We narrowed to 9,326 results for: sacs
-
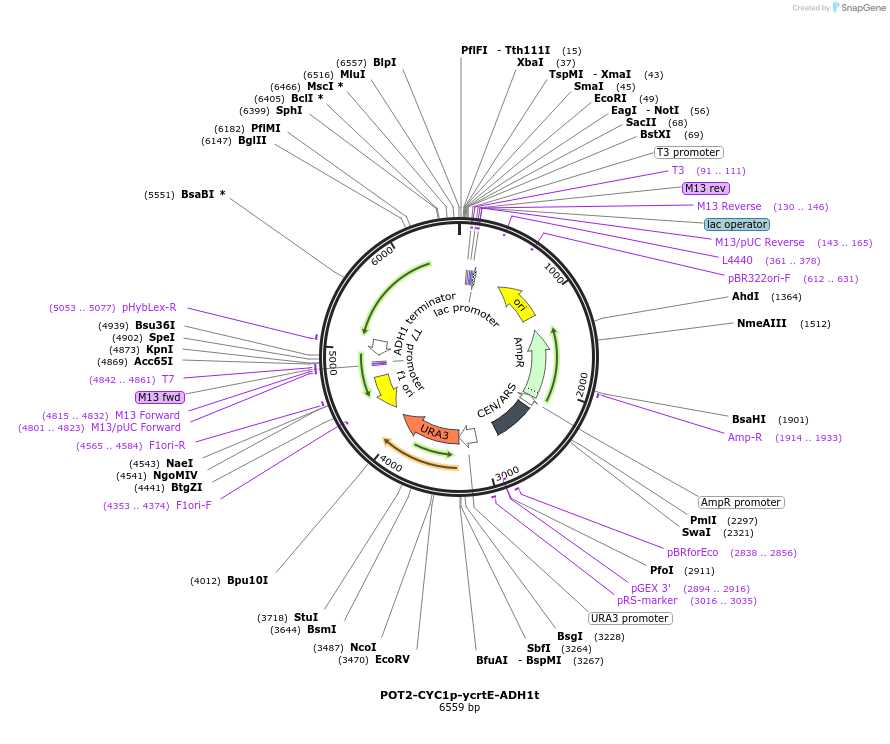
Plasmid#65354PurposeTU for combinatorial construction of ?-carotene pathwayDepositorInsertCYC1p-ycrtE-ADH1t
UseUnspecifiedAvailable SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
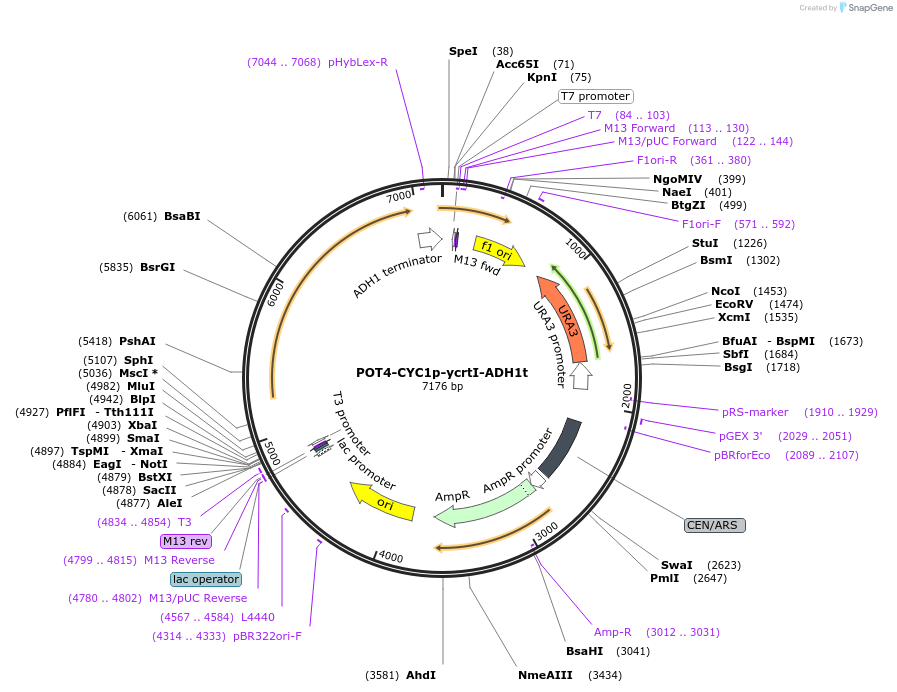
POT4-CYC1p-ycrtI-ADH1t
Plasmid#65357PurposeTU for combinatorial construction of ?-carotene pathwayDepositorInsertCYC1p-ycrtI-ADH1t
UseUnspecifiedAvailable SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
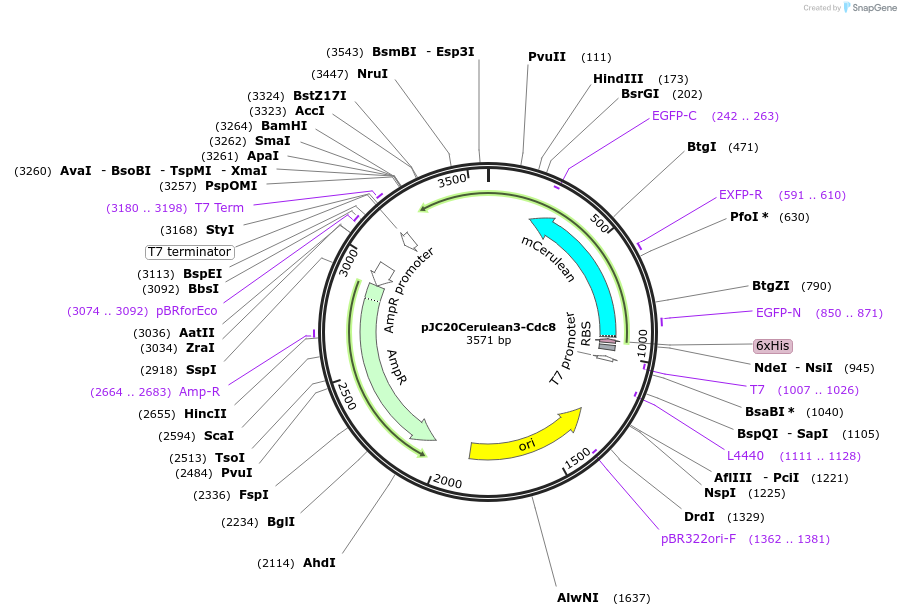
pJC20Cerulean3-Cdc8
Plasmid#99357PurposeBacterial expression of Cerulean3 fused to the 5' of cDNA of the fission yeast tropomyosin, cdc8.DepositorAvailable SinceAug. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
pJRL2 PHO5(TATA-A1T6)pr ECFP (EB#1560)
Plasmid#14846DepositorInsertPHO5 promoter TATA-A1T6 (PHO5 Budding Yeast)
TagsECFPExpressionYeastMutationTATA-A1T6. TATA box mutated from TATATAAG to AATA…Available SinceMay 25, 2007AvailabilityAcademic Institutions and Nonprofits only -
pGST1-yGGA1-VHS
Plasmid#44421DepositorAvailable SinceJuly 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
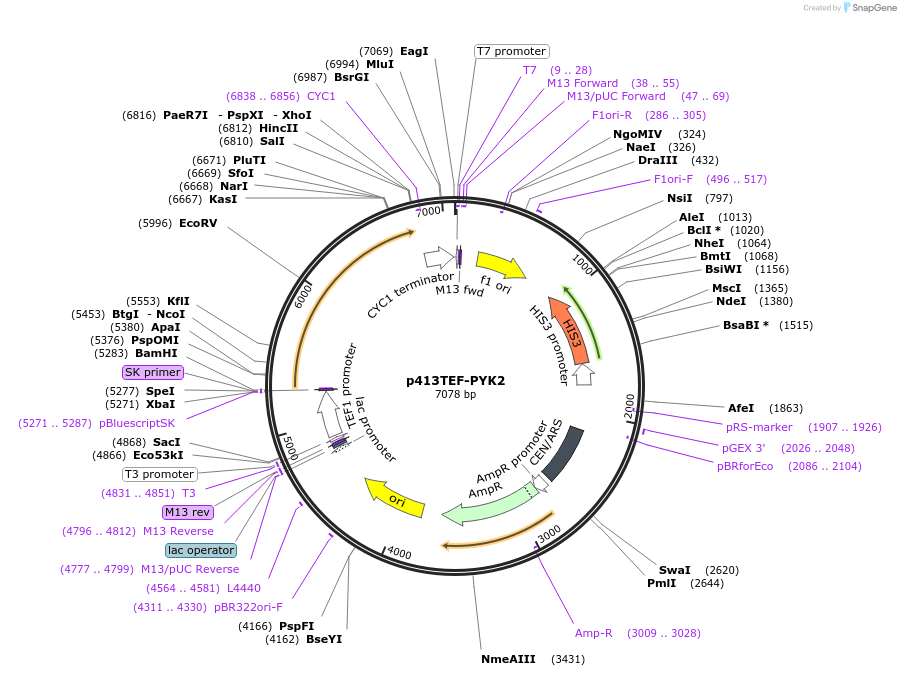
p413TEF-PYK2
Plasmid#34606DepositorAvailable SinceFeb. 16, 2012AvailabilityAcademic Institutions and Nonprofits only -
pAG416GPD-Cerulean-YPT6
Plasmid#18845DepositorAvailable SinceNov. 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
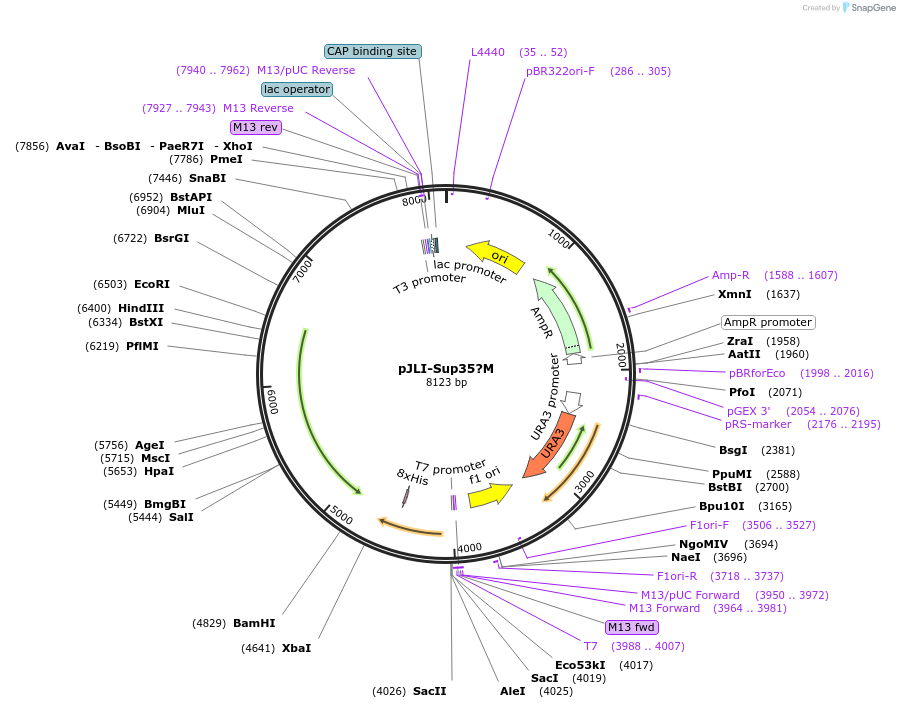
pJLI-Sup35?M
Plasmid#1225DepositorInsertSUP35 (SUP35 Budding Yeast)
UseYeast integrative plasmidMutationdeletion of aa 124-253 of SUP35Available SinceJan. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
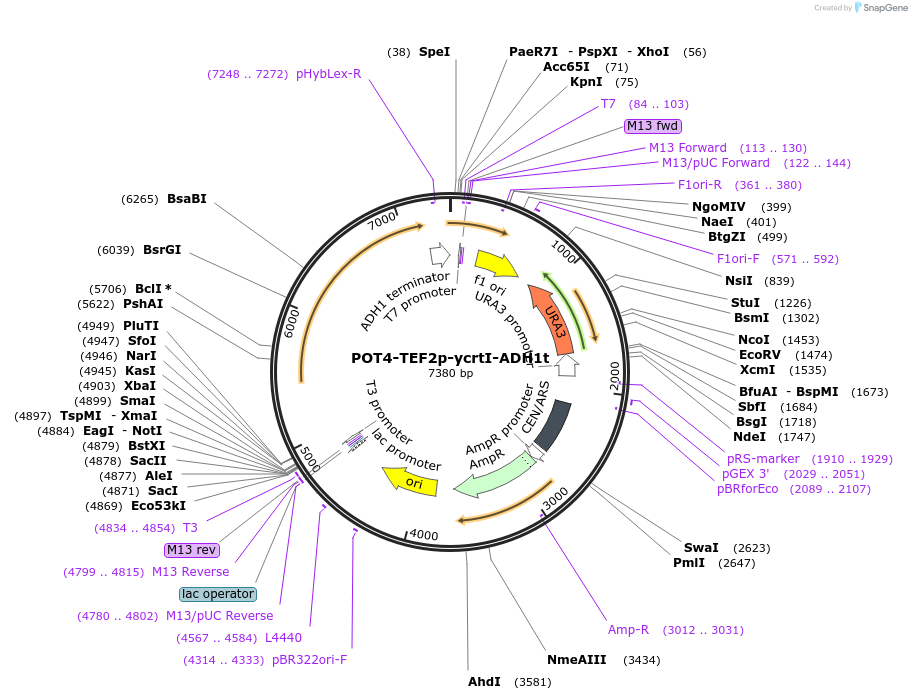
POT4-TEF2p-ycrtI-ADH1t
Plasmid#65356PurposeTU for combinatorial construction of ?-carotene pathwayDepositorInsertTEF2p-ycrtI-ADH1t
UseUnspecifiedAvailable SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
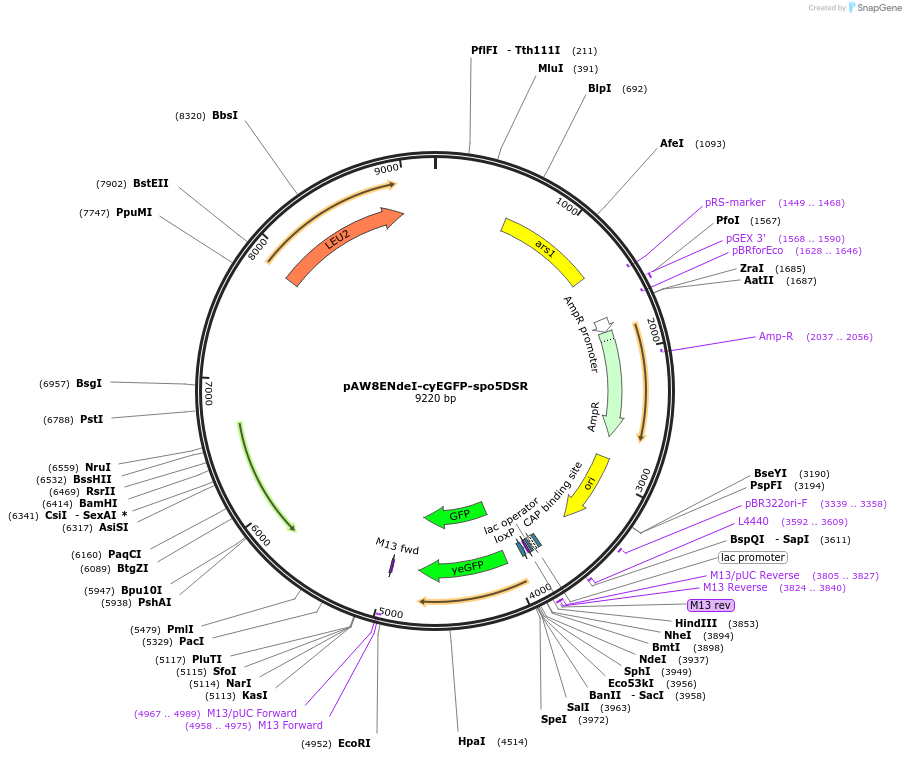
pAW8ENdeI-cyEGFP-spo5DSR
Plasmid#49867Purposefor Cre-lox cassette exchange of C-terminal yEGFP tagged gene sequences together with the S. pombe spo5 gene DSR element into the S. pombe urg1 locusDepositorTypeEmpty backboneUseCre/LoxTagsyEGFPAvailable SinceDec. 20, 2013AvailabilityAcademic Institutions and Nonprofits only -
sup35-R320I
Plasmid#1199DepositorAvailable SinceMay 25, 2005AvailabilityAcademic Institutions and Nonprofits only -
pME-basGal80
Plasmid#51621Purposevertebrate expression of Gal80 preceded by a basal promoterDepositorAvailable SinceMarch 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
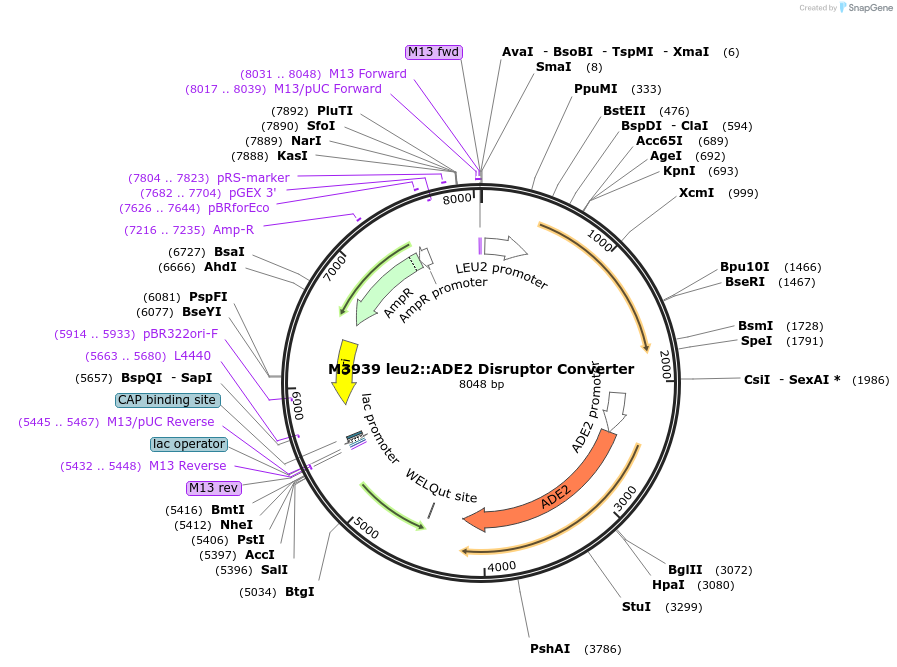
M3939 leu2::ADE2 Disruptor Converter
Plasmid#51672PurposeYeast marker swap plasmid for converting LEU2 to ADE2DepositorAvailable SinceJune 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
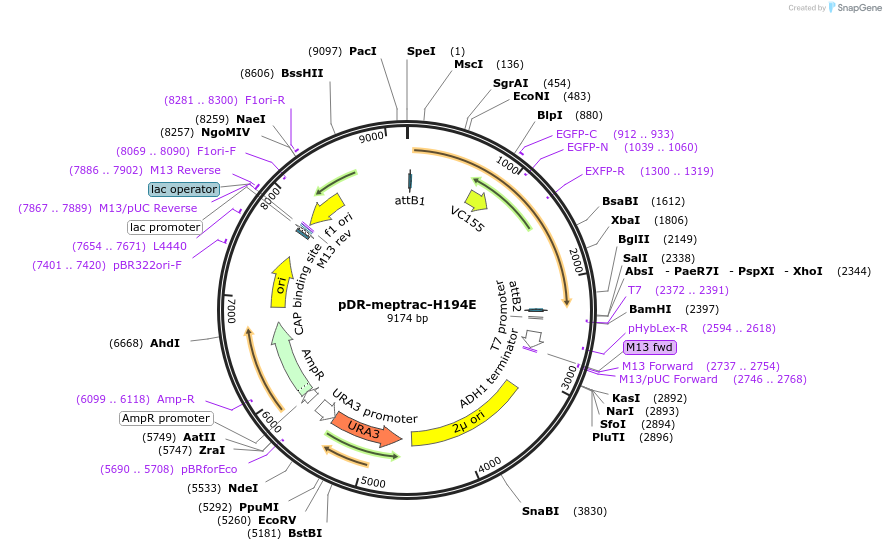
pDR-meptrac-H194E
Plasmid#47764Purposeexpresses MepTrac-H194E in yeastDepositorInsertMepTrac-H194E
TagsmcpGFPExpressionYeastMutationH194EPromoterPMAAvailable SinceSept. 16, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
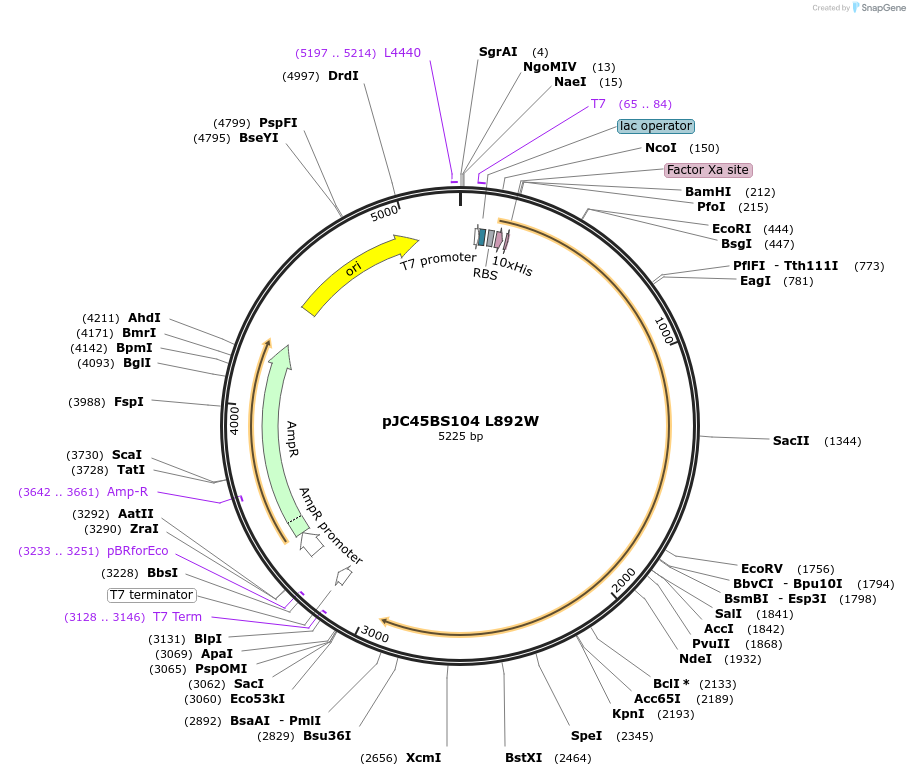
pJC45BS104 L892W
Plasmid#1308DepositorInsertHsp104 L892W (HSP104 Budding Yeast)
Tags10X His tagExpressionBacterialMutationChanged Leu 892 to TrpAvailable SinceJan. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
pPS1597
Plasmid#8920DepositorAvailable SinceAug. 19, 2005AvailabilityAcademic Institutions and Nonprofits only -
pRS414-54
Plasmid#23084DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysines 4 and 9 to Glutamines,…Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
pJD100
Plasmid#23085DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysine 4 to Glutamine, H3 K4Q.Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
pJD101
Plasmid#23086DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysine 4 to Arginine, H3 K4R.Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only