We narrowed to 11,805 results for: NSI
-
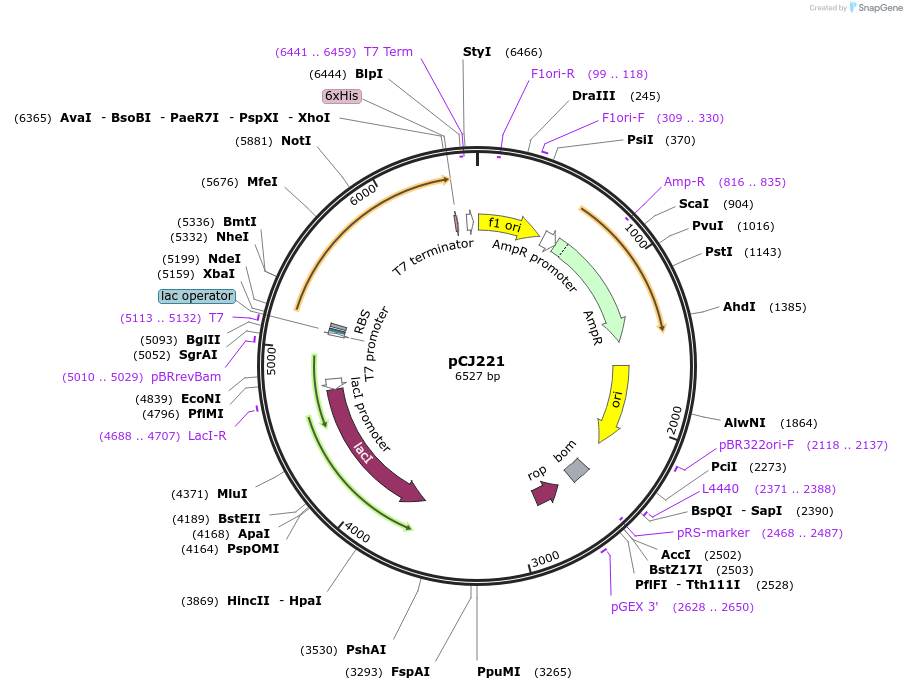
Plasmid#162687PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224H and C529F mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with lid deletion and C224H and C529F mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224H, C529F; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
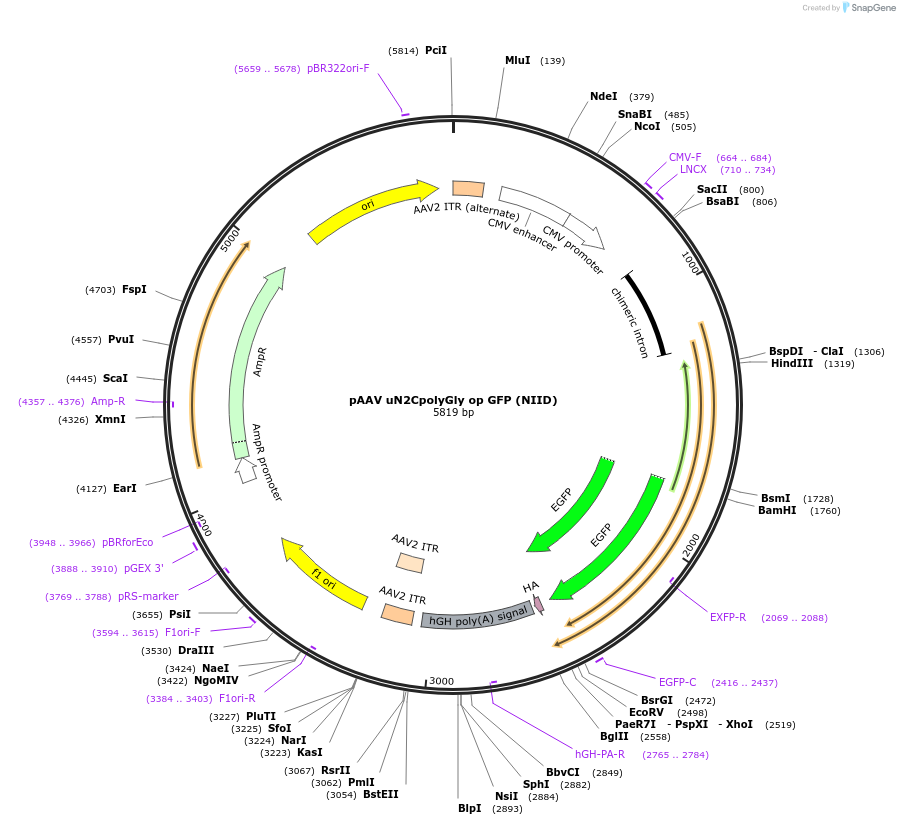
pAAV uN2CpolyGly op GFP (NIID)
Plasmid#224356PurposeExpress GFP tagged upsteram ORF of NOTCH2NLC with an expansion (100x) of GGC repeats with codon optimization (NOT pure GGC repeats)DepositorInsertuN2C with a GGC repeat expansion (100x)
UseAAVTagseGFPExpressionMammalianMutationCodon-optimized for expression in human, so no pu…PromoterCMV + chimeric intron (CAG)Available SinceFeb. 5, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
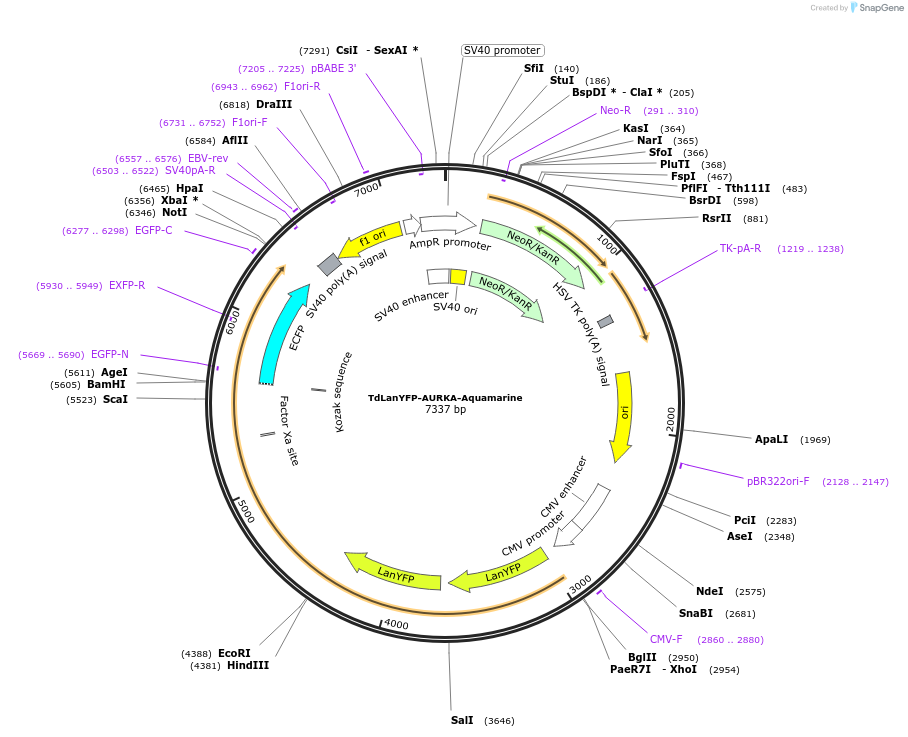
TdLanYFP-AURKA-Aquamarine
Plasmid#228562PurposeEncodes the AURKA biosensor where LC3B is flanked by the Aquamarine/TdLanYFP donor/acceptor FRET pair and under the control of the CMV promoter.DepositorAvailable SinceJan. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
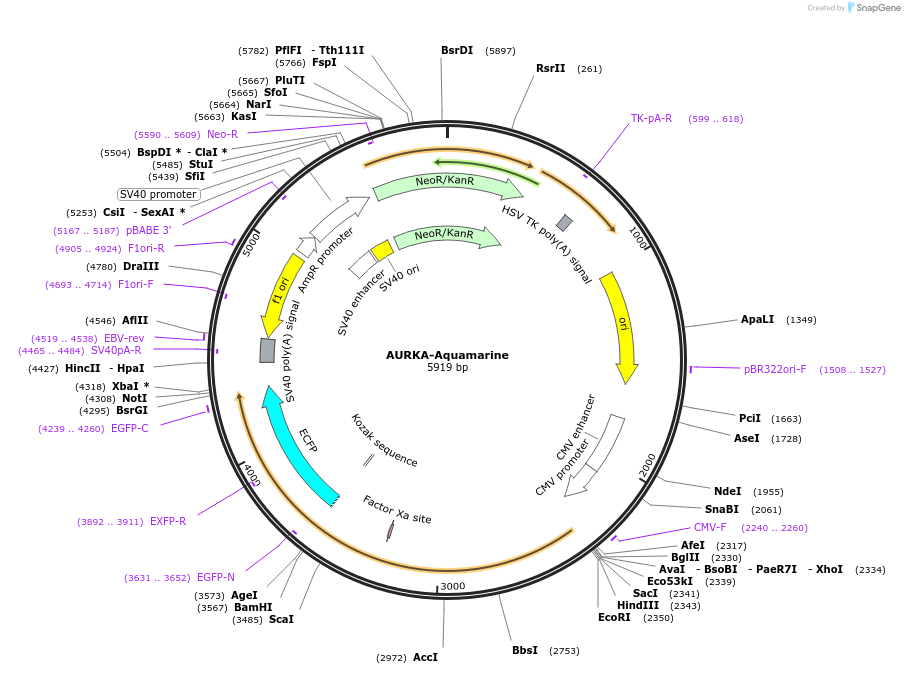
AURKA-Aquamarine
Plasmid#228564PurposeEncodes a donor-only, Aquamarine-tagged version of the AURKA biosensor under the control of a CMV promoterDepositorAvailable SinceJan. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
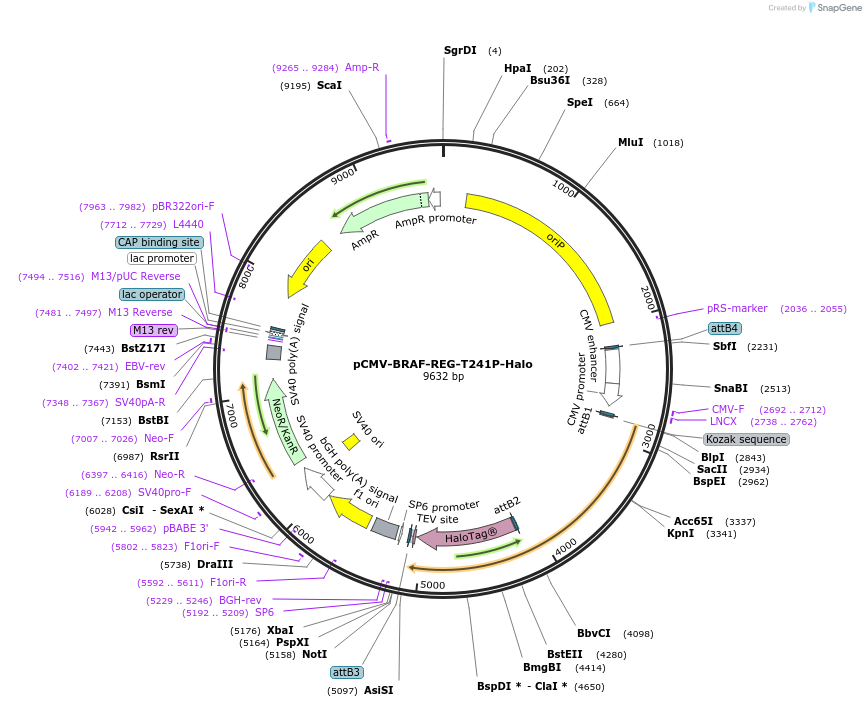
pCMV-BRAF-REG-T241P-Halo
Plasmid#202549PurposeMammalian expression construct encoding the BRAF regulatory (REG) domain (AA 1-435) with a C-terminal HaloTag. The REG domain contains the RASopathy mutation, T241P, within its cysteine-rich domain.DepositorInsertBRAF regulatory (REG) domain (AA 1-435) (BRAF Human)
TagsHaloTagExpressionMammalianMutationThreonine 241 mutated to prolinePromoterCMVAvailable SinceJuly 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
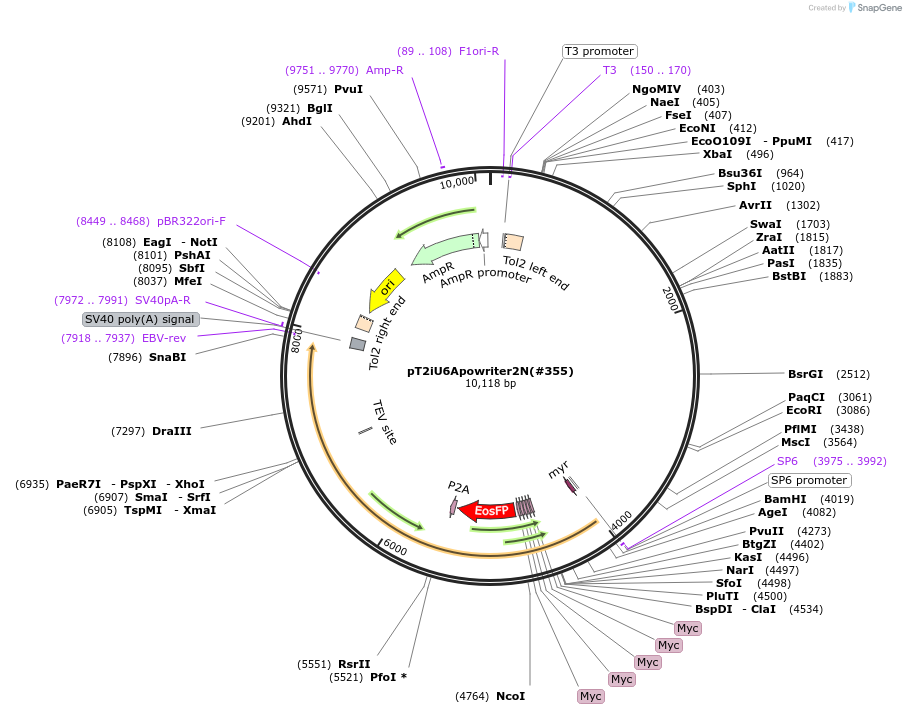
pT2iU6Apowriter2N(#355)
Plasmid#184106Purposecyclofen-inducible apoptosis activation and imaging (tEosFP) by zebrafish transient transgenesis or mRNA synthesisDepositorInsertmyr-DIAP1-5myc-tEosFP-nls'-P2A-Casp9-ERT2
UseZebrafish transient transgenesis + in vitro trans…Tagsinternal tag: 5mycMutationDIAP1: deleted 1-2, N19G, N20V, deleted 146-438; …PromoterZf-Ubi+SP6Available SinceJune 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
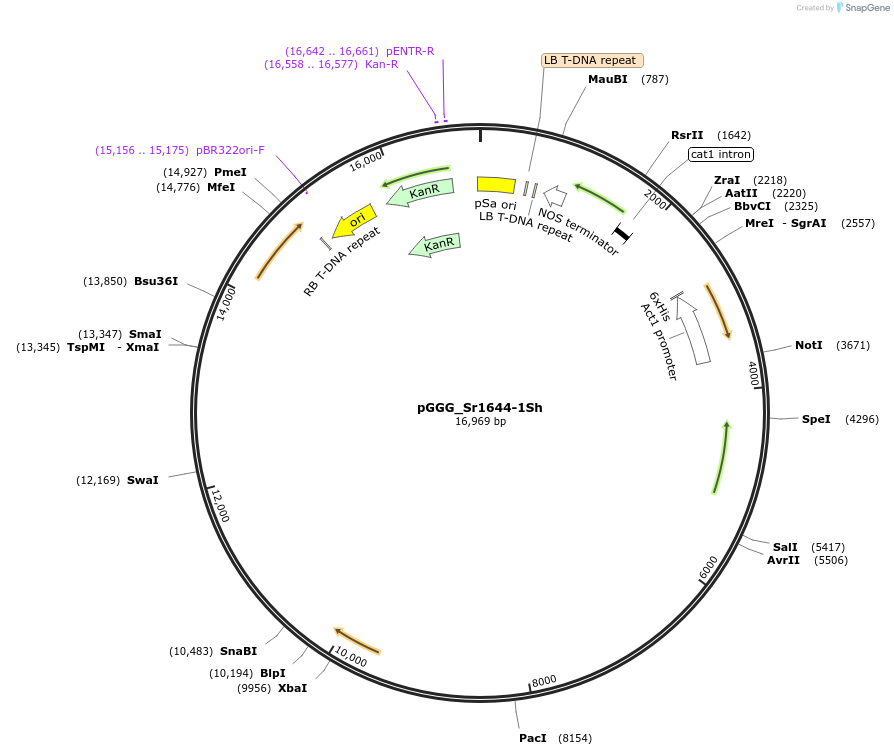
pGGG_Sr1644-1Sh
Plasmid#164087PurposeExpresses pGGG_Sr1644-1Sh in plantsDepositorInsertpGGG_Sr-1644-1Sh
TagsnoneExpressionBacterial and PlantPromoterNative promoterAvailable SinceMay 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
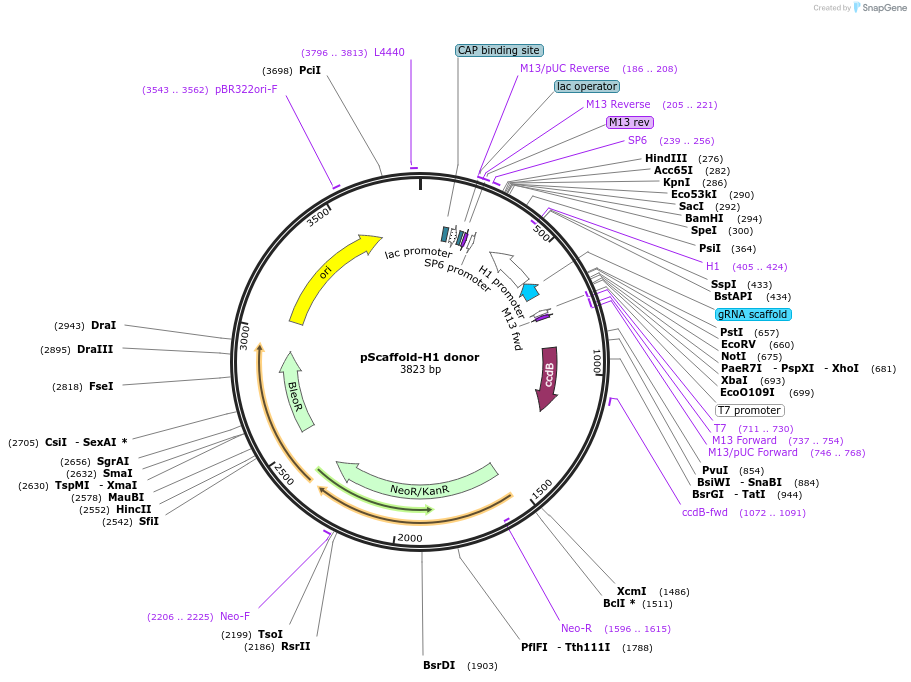
pScaffold-H1 donor
Plasmid#118152PurposePCR template for dual guide RNA cloning, guide RNA scaffold for the Streptococcus pyogenes CRISPR/Cas9 system - H1 promoterDepositorTypeEmpty backboneExpressionMammalianAvailable SinceJuly 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
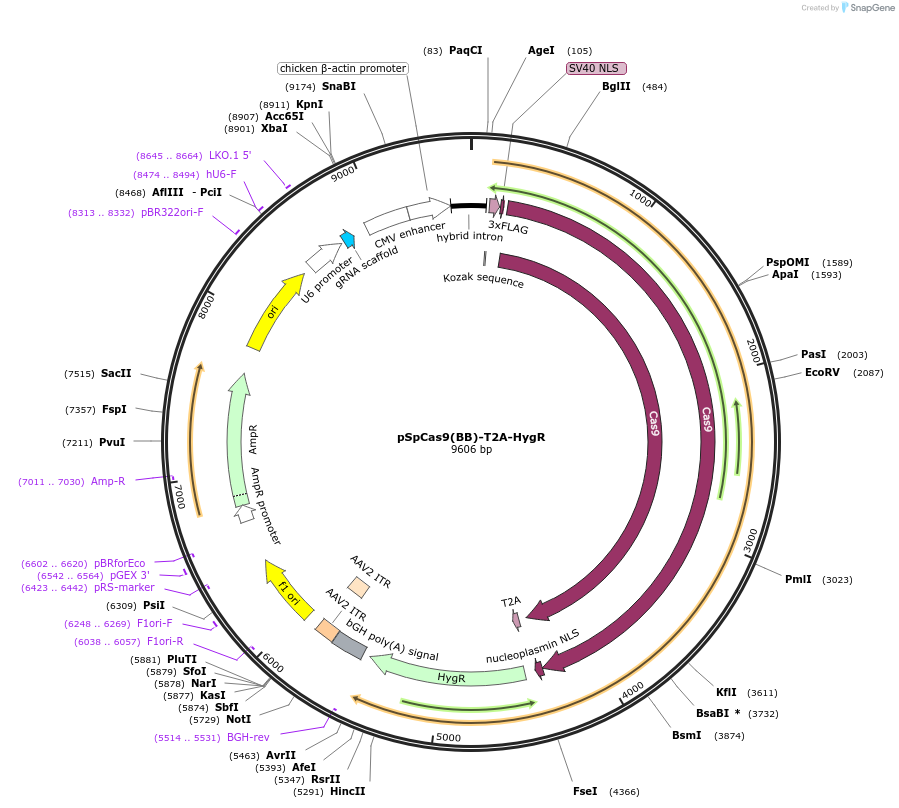
pSpCas9(BB)-T2A-HygR
Plasmid#118153PurposeSpCas9 expression vectorDepositorInserthSpCas9-T2A-HygR
UseCRISPRTags3XFLAGExpressionMammalianPromoterCAG and U6Available SinceMarch 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
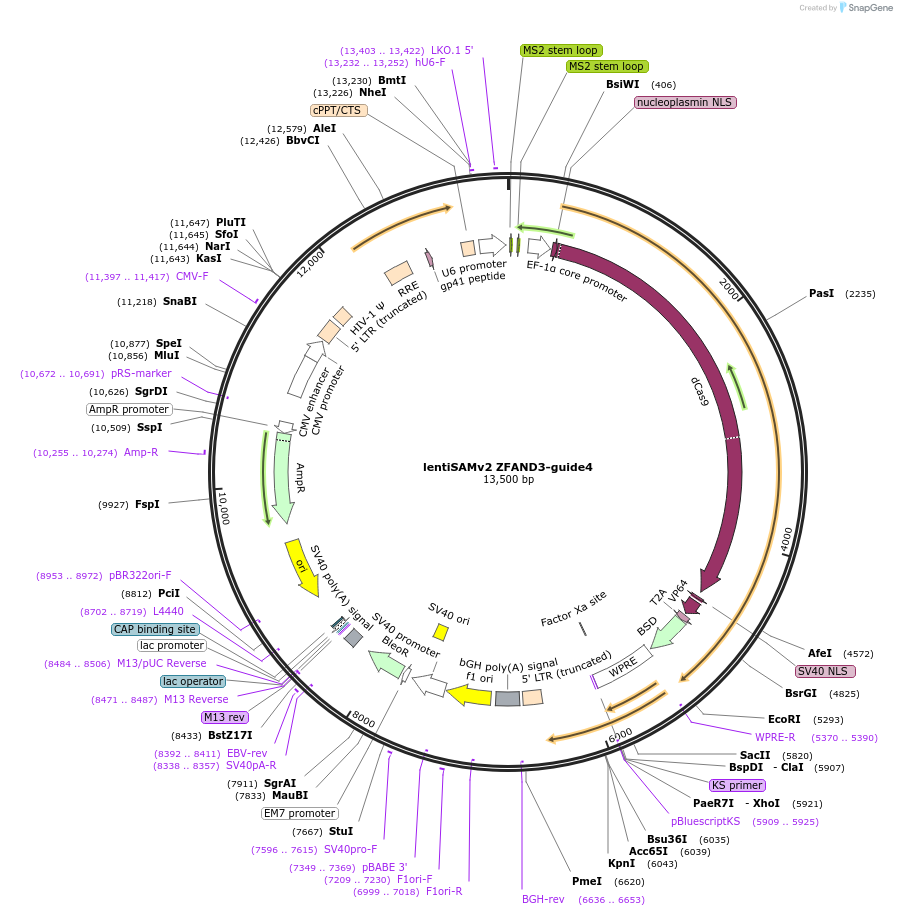
lentiSAMv2 ZFAND3-guide4
Plasmid#125516PurposeCRISPR-mediated activation of ZFAND3. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
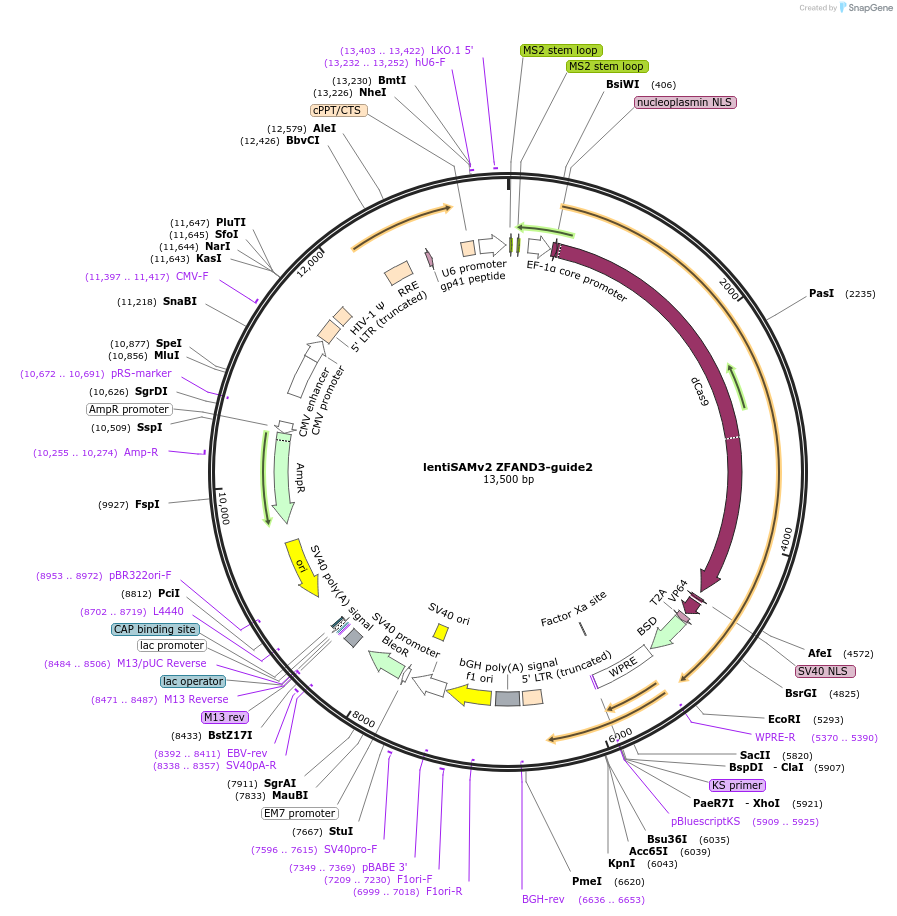
lentiSAMv2 ZFAND3-guide2
Plasmid#125514PurposeCRISPR-mediated activation of ZFAND3. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
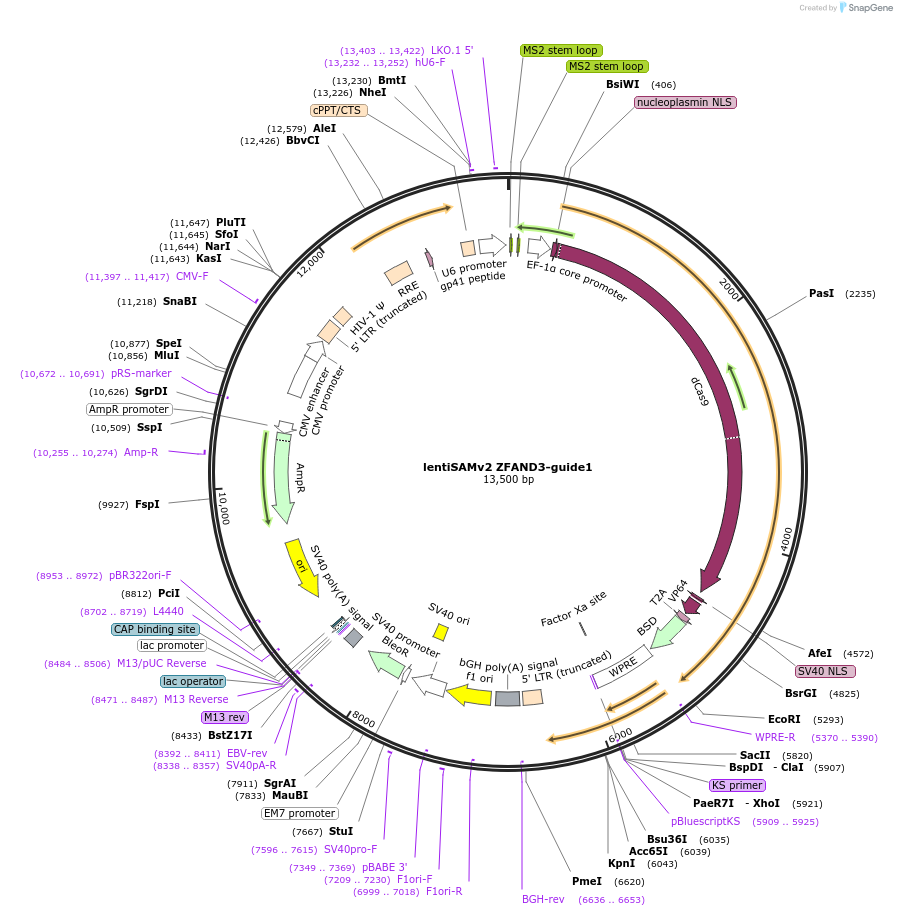
lentiSAMv2 ZFAND3-guide1
Plasmid#125513PurposeCRISPR-mediated activation of ZFAND3. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
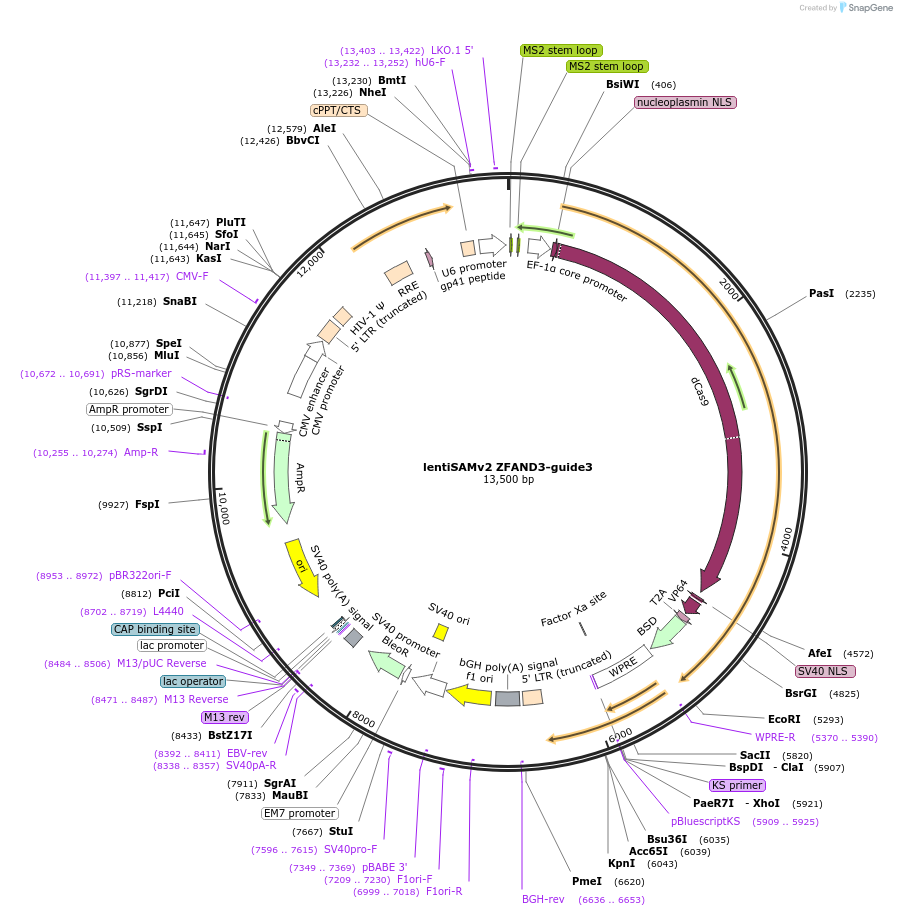
lentiSAMv2 ZFAND3-guide3
Plasmid#125515PurposeCRISPR-mediated activation of ZFAND3. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
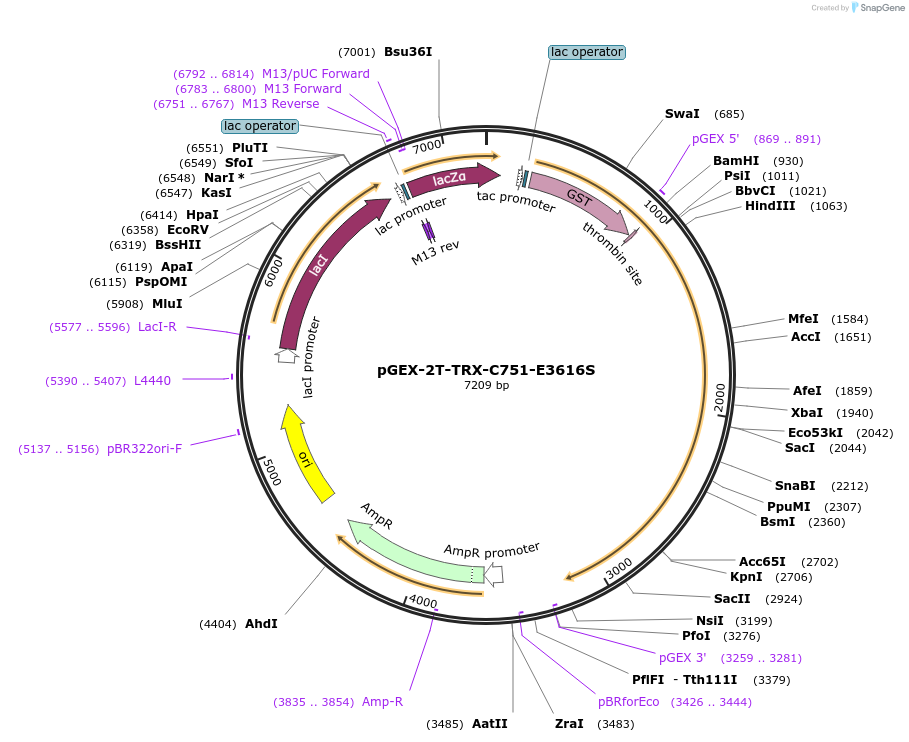
pGEX-2T-TRX-C751-E3616S
Plasmid#182844Purposemutation E3616S (GAA/TCA), PCR product coding C-terminal residues 2976-3726 of TRX was inserted into modified pGEX-2T by Nde I-Nsi I sites. Expression in E. coli. by IPTG inductionDepositorAvailable SinceApril 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
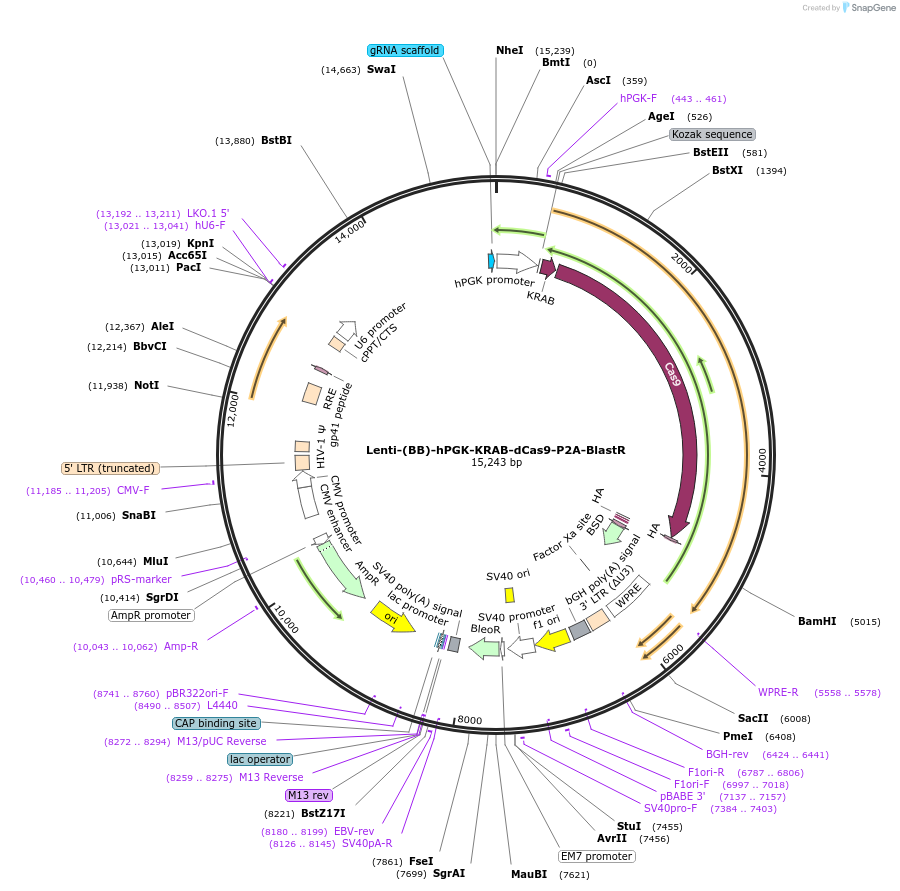
Lenti-(BB)-hPGK-KRAB-dCas9-P2A-BlastR
Plasmid#118155PurposeCatalytically inactive Cas9 from S. pyogenes with P2A-BlastR under the hPGK promoter, and cloning backbone for sgRNA. Contains BsmBI sites for insertion of spacer sequences.DepositorInsertKRAB-dCas9-P2A-BlastR
UseCRISPR and LentiviralTagsHAExpressionMammalianPromoterhPGK and U6Available SinceMay 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
lentiSAMv2 OPTN TSS-guide4
Plasmid#118199PurposeCRISPR-mediated activation of OPTN. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceApril 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
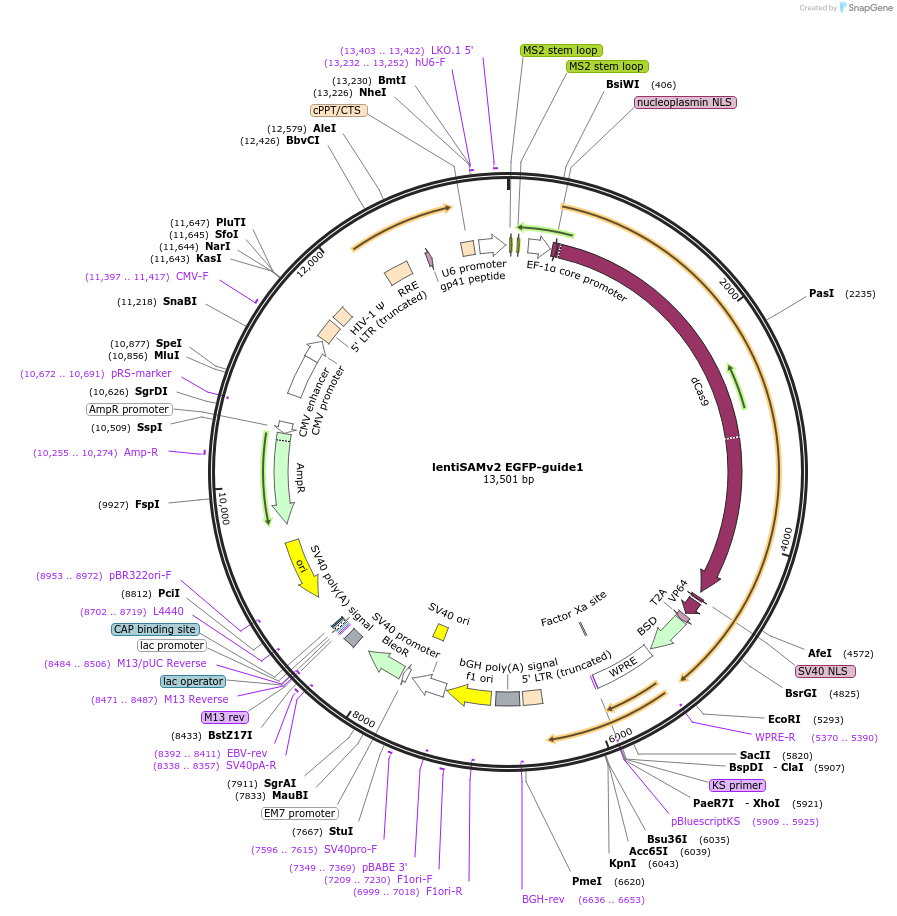
lentiSAMv2 EGFP-guide1
Plasmid#118166PurposeCRISPRa negative control. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR, and sgRNA1 against the EGFP CDSDepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceApril 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
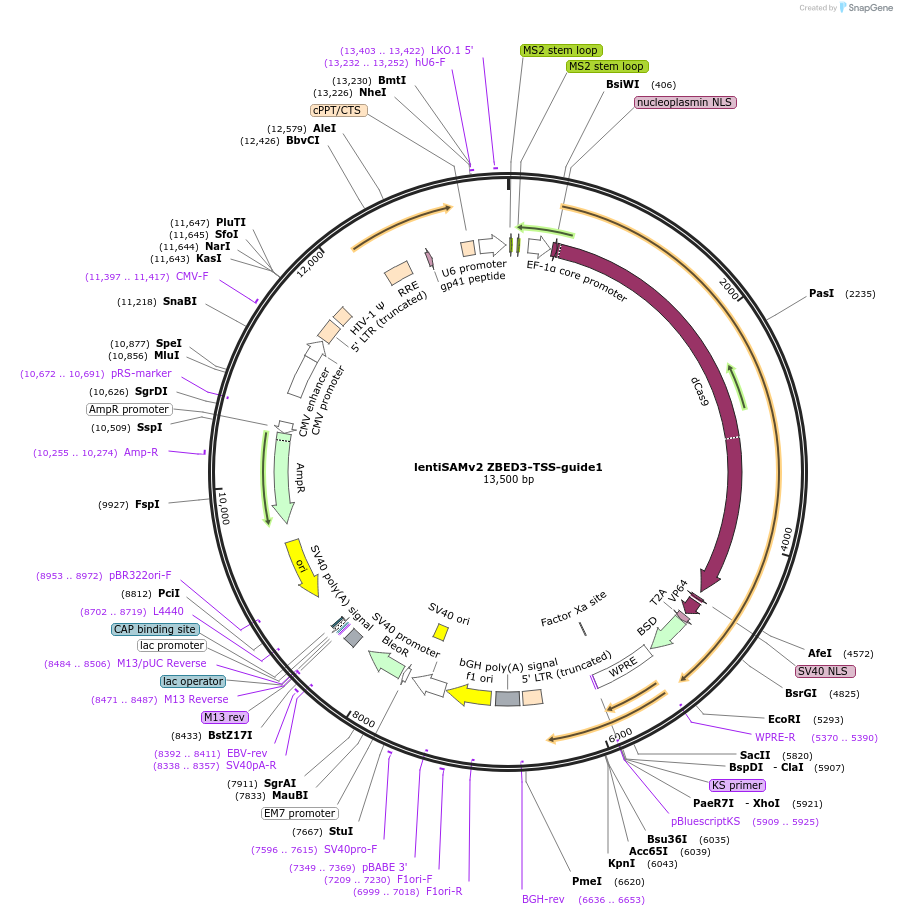
lentiSAMv2 ZBED3-TSS-guide1
Plasmid#125506PurposeCRISPR-mediated activation of ZBED3. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
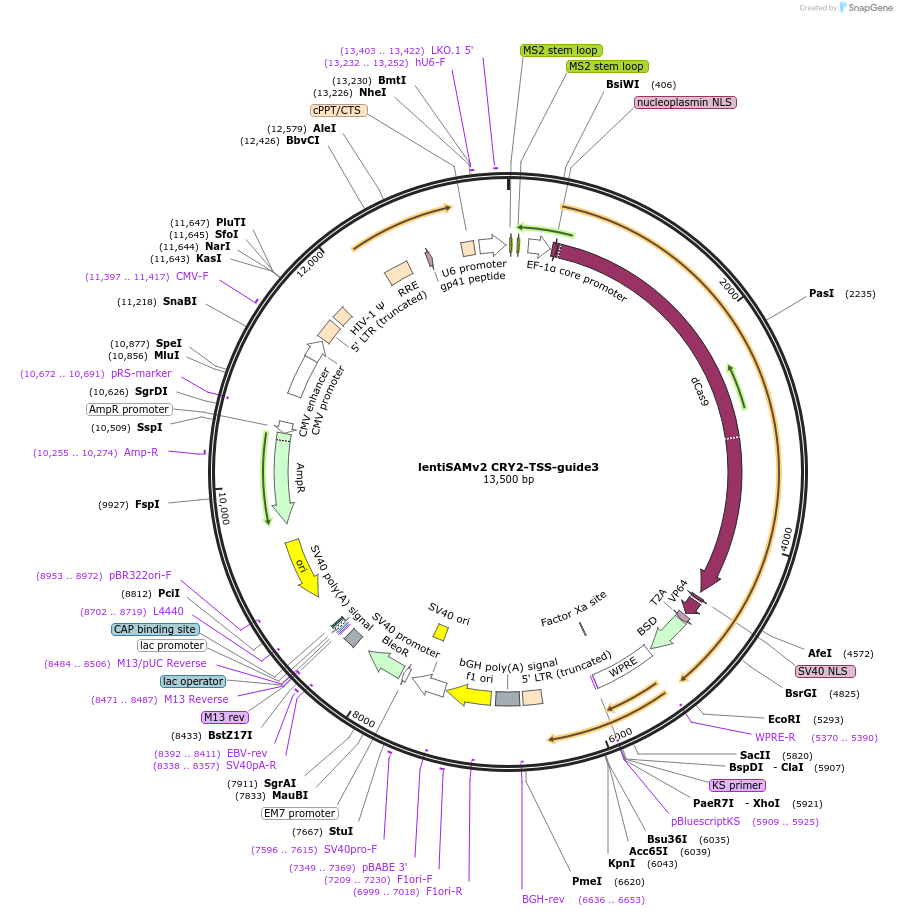
lentiSAMv2 CRY2-TSS-guide3
Plasmid#125491PurposeCRISPR-mediated activation of CRY2. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
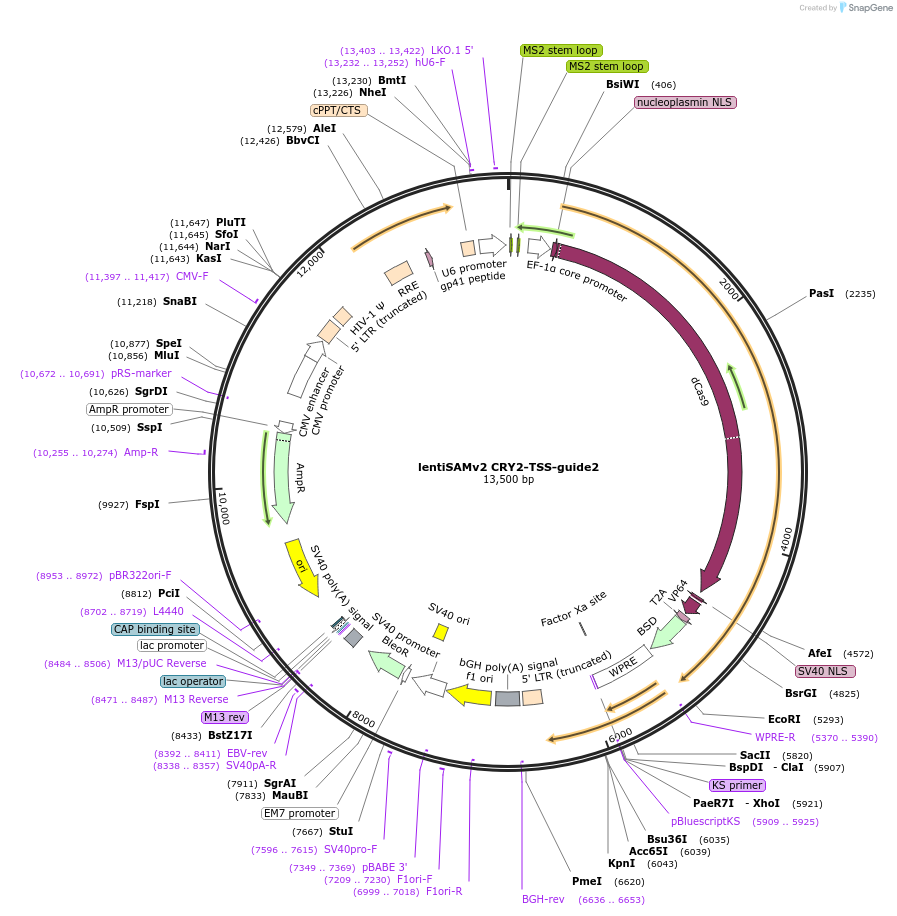
lentiSAMv2 CRY2-TSS-guide2
Plasmid#125490PurposeCRISPR-mediated activation of CRY2. Catalytically inactive Cas9 from S. pyogenes and VP64 with 2A-BlastR.DepositorInsertdCas9-VP64-2A-BlastR
UseCRISPR and LentiviralExpressionMammalianPromoterEF1a core and U6Available SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only