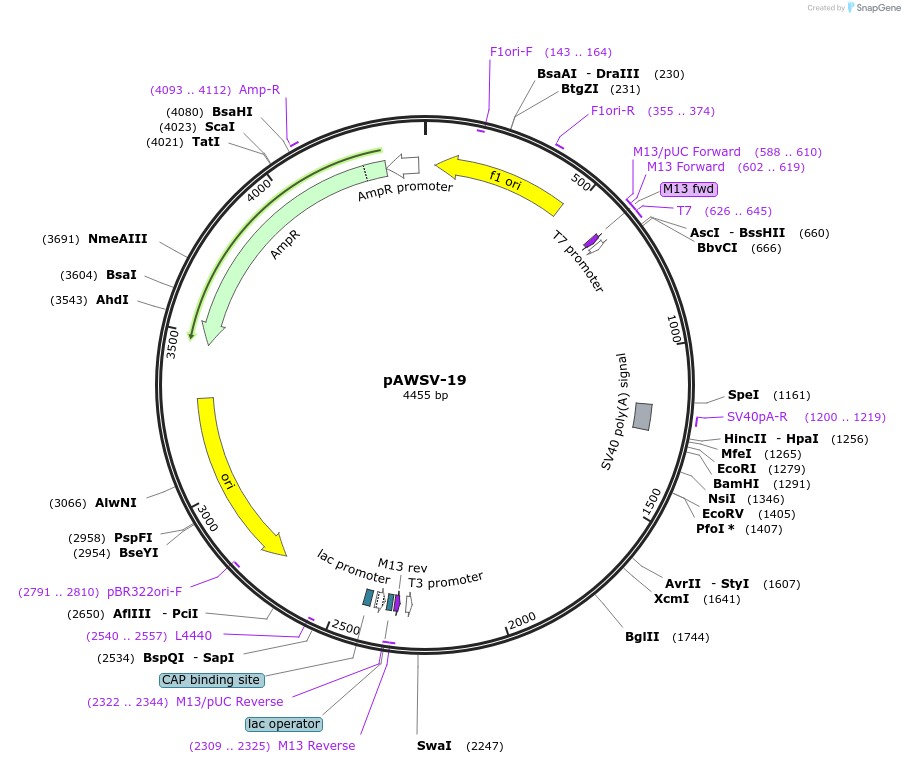
pAWSV-19
Plasmid#72772PurposepBT264-[miR-211]DepositorInsert[miR-211]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
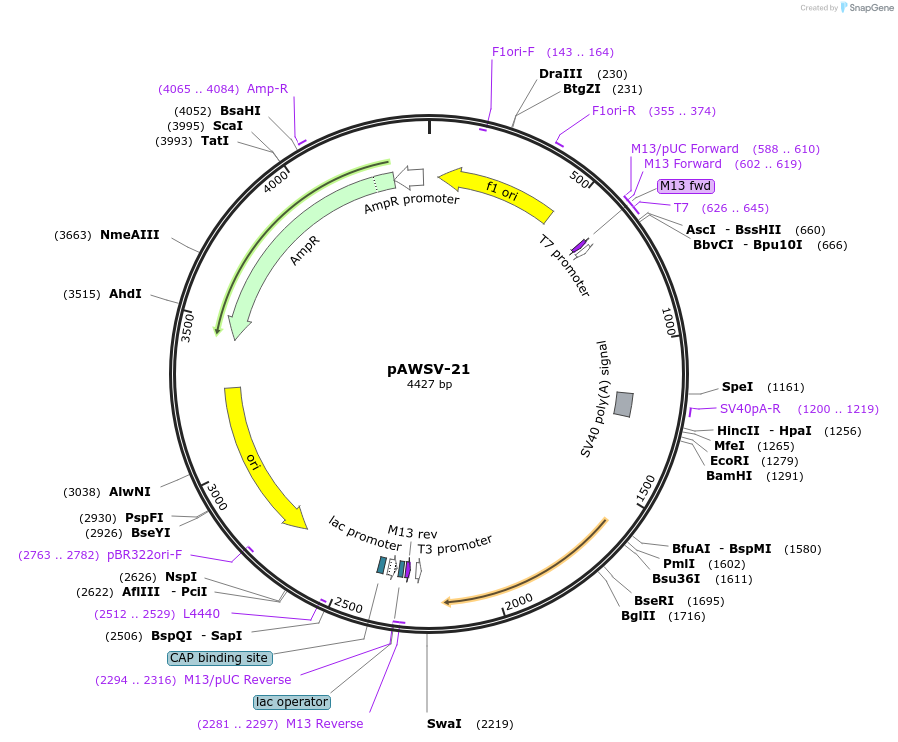
pAWSV-21
Plasmid#72774PurposepBT264-[miR-216]DepositorInsert[miR-216]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
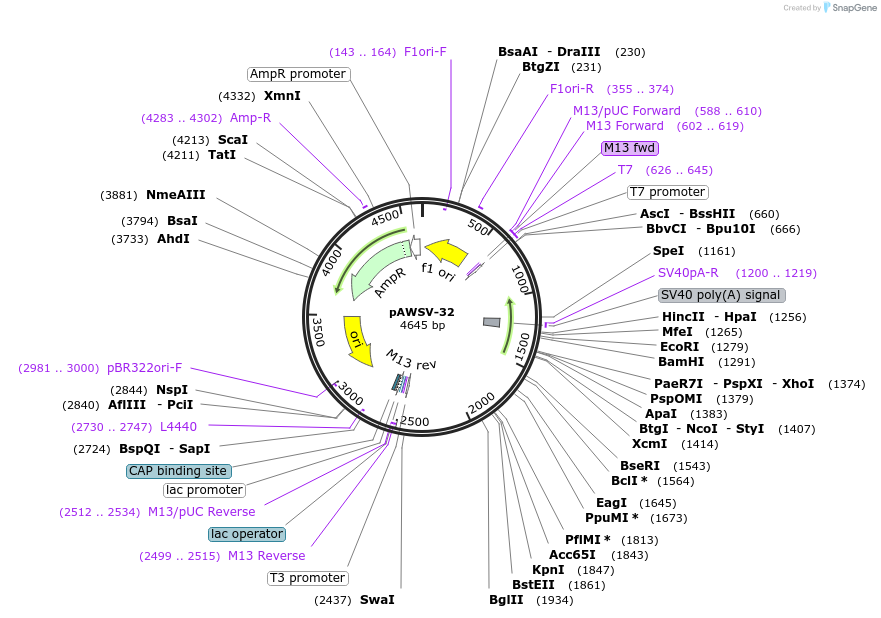
pAWSV-32
Plasmid#72785PurposepBT264-[miR-429]DepositorInsert[miR-429]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
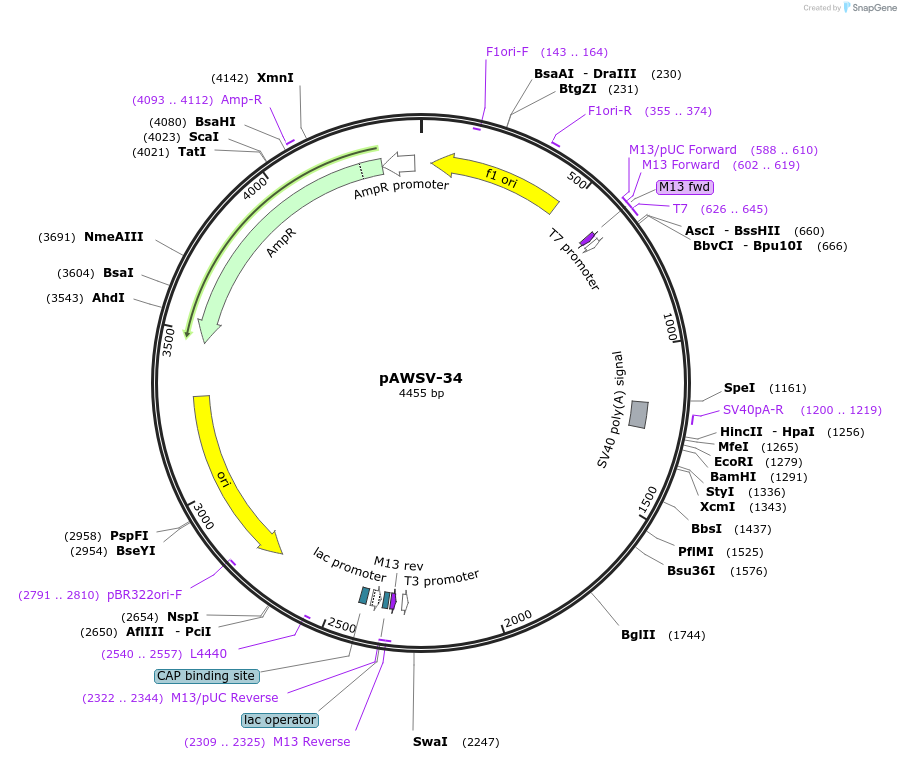
pAWSV-34
Plasmid#72787PurposepBT264-[miR-488]DepositorInsert[miR-488]
Available SinceFeb. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
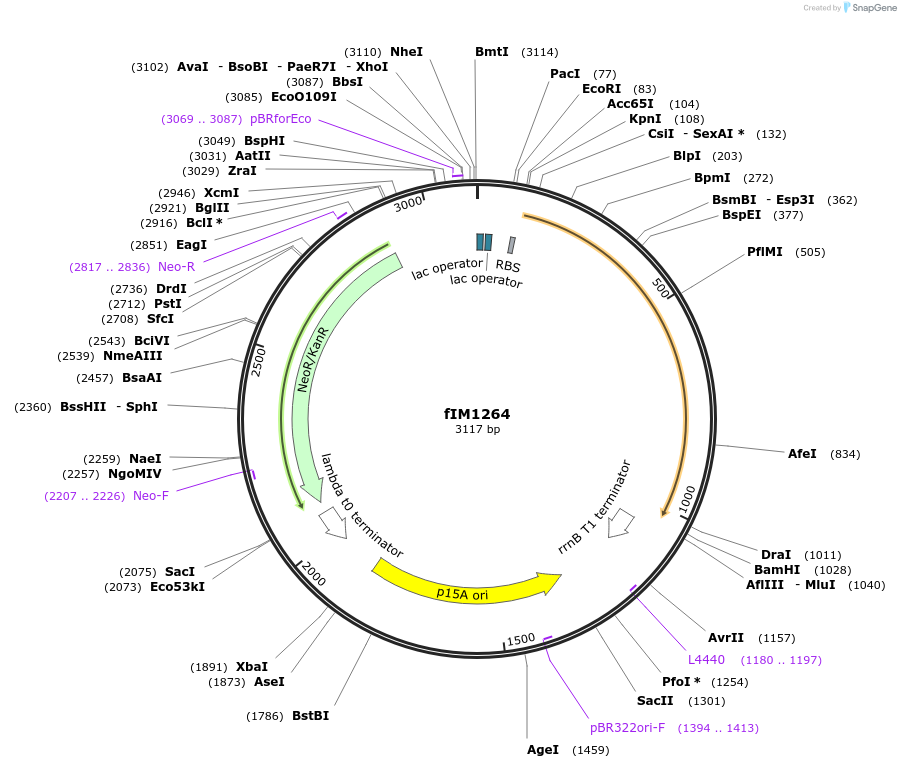
fIM1264
Plasmid#132571PurposepZA2-pLlacO-oxyRDepositorAvailabilityAcademic Institutions and Nonprofits only -
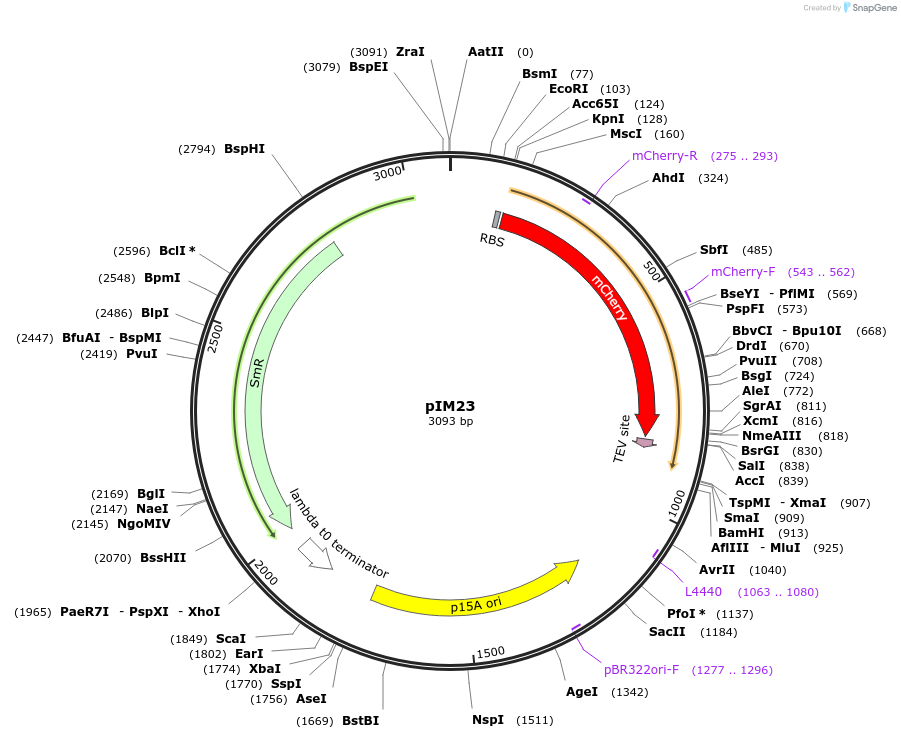
pIM23
Plasmid#132586PurposepZA4-oxySp-mCherry-TEVrs-LAADepositorInsertsoxySp-mCherry-TEVrs-LAA-TermT1
None
ExpressionBacterialPromoternone and oxySpAvailabilityAcademic Institutions and Nonprofits only -
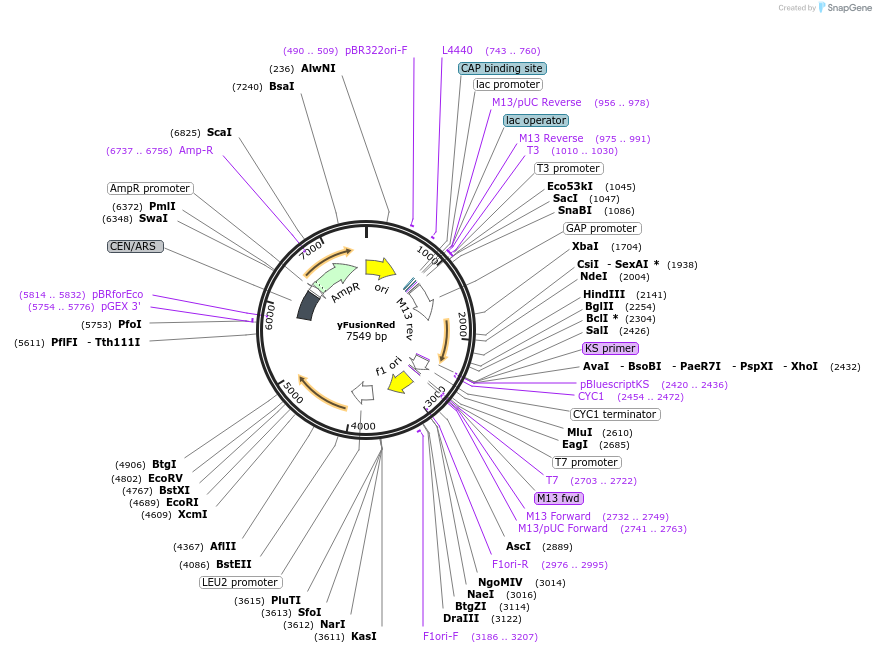
yFusionRed
Plasmid#111916Purposeyeast optimized FusionRedDepositorInsertyFusionRed
ExpressionYeastPromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
AByI
Plasmid#164153PurposeIntegrates marker gene onto D. melanogaster Y chromosomeDepositorInsertY-Left-GypSy_3xp3-optimized-tdtomato-Attp-CTCF-Y-Right
AvailabilityAcademic Institutions and Nonprofits only -
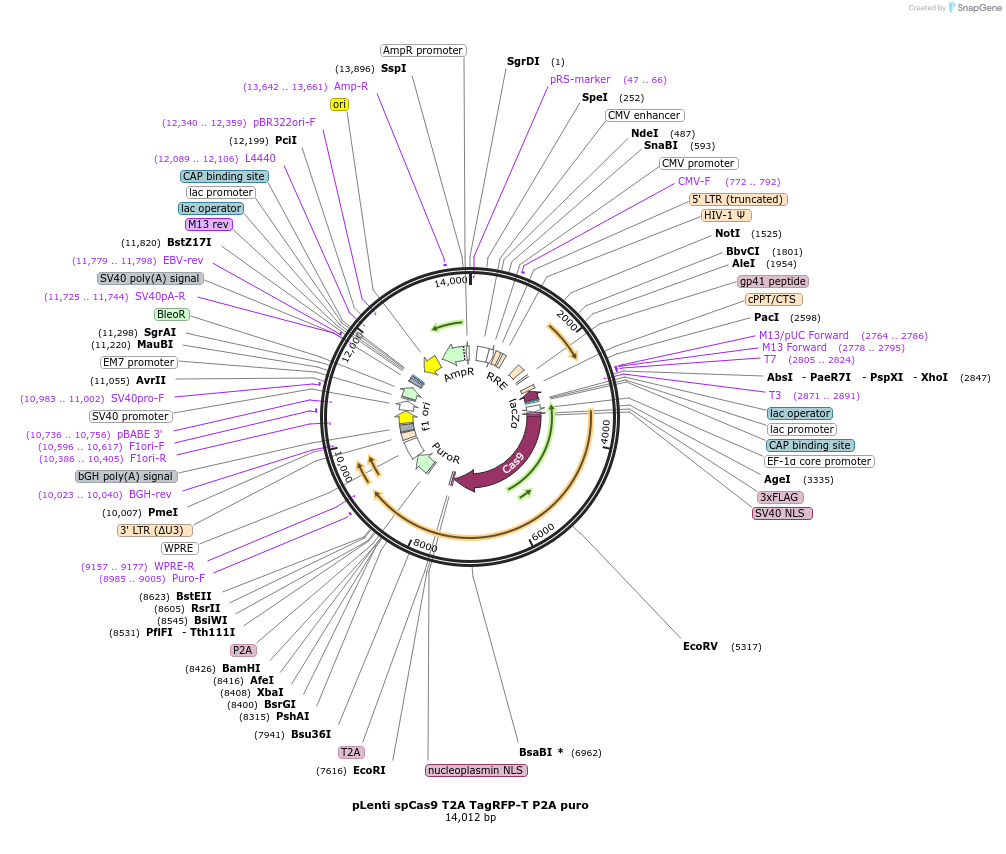
pLenti spCas9 T2A TagRFP-T P2A puro
Plasmid#122200PurposeThe plasmid codes for a Flag-spCas9 protein, a TagRFP-T fluorescent protein and a puromycin resistance. The plasmid has two BsmBI acceptor sites to insert gRNA expressing sequences.DepositorInsertsspCas9
TagRFP-T
UseLentiviralTagsT2AExpressionMammalianAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
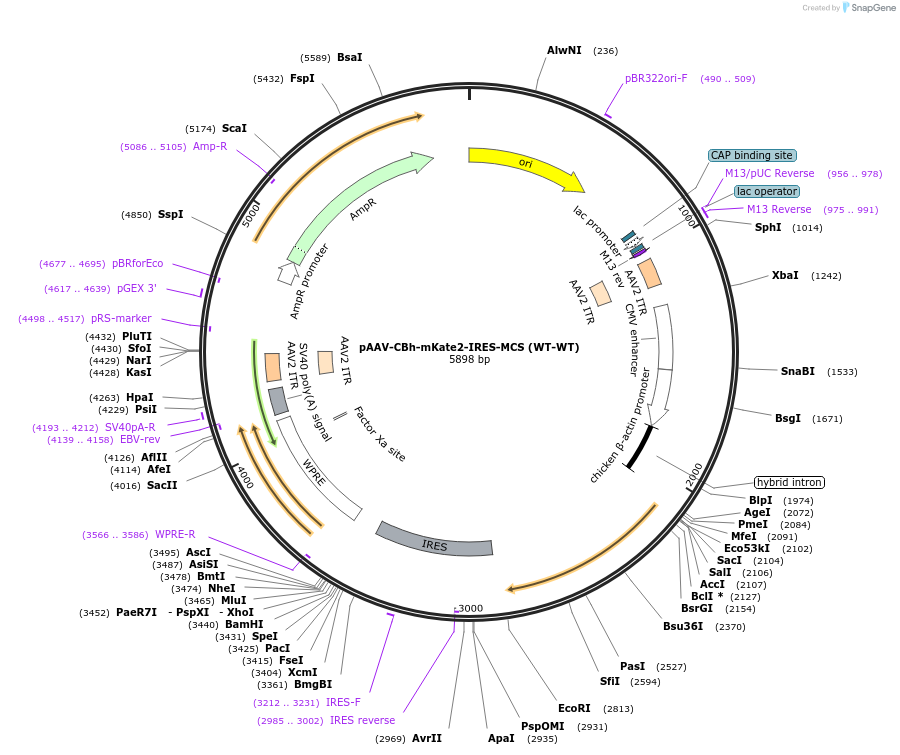
pAAV-CBh-mKate2-IRES-MCS (WT-WT)
Plasmid#105921PurposeAAV-mediated gene expression from CBh promoter. Contains canonical left and right AAV2 ITRsDepositorHas ServiceAAV2InsertsmKate2
IRES-Multiple cloning site
UseAAVMutationcodon optimizedPromoterCBh and same CBh promoter as mKate2Available SinceFeb. 15, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
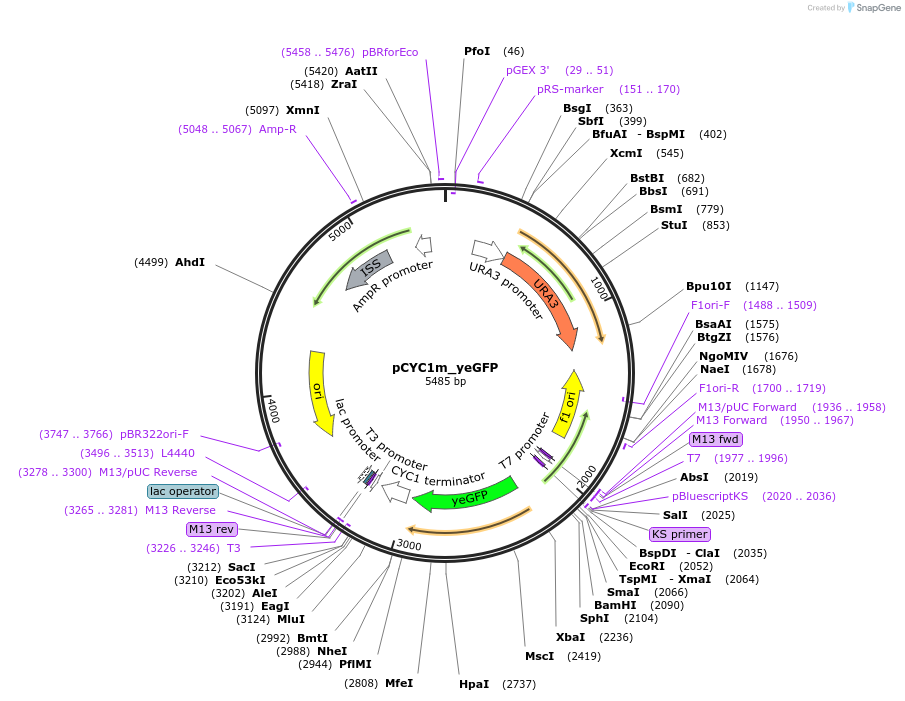
pCYC1m_yeGFP
Plasmid#64389Purposeencodes yeast enhanced GFP under control of minimal pCYC1 promoterDepositorInsertyeast enhanced GFP
ExpressionYeastPromoterpCYC1Available SinceMay 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
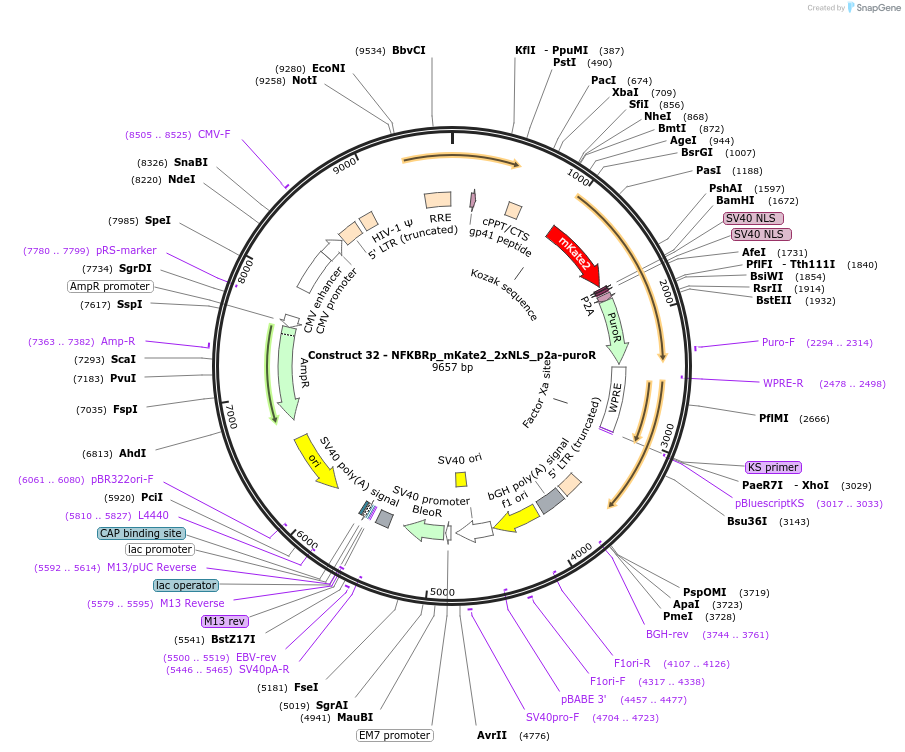
Construct 32 - NFKBRp_mKate2_2xNLS_p2a-puroR
Plasmid#82024PurposeLentiviral construct for building stable cell lines expressing mKate2 in the presence of TNFa via puromycin selectionDepositorInsertmKate2-2xNLS
UseLentiviral and Synthetic BiologyExpressionMammalianAvailable SinceSept. 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
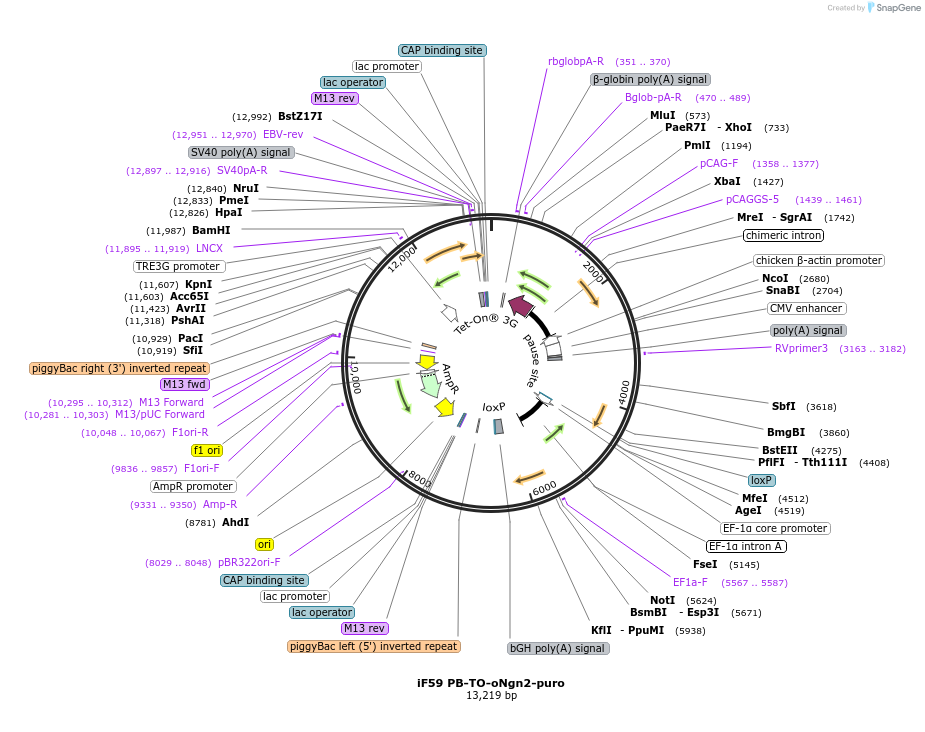
iF59 PB-TO-oNgn2-puro
Plasmid#204734PurposeImproved Dox-inducible neurogenin-2; Piggybac Tet-ON plasmid for differentiating hiPSCs into glutamatergic neurons via NGN2 expression. oNGN2 is a Phospho-mutant which avoids inhibition by cdk kinase.InsertOptimized neurogenin-2
ExpressionMammalianAvailable SinceAug. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
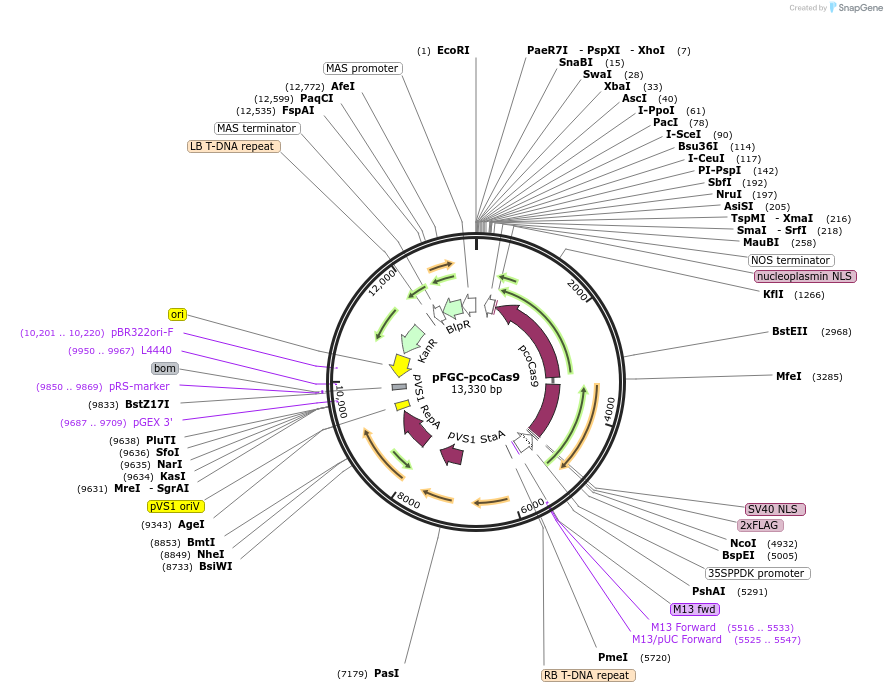
pFGC-pcoCas9
Plasmid#52256PurposeA binary plasmid containing 35SPPDK:pcoCas9:NOS cassette for plant expression of Cas9 and multiple cloning sites for insertion of guide RNADepositorInsert35SPPDK:pcoCas9:NOS cassette
UseCRISPR; Plant expressionPromoterconstitutive 35SPPDK promoterAvailable SinceMarch 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
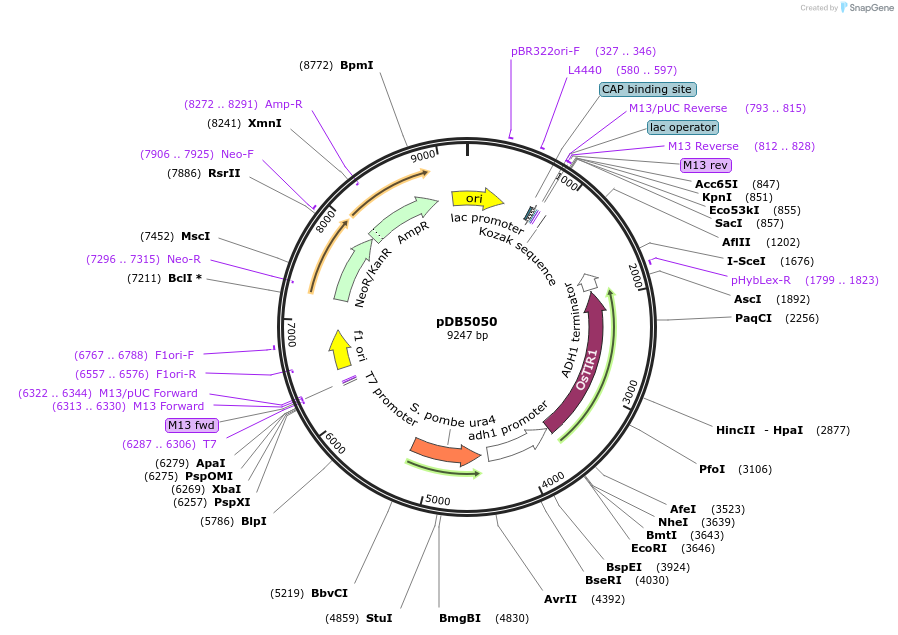
pDB5050
Plasmid#171126PurposeA plasmid containing the codon-optimized OsTIR1, which is under the control of adh1 promoter. It served as one of the F-box protein expression plasmids in AID system.DepositorInsertOsTIR1
UseOtherMutationnon applicableAvailable SinceAug. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
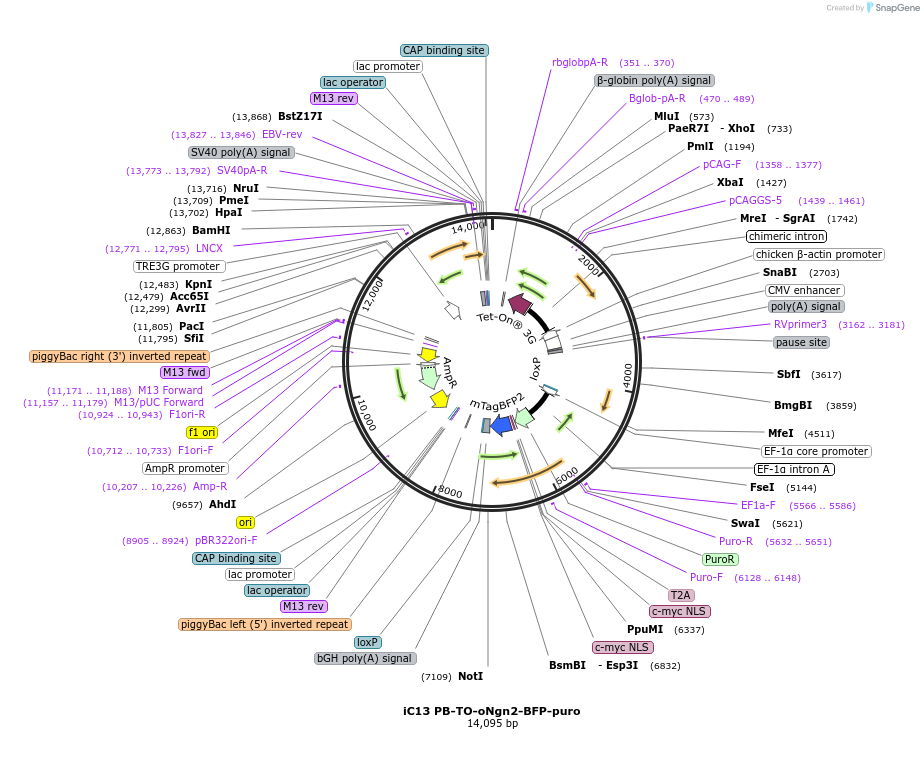
iC13 PB-TO-oNgn2-BFP-puro
Plasmid#204733PurposeImproved Dox-inducible neurogenin-2; Piggybac Tet-ON plasmid for differentiating hiPSCs into glutamatergic neurons via NGN2 expression. oNGN2 is a Phospho-mutant which avoids inhibition by cdk kinase.InsertOptimized neurogenin-2
ExpressionMammalianAvailable SinceAug. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
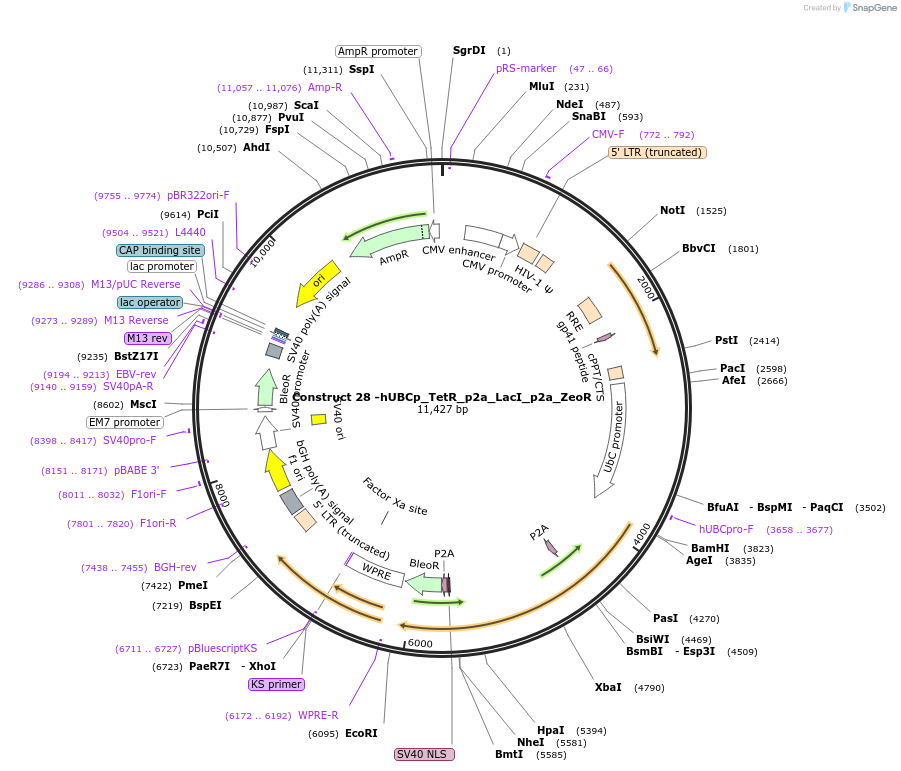
Construct 28 -hUBCp_TetR_p2a_LacI_p2a_ZeoR
Plasmid#81253PurposeLentiviral construct for building stable cell lines expressing TetR and LacI via Zeocin selectionDepositorInsertTetR and LacI
UseLentiviral and Synthetic BiologyExpressionMammalianAvailable SinceSept. 20, 2016AvailabilityAcademic Institutions and Nonprofits only