We narrowed to 81,061 results for: myc
-
Plasmid#133668PurposeIntegration of COX4-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only
-
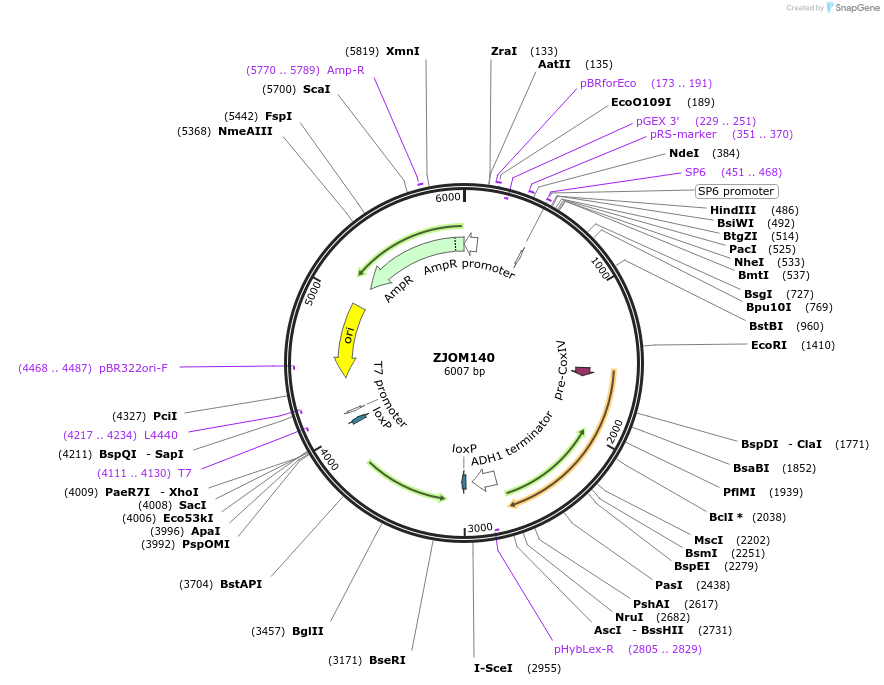
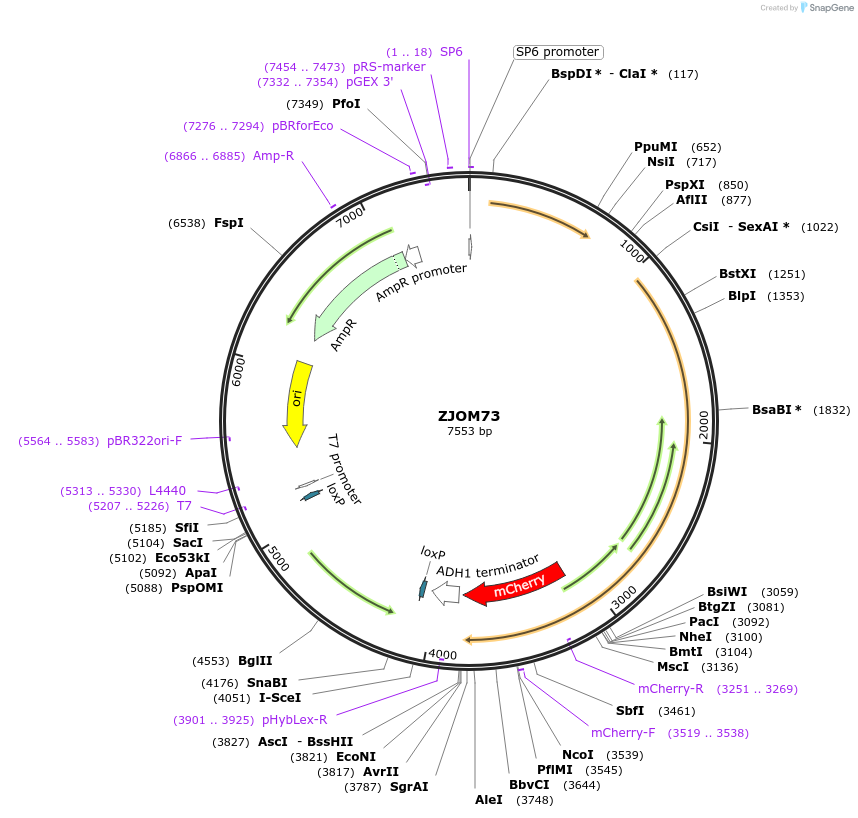
ZJOM73
Plasmid#133651PurposeIntegration of CHS5-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
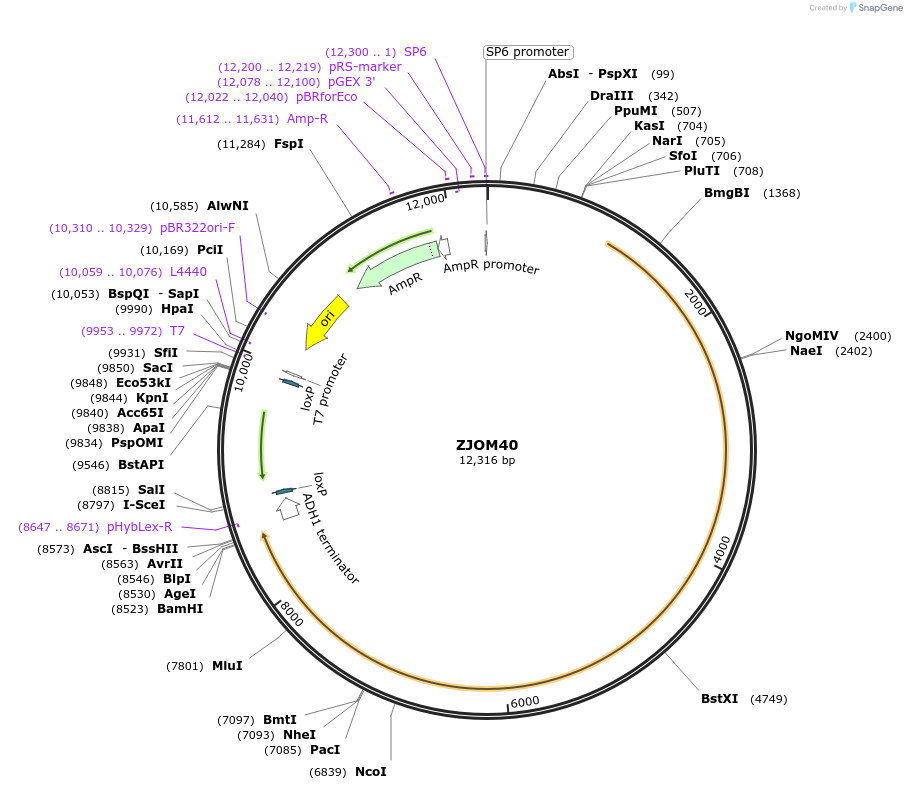
ZJOM40
Plasmid#133650PurposeIntegration of SEC7-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
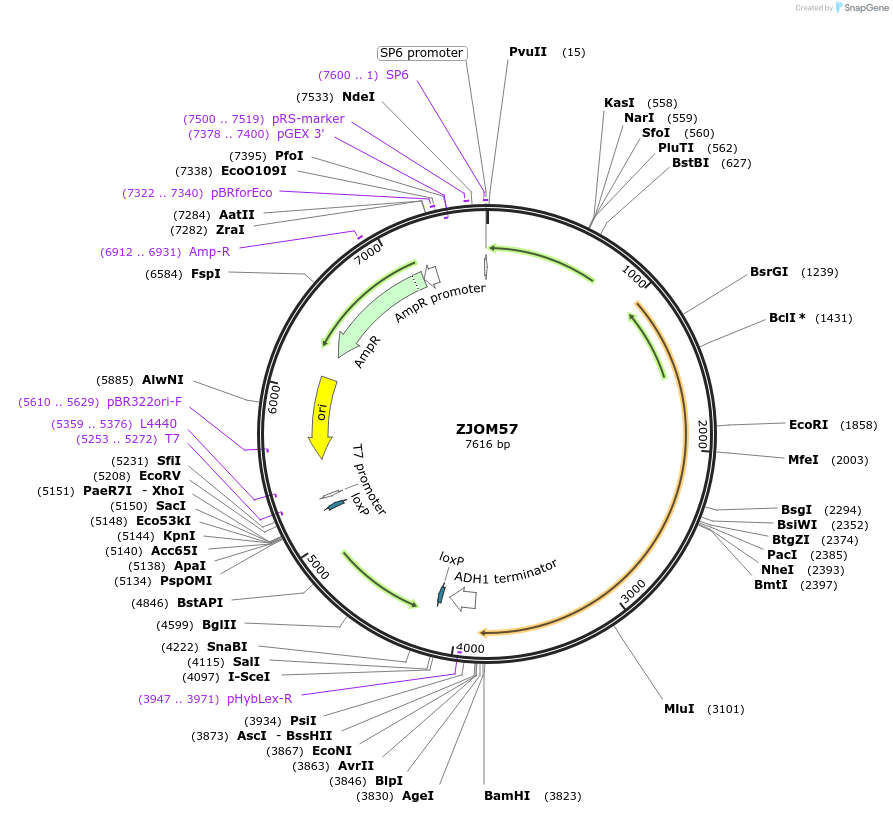
ZJOM57
Plasmid#133656PurposeIntegration of PEX3-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast peroxisome.DepositorAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
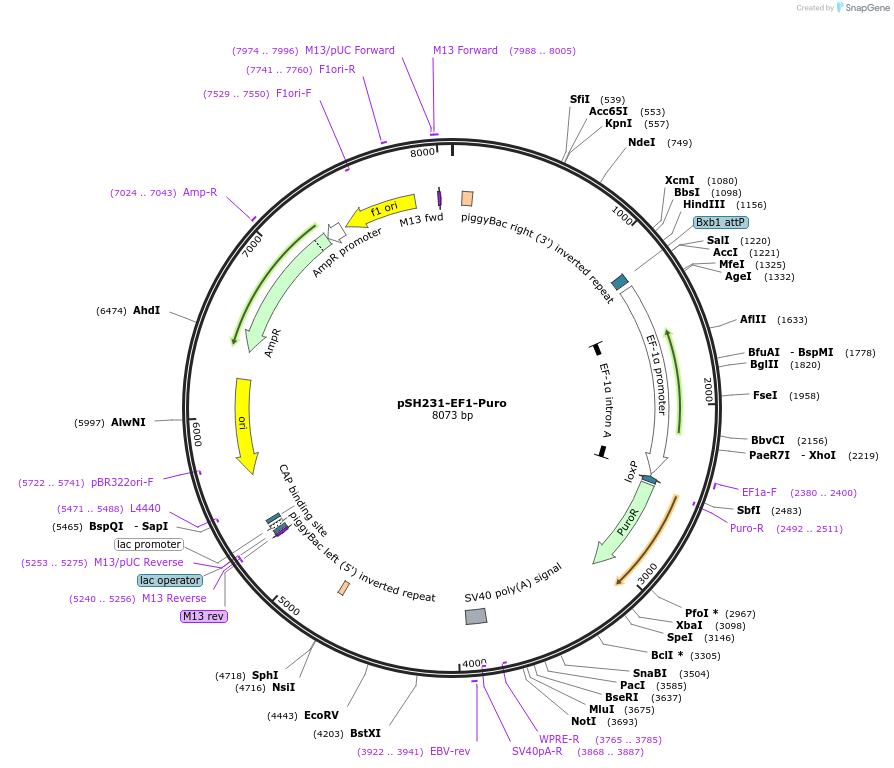
pSH231-EF1-Puro
Plasmid#115143PurposeSafe harbor site 231 knock-in vector with puromycin resistance selection cassetteDepositorInsertPuromycin
ExpressionMammalianPromoterEF1aAvailable SinceOct. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
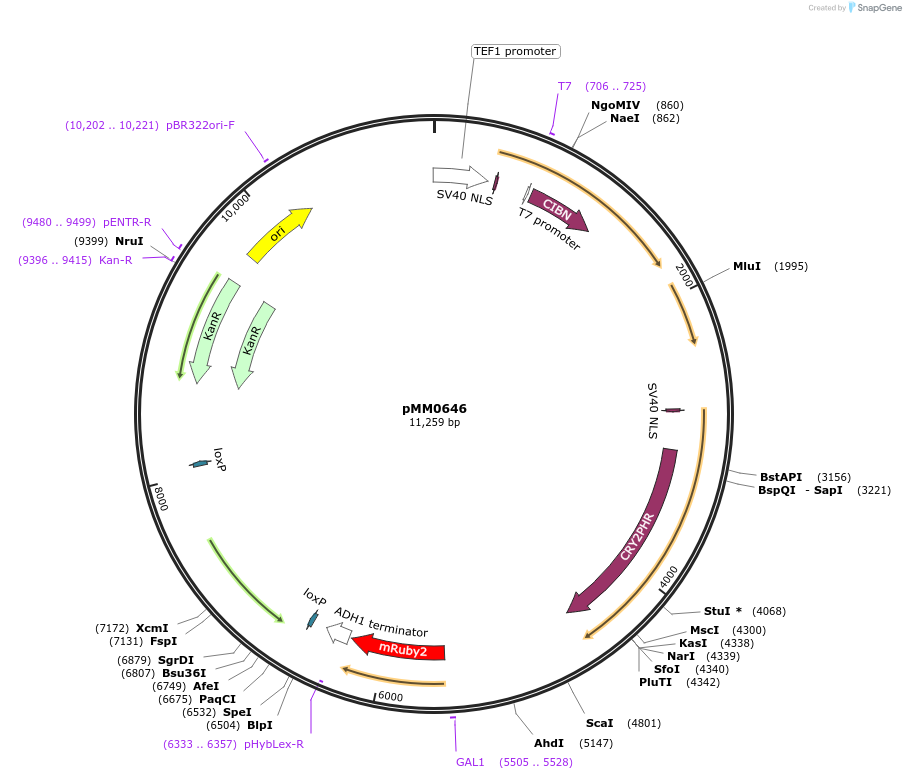
pMM0646
Plasmid#129007PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2 with loxable KIURA3 markerDepositorInsertKanR-ColE1 URA 5'-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-loxP-KIURA3-loxP-URA3 3'
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
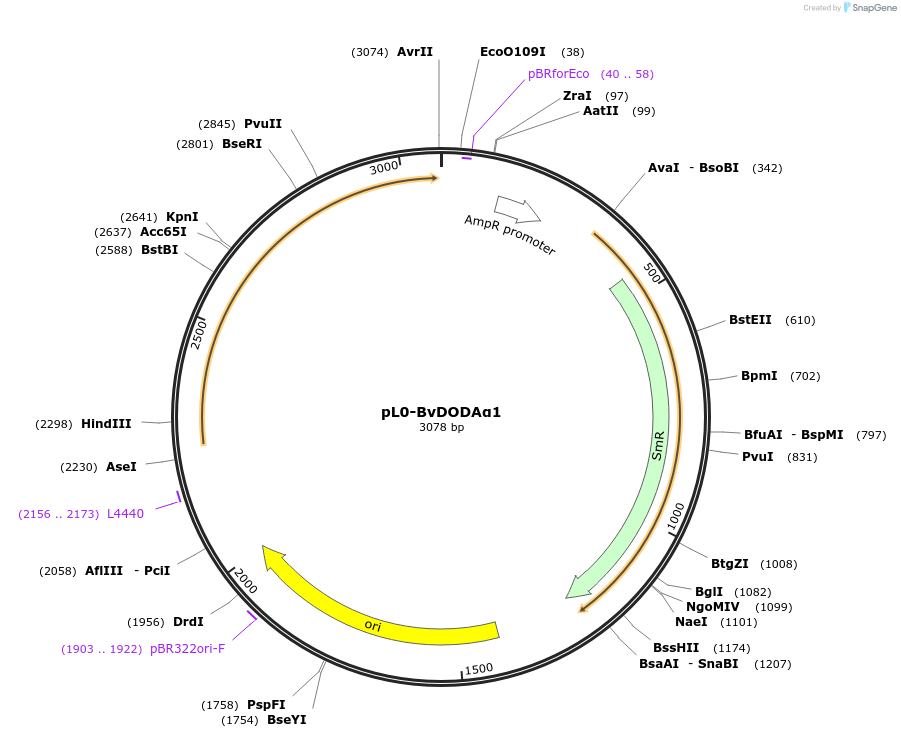
pL0-BvDODAα1
Plasmid#162528PurposeBeta vulgaris 4,5-DOPA dioxygenase alpha 1 coding sequence in pICH41308 MoClo Golden Gate level 0 acceptor for CDS1 modules.DepositorInsertDODAα1
UsePuc19-derivedExpressionBacterialAvailable SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
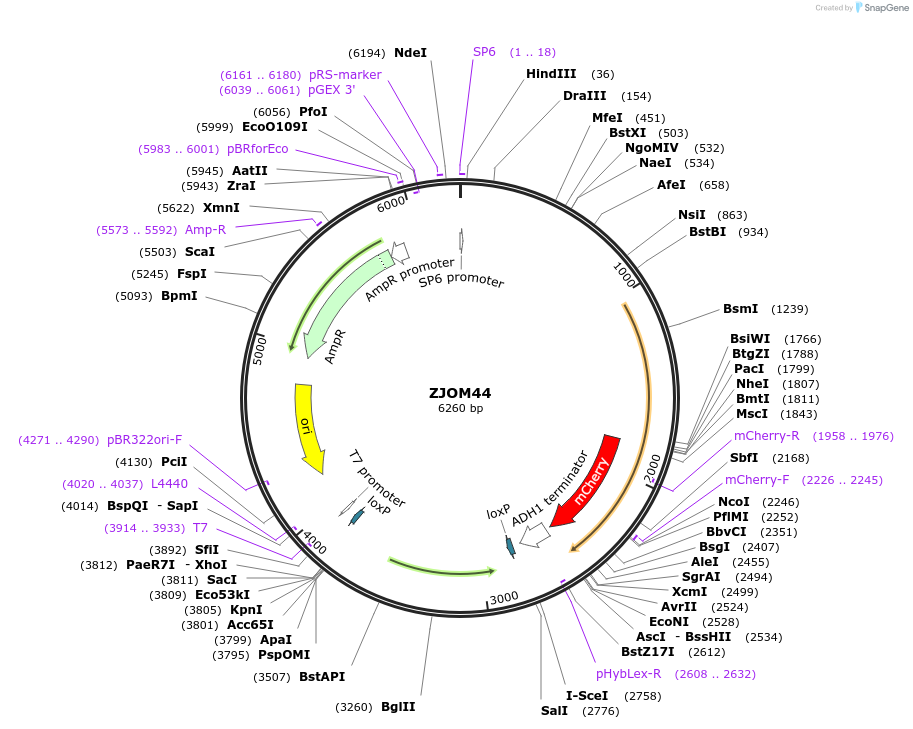
ZJOM44
Plasmid#133653PurposeIntegration of SNF7-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
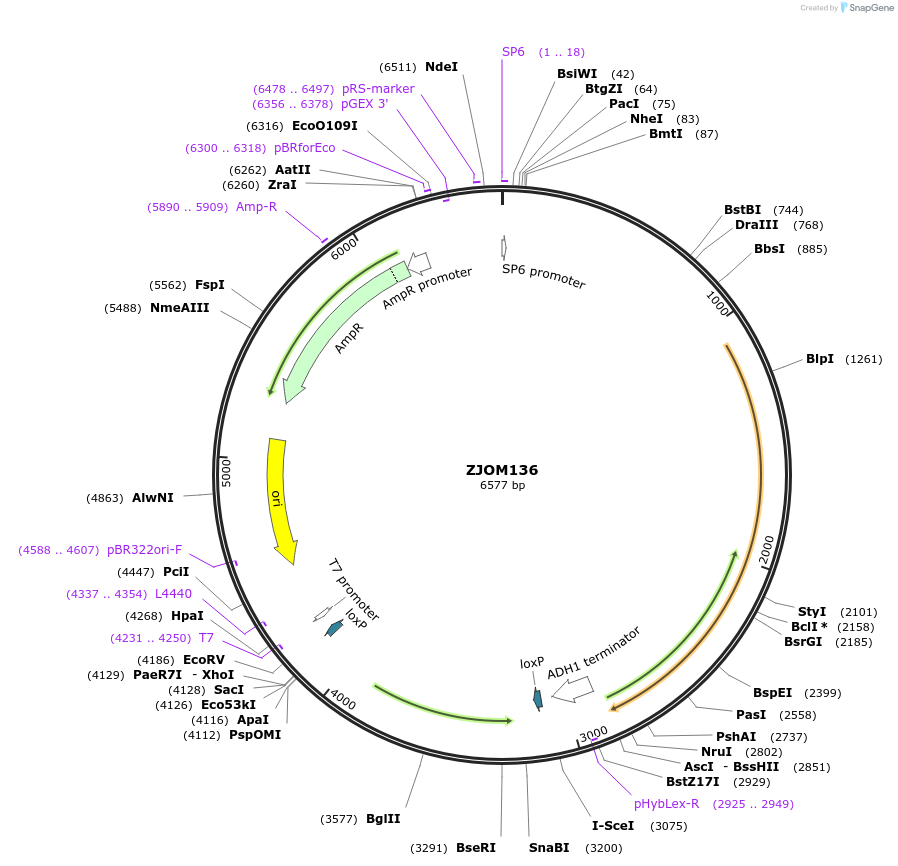
ZJOM136
Plasmid#133660PurposeIntegration of ELO3-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
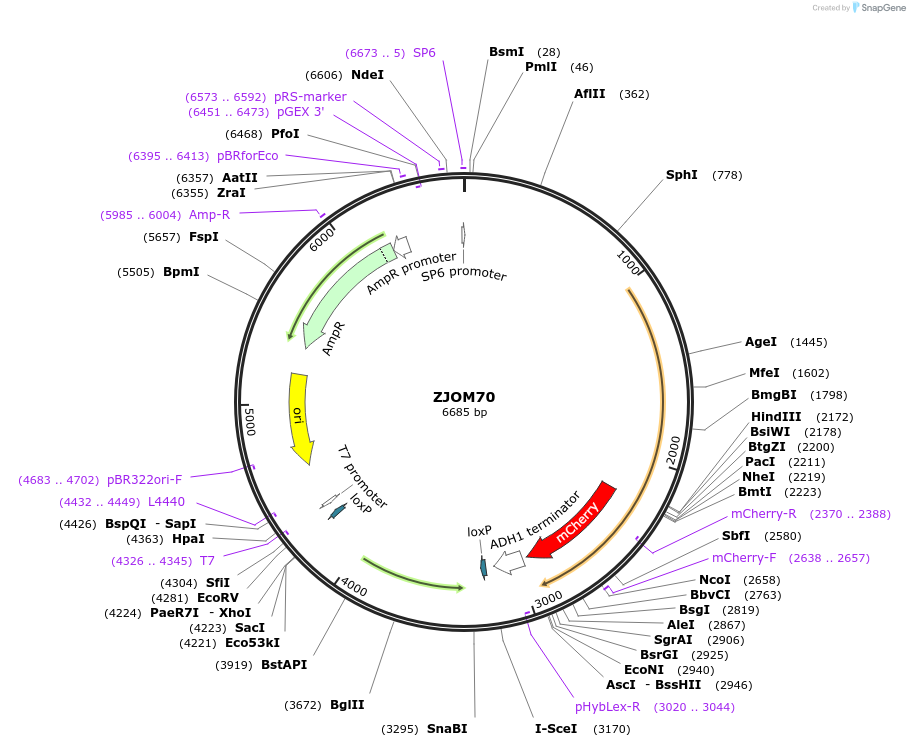
ZJOM70
Plasmid#133658PurposeIntegration of ERG6-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
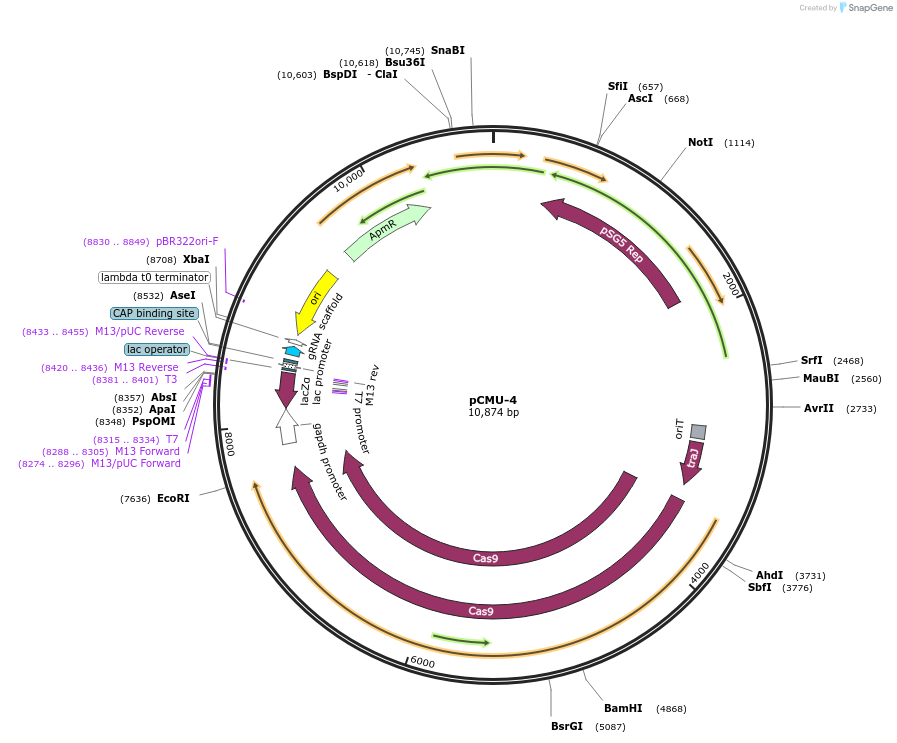
pCMU-4
Plasmid#172353PurposeCRISPR gene editing in Streptomyces, codon-optimized Cas9 behind theophylline riboswitch and D4 promoter, with custom gRNADepositorInsertD4-TheoR-cas9
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterD4Available SinceDec. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
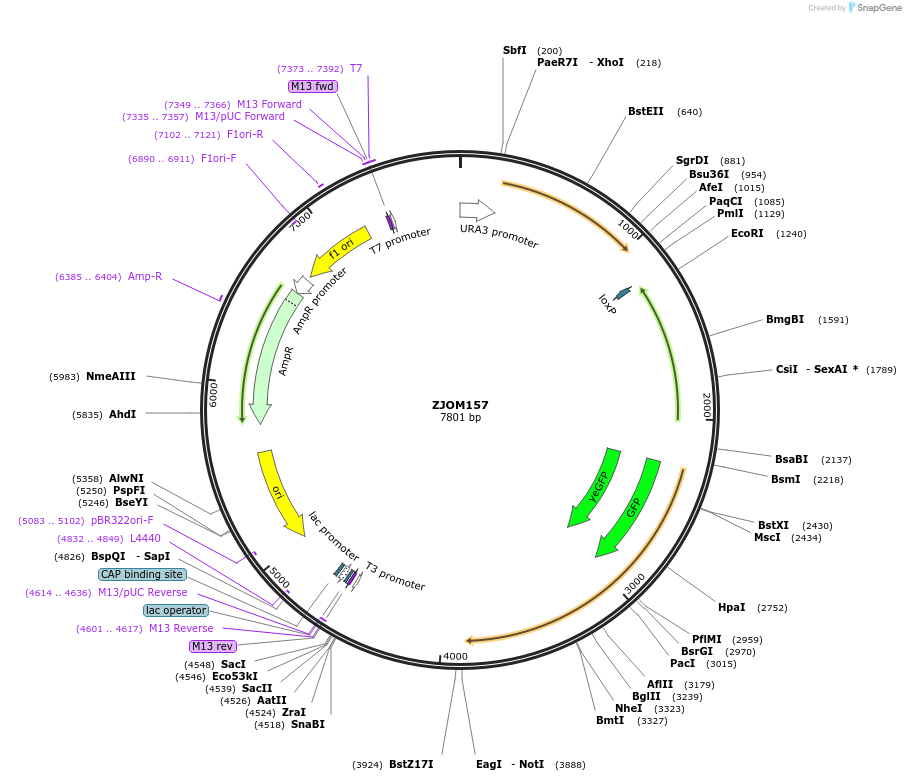
ZJOM157
Plasmid#133640PurposeIntegration of GFP-PEP12 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis).Marker for yeast late endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
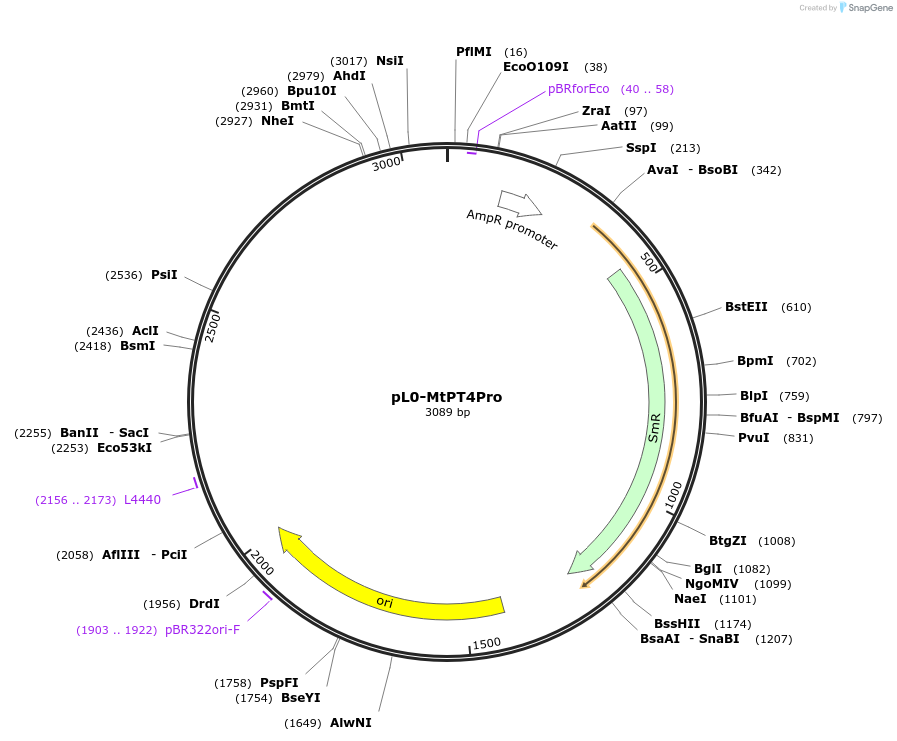
pL0-MtPT4Pro
Plasmid#159414PurposeMedicago truncatula PT4 promoter sequence (836 bp immediately upstream of start codon) in pICH41295 MoClo Golden Gate level 0 acceptor for Pro+5U modules.DepositorInsertMtPT4 Promoter
UsePuc19-derivedExpressionBacterialAvailable SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
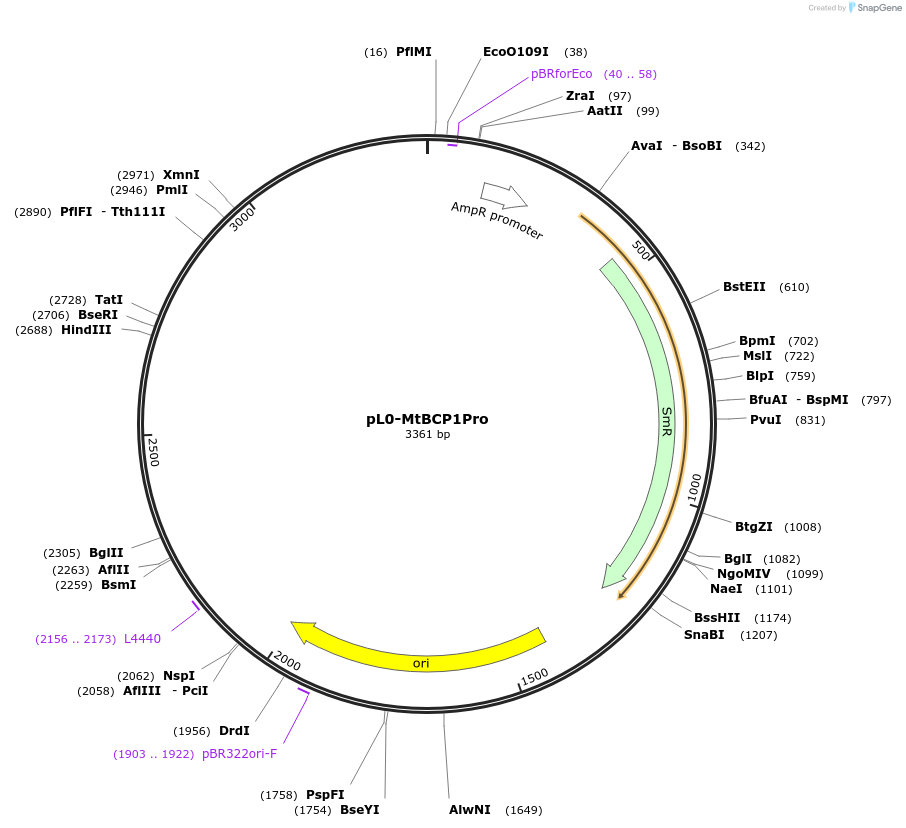
pL0-MtBCP1Pro
Plasmid#159415PurposeMedicago truncatula BCP1 promoter sequence (1108 bp immediately upstream of start codon) in pICH41295 MoClo Golden Gate level 0 acceptor for Pro+5U modules.DepositorInsertMtBCP1 Promoter
UsePuc19-derivedExpressionBacterialAvailable SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
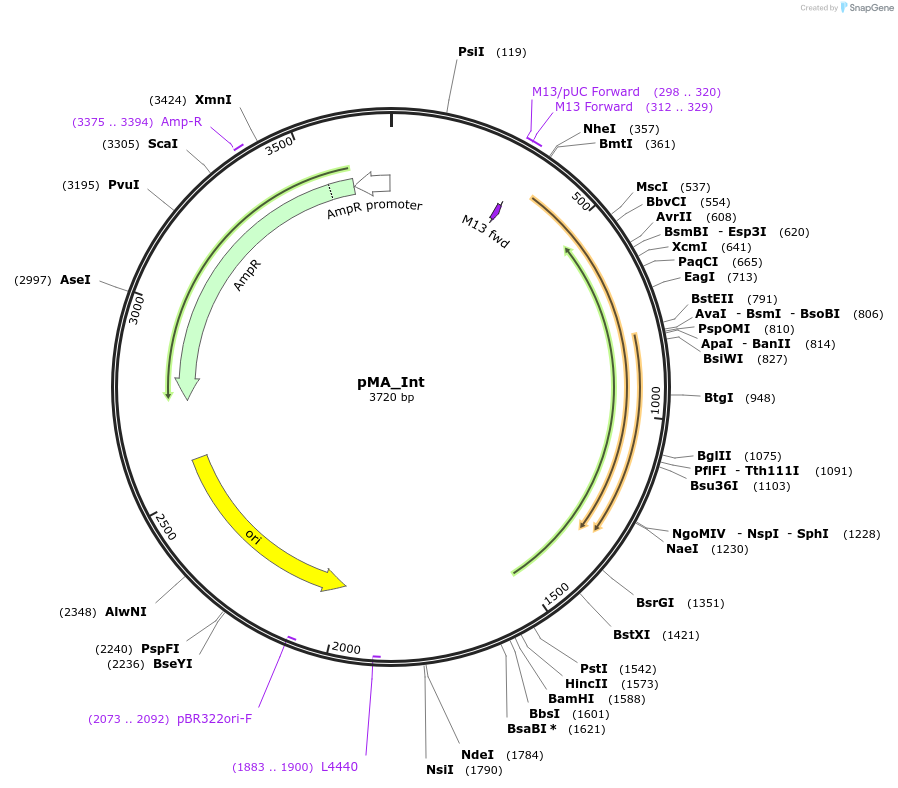
pMA_Int
Plasmid#110096PurposeE. coli replicative vector which contains the mycophage integrase gene required for the attP/attB integration. Is co-transformed in trans together with pFLAG_attP, but cannot replicate in mycobacteria, and is thus lost shortly after electroporation.DepositorInsertmycophage integrase
UseRequired for transformation of attb integrating v…Available SinceJune 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
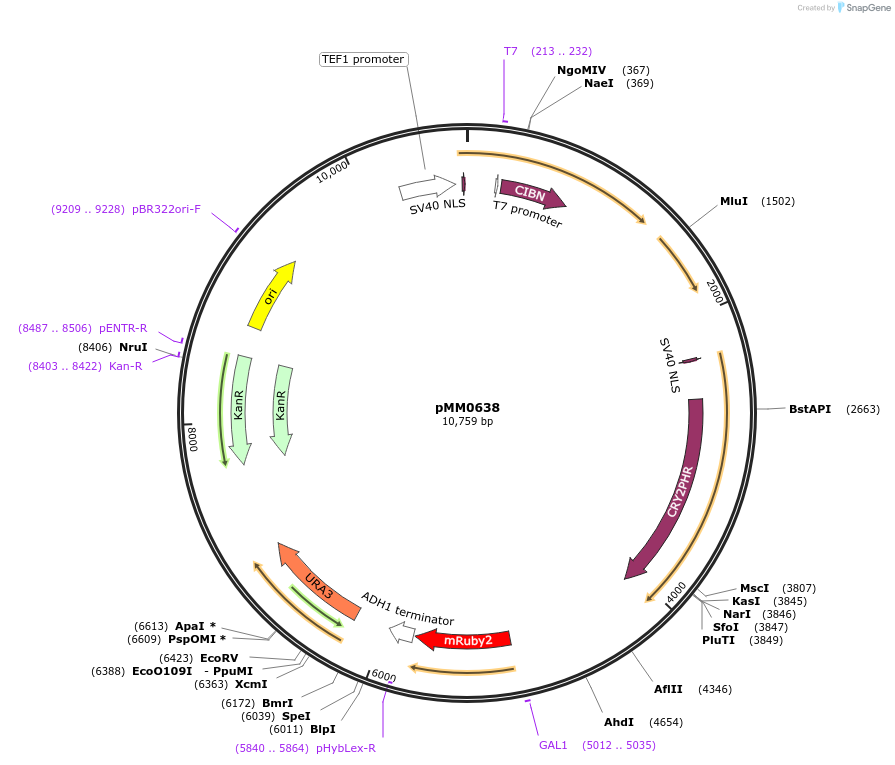
pMM0638
Plasmid#129005PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2DepositorInsert5' URA3 -pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-URA3 URA3 3'- KanR-ColE1
ExpressionYeastAvailable SinceOct. 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
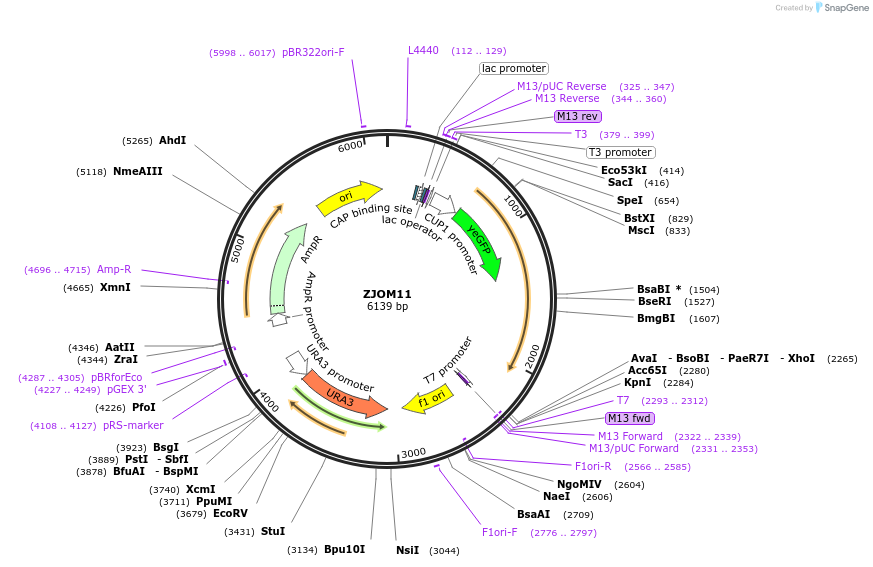
ZJOM11
Plasmid#133637PurposeIntegration of GFP-TLG1 as an additional copy in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
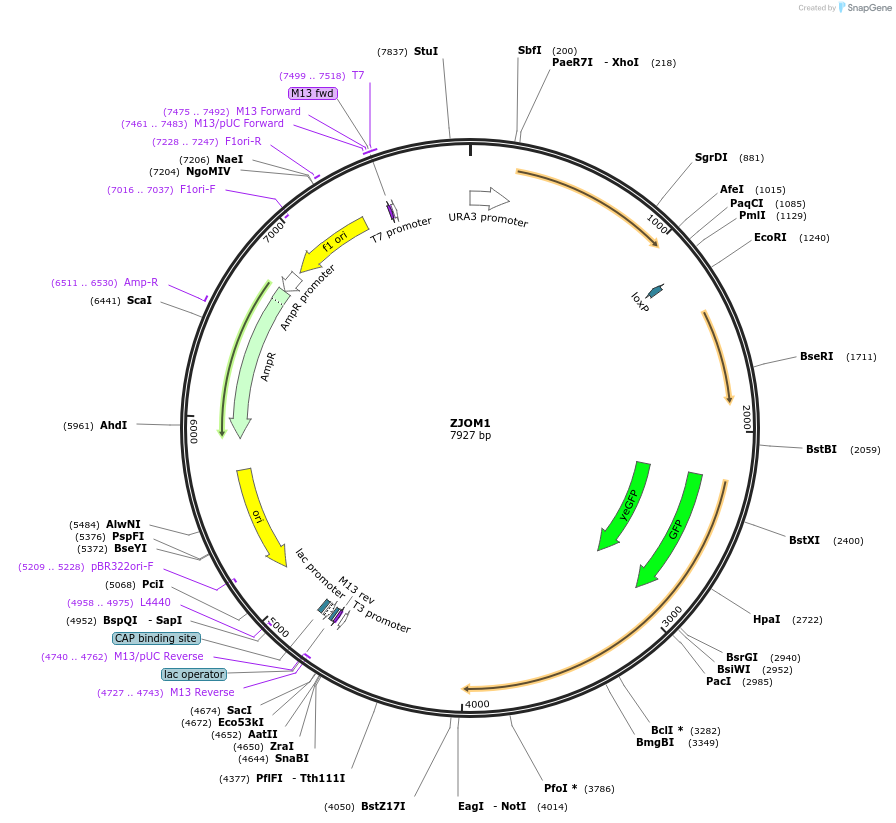
ZJOM1
Plasmid#133632PurposeIntegration of GFP-SED5 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
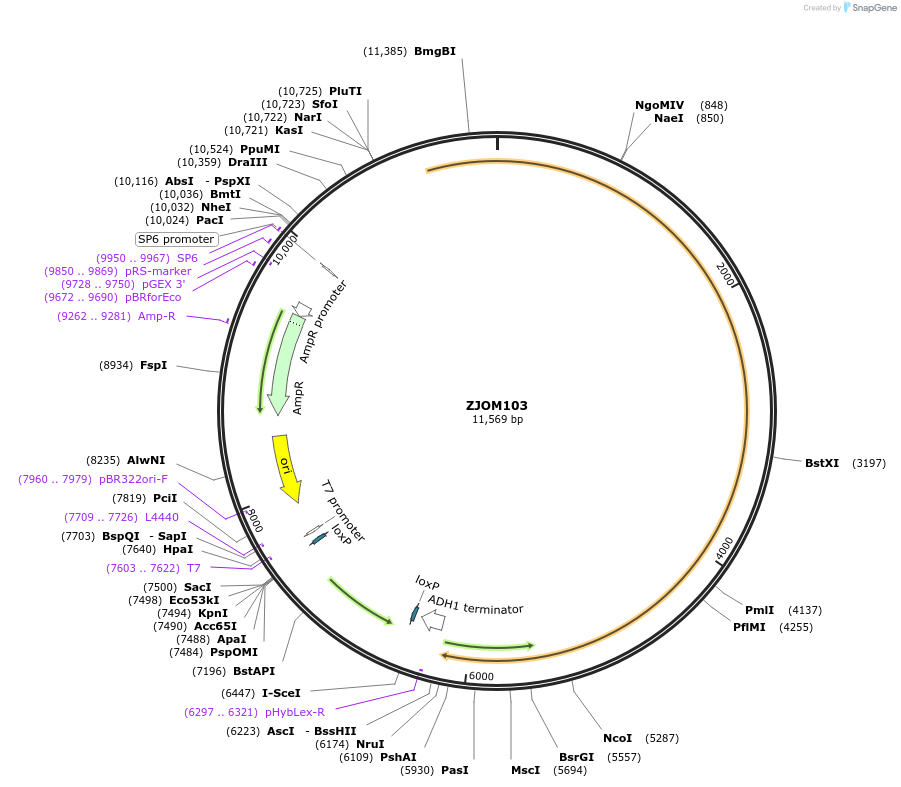
ZJOM103
Plasmid#133664PurposeIntegration of SEC7-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
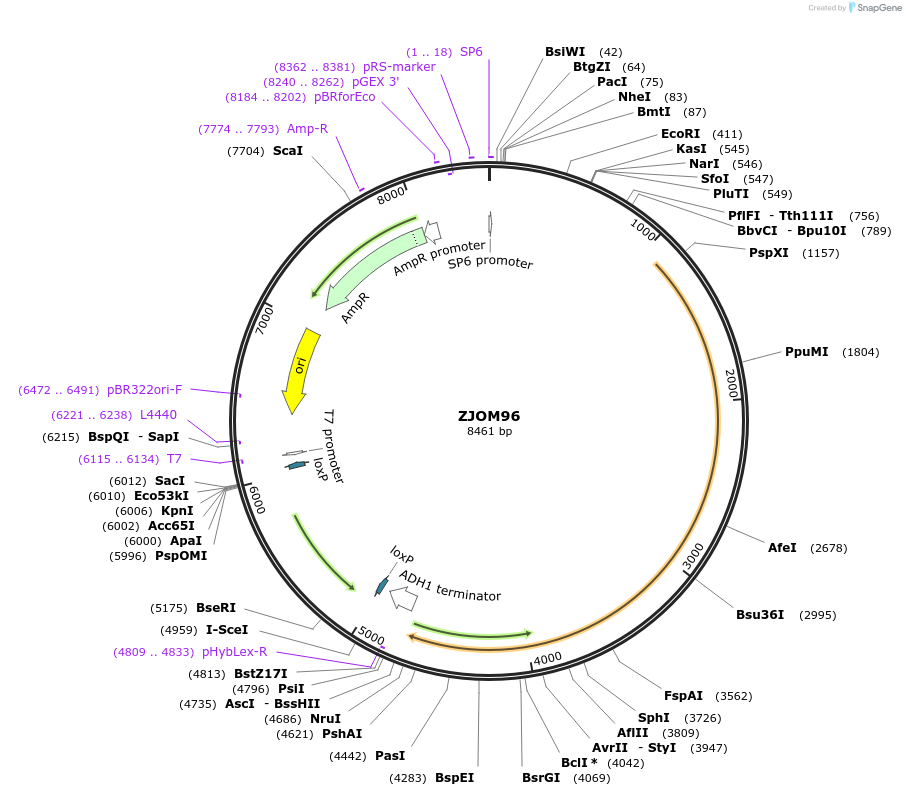
ZJOM96
Plasmid#133663PurposeIntegration of SEC26-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only