We narrowed to 22,191 results for: his
-
Plasmid#190964PurposeTest the U-to-C editing activity of KP4 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP4
UseTagsHisExpressionBacterialMutationPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-PLS-KP6-E67A_rpoA-C
Plasmid#190972PurposeTest the C-to-U editing activity of KP6:E67A on AtrpoA editing site, in E. coliDepositorInsertPLS-KP6
UseTagsHisExpressionBacterialMutationDYW-KP:E67APromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
PM18033-PLS-KP3-E67A_rpoA-U
Plasmid#190983PurposeTest the U-to-C editing activity of KP3:E67A on AtrpoA editing site, in human cellsDepositorInsertPLS-KP3
UseTagsHisExpressionMammalianMutationDYW-KP:E67APromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
PM18033-PLS-KP4_rpoA-U
Plasmid#190984PurposeTest the U-to-C editing activity of KP4 on AtrpoA editing site, in human cellsDepositorInsertPLS-KP4
UseTagsHisExpressionMammalianMutationPromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-PLS-KP5_rpoA-U
Plasmid#190965PurposeTest the U-to-C editing activity of KP5 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP5
UseTagsHisExpressionBacterialMutationPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
PM18033-PLS-KP3-A66S_rpoA-U
Plasmid#190981PurposeTest the U-to-C editing activity of KP3:A66S on AtrpoA editing site, in human cellsDepositorInsertPLS-KP3
UseTagsHisExpressionMammalianMutationDYW-KP:A66SPromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
PM18033-PLS-KP6-E67A_rpoA-U
Plasmid#190993PurposeTest the U-to-C editing activity of KP6:E67A on AtrpoA editing site, in human cellsDepositorInsertPLS-KP6
UseTagsHisExpressionMammalianMutationDYW-KP:E67APromoterCMVAvailable SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETM20-9S-S2 [rpoA]
Plasmid#176217PurposeExpression of TRX-9S-S2 [rpoA]DepositorInsertTRX-9S-S2 [rpoA]
UseSynthetic BiologyTagsHIS and TRXExpressionMutationPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETM20-9P-S2 [rpoA]
Plasmid#176218PurposeExpression of TRX-9P-S2 [rpoA]DepositorInsertTRX-9P-S2 [rpoA]
UseSynthetic BiologyTagsTRX-HisExpressionMutationPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETM20-9S-DYW [rpoA]
Plasmid#176220PurposeExpression of TRX-9S-DYW [rpoA]DepositorInsertTRX-9S-DYW [rpoA]
UseSynthetic BiologyTagsTRX-HisExpressionMutationPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pETM20-avi-9S-S2 [rpoA]
Plasmid#176223PurposeExpression of TRX-AviTag-9S-S2 [rpoA]DepositorInsertTRX-Avi-9S-S2 [rpoA]
UseSynthetic BiologyTagsTRX-His-AviTagExpressionMutationPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
pBXPHM3-NB-D6
Plasmid#167989Purposeexpression of a nanobody against rat PepT2 as a n terminal MBP fusionDepositorInsertNanobodyD6
UseTagsHis-MBPExpressionBacterialMutationPromoterpBADAvailable SinceOct. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Pythium oligandrum PRL (Δprenylation motif)
Plasmid#166423PurposeExpress pET-15b(+) Pythium oligandrum PRL (Δprenylation motif)DepositorInsertPythium oligandrum PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
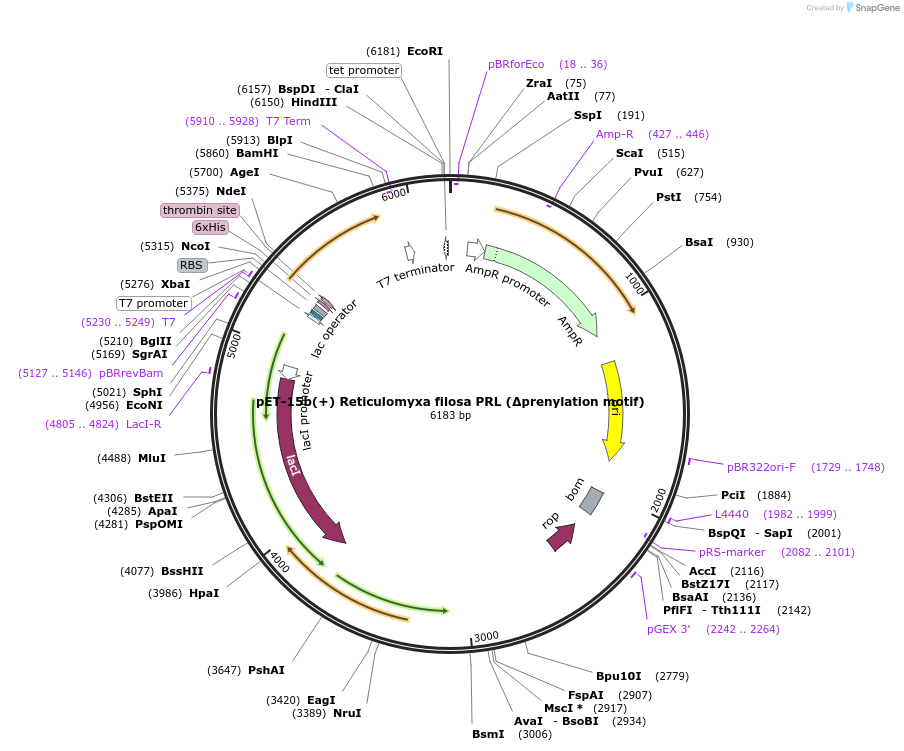
pET-15b(+) Reticulomyxa filosa PRL (Δprenylation motif)
Plasmid#166424PurposeExpress pET-15b(+) Reticulomyxa filosa PRL (Δprenylation motif)DepositorInsertReticulomyxa filosa PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Sphaeforma arctica PRL (Δprenylation motif)
Plasmid#166425PurposeExpress pET-15b(+) Sphaeforma arctica PRL (Δprenylation motif)DepositorInsertSphaeforma arctica PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Acytostelium subglobosum PRL (Δprenylation motif)
Plasmid#166415PurposeExpress pET-15b(+) Acytostelium subglobosum PRL (Δprenylation motif)DepositorInsertAcytostelium subglobosum PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Caenorhabditis elegans PRL (Δprenylation motif)
Plasmid#166416PurposeExpress pET-15b(+) Caenorhabditis elegans PRL (Δprenylation motif)DepositorInsertcaenorhabditis elegans PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Galdieria sulphuraria PRL (Δprenylation motif)
Plasmid#166419PurposeExpress pET-15b(+) Galdieria sulphuraria PRL (Δprenylation motif)DepositorInsertGaldieria sulphuraria PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET-15b(+) Monosiga brevicollis PRL (Δprenylation motif)
Plasmid#166420PurposeExpress pET-15b(+) Monosiga brevicollis PRL (Δprenylation motif)DepositorInsertMonosiga brevicollis PRL
UseTagsN-terminal His-tagExpressionBacterialMutationDeleted nucleotides coding for 4 C-terminal amino…PromoterAmpR, lac1Available SinceSept. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET28a-eGFP-P(11)4
Plasmid#185786PurposeEncodes enhanced green fluorescent protein linked to P(11)4 self-assembling peptide via a GS-linker sequenceDepositorInserteGFP-P(11)4
UseTagsHis-tagExpressionBacterialMutationPromoterT7Available SinceAug. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET28a-ATA-P(11)4
Plasmid#185787PurposeEncodes enzyme ω-aminotransferase linked to P(11)4 self-assembling peptide via a GS-linker sequenceDepositorInsertATA-P(11)4
UseTagsHis-tagExpressionBacterialMutationPromoterT7Available SinceAug. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
Stx3
Plasmid#186685PurposeExpresses Syntaxin 3 in E. ColiDepositorInsertSyntaxin 3
UseTags6 his tagExpressionBacterialMutationPromoterAvailable SinceAug. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
pET28a-GST-P(11)4
Plasmid#185789PurposeEncodes glutathione S-transferase linked to P(11)4 self-assembling peptide via a GS-linker sequenceDepositorInsertGST-P(11)4
UseTagsHis-tagExpressionBacterialMutationPromoterT7Available SinceJuly 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
pEUK035
Plasmid#180827PurposeT7-inducible expression construct to produce SwGdmB (HY78_09235) in E. coliDepositorArticleInsertSwGdmB
UseTags6X HisExpressionBacterialMutationPromoterT7Available SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pEUK036
Plasmid#180828PurposeT7-inducible expression construct to produce CnGdmA (CNE_RS35785) in E. coliDepositorArticleInsertCnGdmA
UseTags6X HisExpressionBacterialMutationPromoterT7Available SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
pEUK066
Plasmid#180830PurposeT7-inducible expression construct to produce NaGdmA (SARO_RS07455) in E. coliDepositorArticleInsertNaGdmA
UseTags6X HisExpressionBacterialMutationPromoterT7Available SinceJune 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
IBMc397
Plasmid#161955PurposeDemonstrates chemically induced dimmerization of acVHH. Expression plasmid for split, evolved T7 RNA polymerase C-lobe fused to acVHHDepositorInsertpSB3K3-TT-PBAD(SapI-)-B30-His-acVHH-15aa-T7RNAP(181+)-T
UseTagsExpressionBacterialMutationPromoterPBADAvailable SinceOct. 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
bdSUMO-5.3-GFP-HiBit
Plasmid#171014PurposeFor bacterial expression of bdSUMO tagged muGFP with 5.3 peptide and NanoLuc complementation peptide variant #86DepositorInsert14x-His-bdSUMO-5.3-muGFP-HiBiT
UseLuciferaseTagsExpressionBacterialMutationPromoterT7Available SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
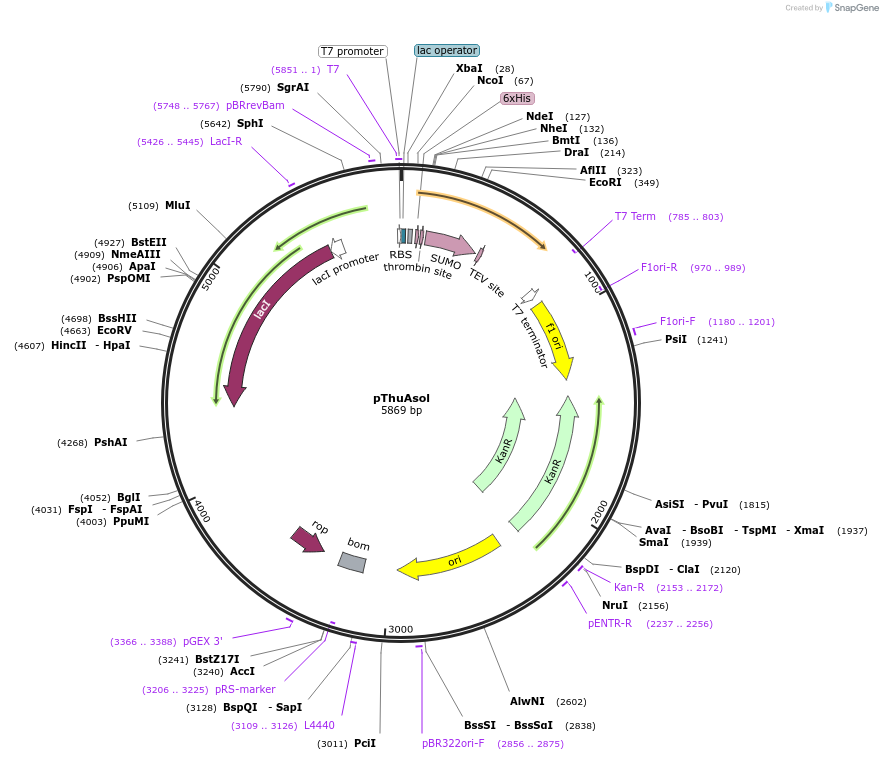
pThuAsol
Plasmid#171675PurposepThuAsol expression in E. coliDepositorInsertSUMO-TEV-ThuAsol
UseTagsHis-tag and SUMO-tagExpressionBacterialMutationPromoterT7Available SinceAug. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
OfCasp3a
Plasmid#166053Purposea caspase of Orbicella faveolata, which has higher sequence similarity with human caspase-3 but has CARD domain and function as an initiator caspaseDepositorInsertOrbicella faveolata caspase-3a
UseTagsHis tagExpressionBacterialMutationPromoterT7 lac promoterAvailable SinceMarch 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
PaCasp7a
Plasmid#166051Purposea caspase of Porites astreoides, which has higher sequence similarity with human caspase-7 but has a CARD domain and function as an initiator caspaseDepositorInsertPorites astreiodes caspase-7a
UseTagsHis tagExpressionBacterialMutationPromoterT7 promoterAvailable SinceMarch 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pFE-sfGFP
Plasmid#162688PurposepFE vector control expressing sfGFPDepositorInsertsfGFP
UseTagsC terminal 6x His tagExpressionBacterialMutationPromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
pDUAL_2_lacZ
Plasmid#155173Purposesee pDUAL_lacZ: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceOct. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pDUAL_araBAD
Plasmid#155172PurposeBaSP and CuCBP co-expression: araBAD promoter controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
pDUAL_2_tacI
Plasmid#155174Purposesee pDUAL_tacI: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pDUAL_lacZ
Plasmid#155169PurposeBaSP and CuCBP co-expression: lacZ promoter controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pBICI_2_medium
Plasmid#155167Purposesee pBICI_medium: exchanged origin of replication (pBR322) and introduced bom siteDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pBICI_strong
Plasmid#155165PurposeBaSP and CuCBP co-expression: strong RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pBICI_medium
Plasmid#155164PurposeBaSP and CuCBP co-expression: medium RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pBICI_weak
Plasmid#155163PurposeBaSP and CuCBP co-expression: weak RBS controls BaSP expressionDepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterAvailable SinceSept. 8, 2020AvailabilityAcademic Institutions and Nonprofits only