We narrowed to 11,014 results for: phen
-
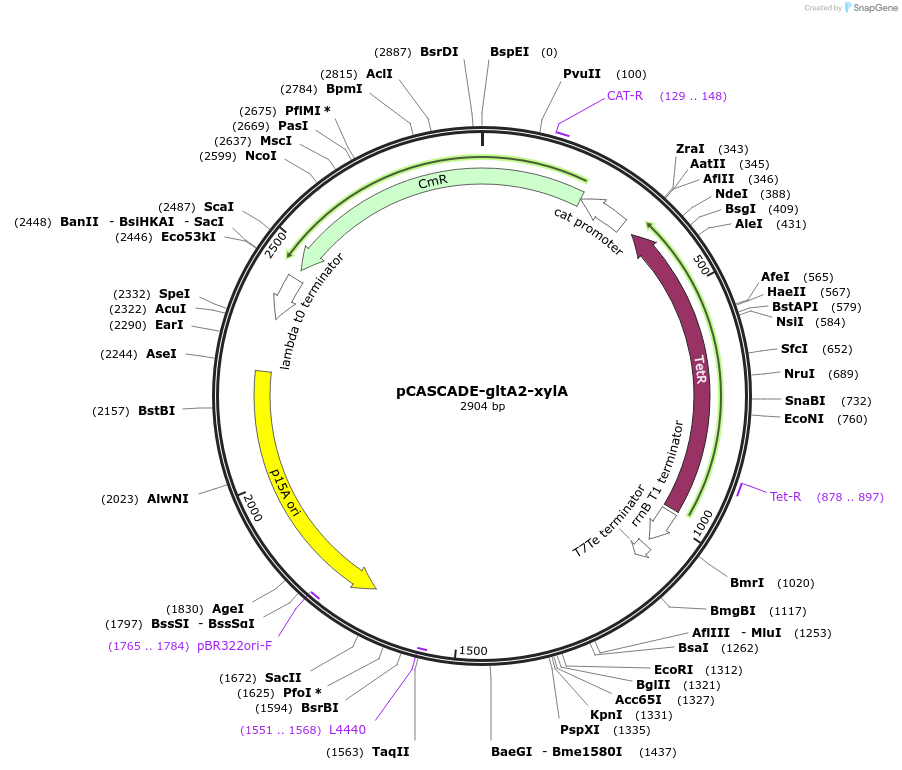
Plasmid#158613PurposeLow phosphate inducible gRNA array to silence the xylA and gltA2 promotersDepositorInsertgltA2-xylA guide array
ExpressionBacterialPromoterE . coli ugpB gene promoter, low phosphate induct…Available SinceJan. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
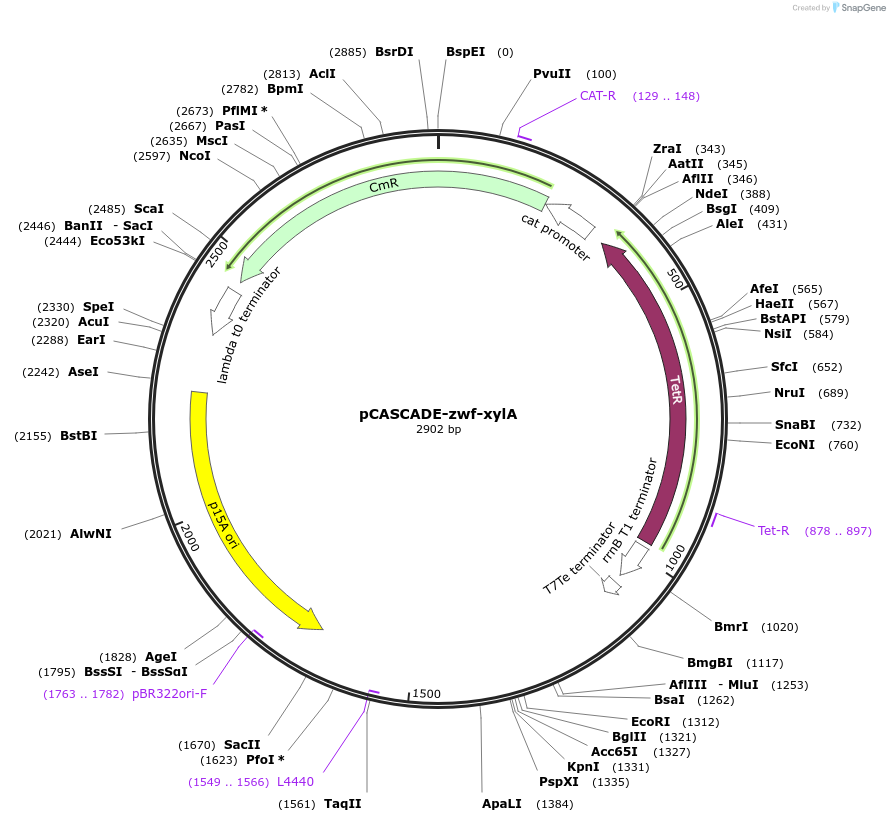
pCASCADE-zwf-xylA
Plasmid#158614PurposeLow phosphate inducible gRNA array to silence the xylA and zwf promotersDepositorInsertzwf-xylA guide array
ExpressionBacterialPromoterE . coli ugpB gene promoter, low phosphate induct…Available SinceJan. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
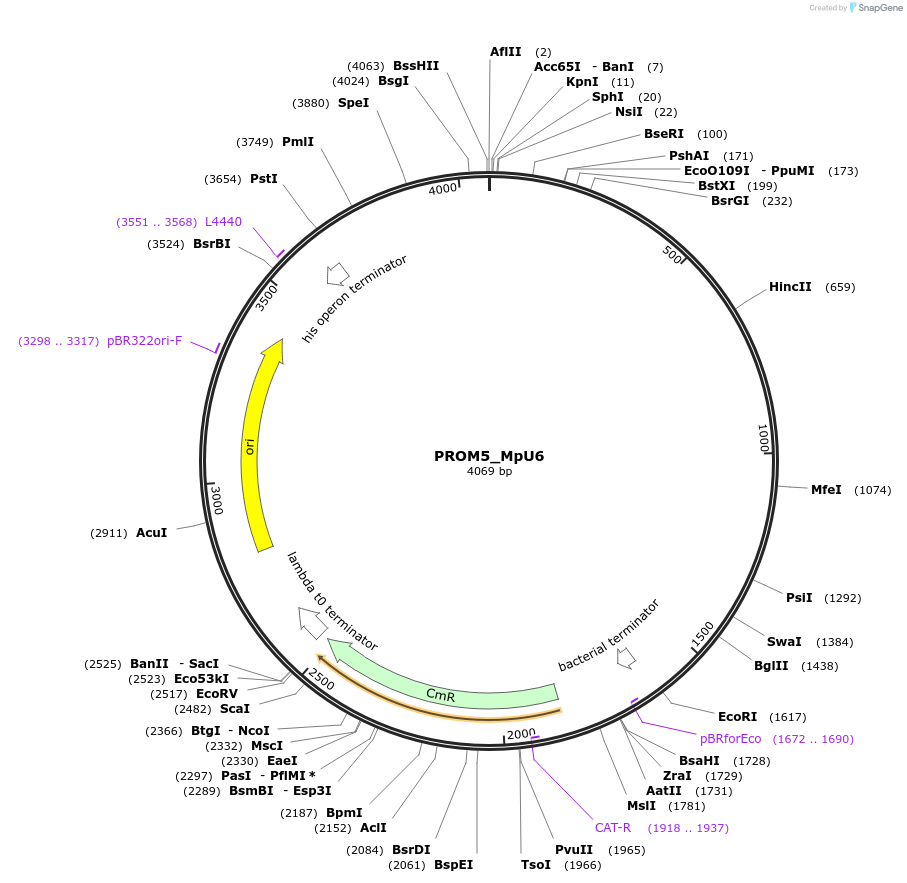
PROM5_MpU6
Plasmid#136120PurposeMp U6 promoter (type III RNA polymerase promoter) to drive expression of gRNA for CRISPR/Cas9DepositorInsertPROM5_MpU6
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
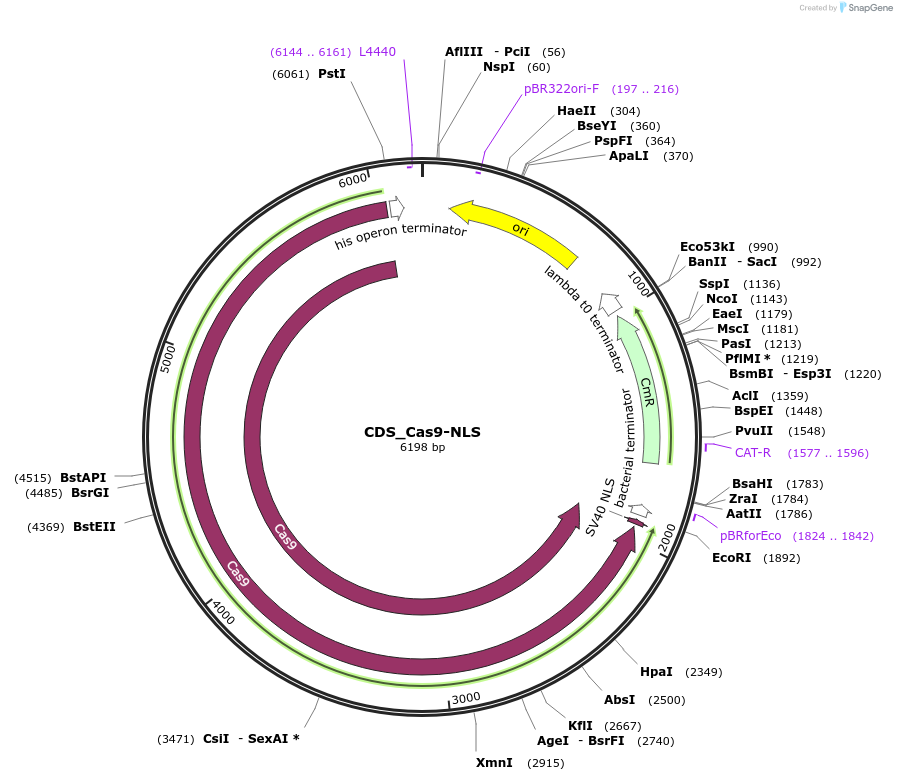
CDS_Cas9-NLS
Plasmid#136121PurposePuchta Arabidopsis codon-optimised Cas9 for CRISPRDepositorInsertCDS_AtcoCas9-NLS
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
CDS12-MpER-Targ
Plasmid#136094PurposeCDS12 with ER targeting peptide to fuse with CTAG containing HDEL ER retention signal at the 3'endDepositorInsertER targeting peptide from predicted chitinase Mapoly0069s0092
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
CTAG_eGFP-GHtag-HDEL
Plasmid#136096PurposeFP with ER retention signal for C-term fusion with CDS12 with ER targeting signal peptide. Contains Gly-His tagDepositorInsertCTAG_eGFP(noATG,noStop)-3xGly5xHys-HDEL
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
pUAP12008
Plasmid#177036PurposeLevel 0 promoter and 5'UTR part for MoClo assembly, GGAG-CCATDepositorInsertCauliflower Mosaic Virus (CaMV) 35S promoter and Tobacco Mosaic Virus (TMV) omega 5' UTR
UseSynthetic BiologyAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
pEPQD0CM0061
Plasmid#177026PurposeLevel 0 CDS (with stop codon) part for MoClo assembly, AATG-GCTTDepositorInsertiridoid oxidase; CYP76A26
UseSynthetic BiologyMutationamplified from cDNA, contains T359S relative to K…Available SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
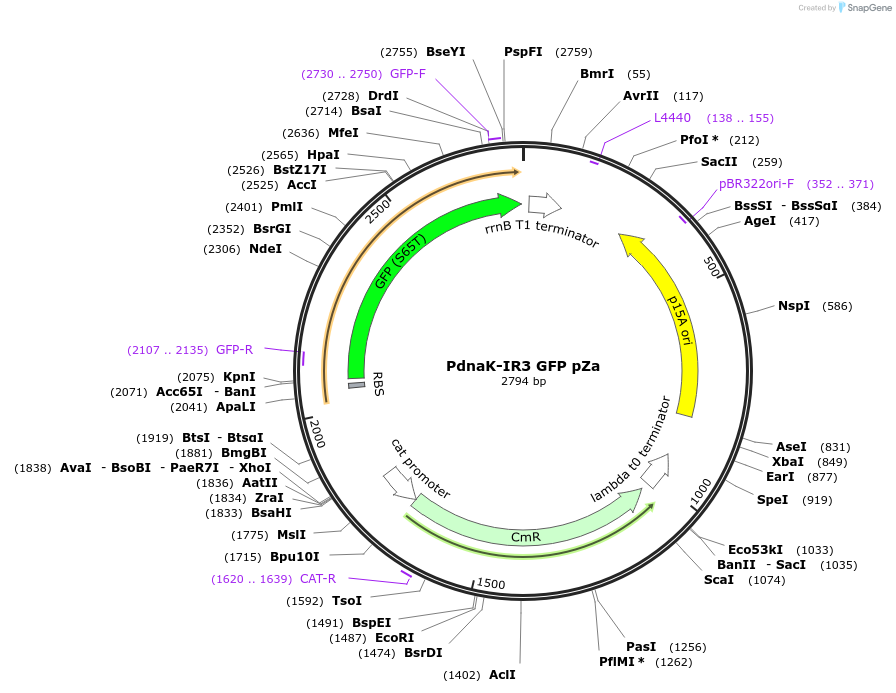
PdnaK-IR3 GFP pZa
Plasmid#170084PurposeGFP expression under the control of E. coli dnaK promoter engineered with IR3 HAIR motif from M. tuberculosisDepositorInsertGreen fluorescent protein
UseSynthetic BiologyExpressionBacterialPromoterPdnaK-IR3Available SinceSept. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
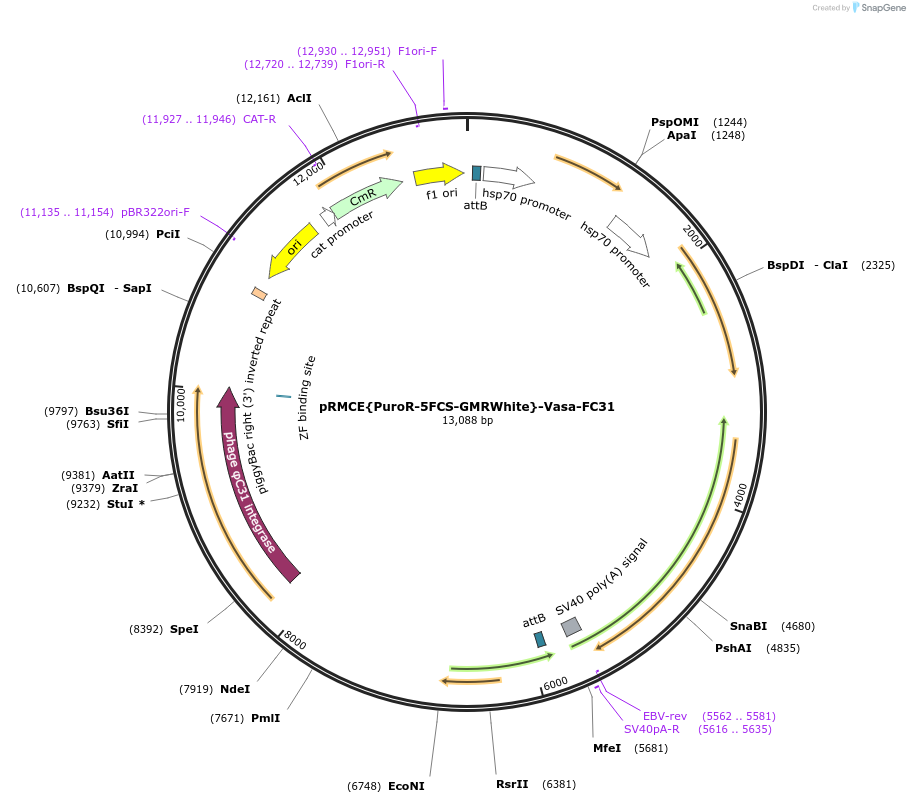
pRMCE{PuroR-5FCS-GMRWhite}-Vasa-FC31
Plasmid#165888PurposePuromycin resistant FC31 integrase mediated RMCE cassette for upgrading MiMic insertionsDepositorInsertAll-in-one FC31 RMCE Cassette
UseSynthetic BiologyAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
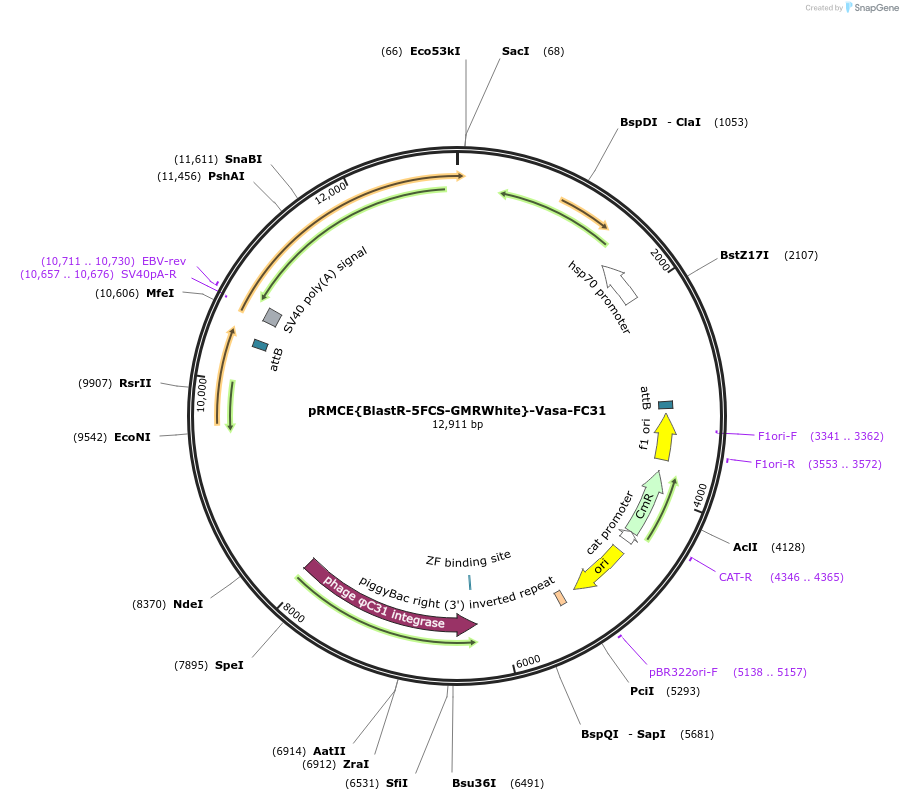
pRMCE{BlastR-5FCS-GMRWhite}-Vasa-FC31
Plasmid#165889PurposeBlasticidin resistant FC31 integrase mediated RMCE cassette for upgrading MiMic insertionsDepositorInsertAll-in-one FC31 RMCE Cassette
UseSynthetic BiologyAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
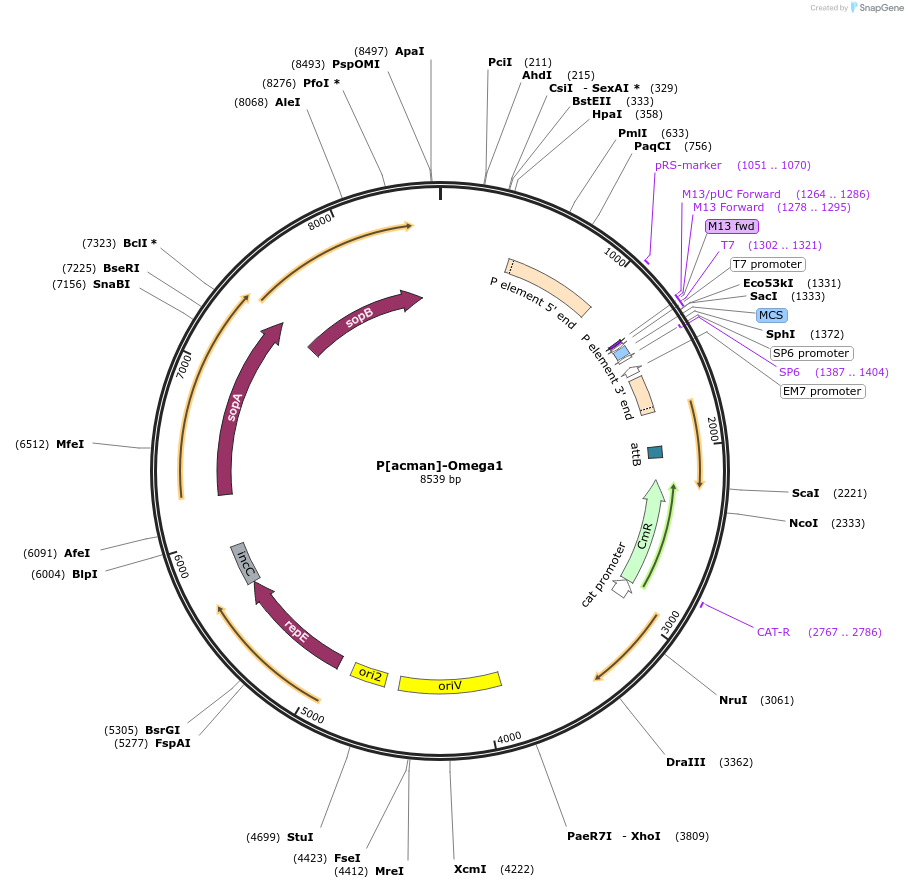
P[acman]-Omega1
Plasmid#165860PurposeOmega1 Basic cloning vector GB2.0 compatible P[acman]-derived backbone. Allows for copy induction from low to high using AutoFOS or similar. Requires EPI300 bacterial strain (DH10B Derived) or similar. uses blue-white screeningDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
PdnaK-IR2 GFP pZa
Plasmid#170081PurposeGFP expression under the control of E. coli dnaK promoter engineered with IR2 HAIR motif from M. tuberculosisDepositorInsertGreen fluorescent protein
UseSynthetic BiologyExpressionBacterialPromoterPdnaK-IR2Available SinceSept. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
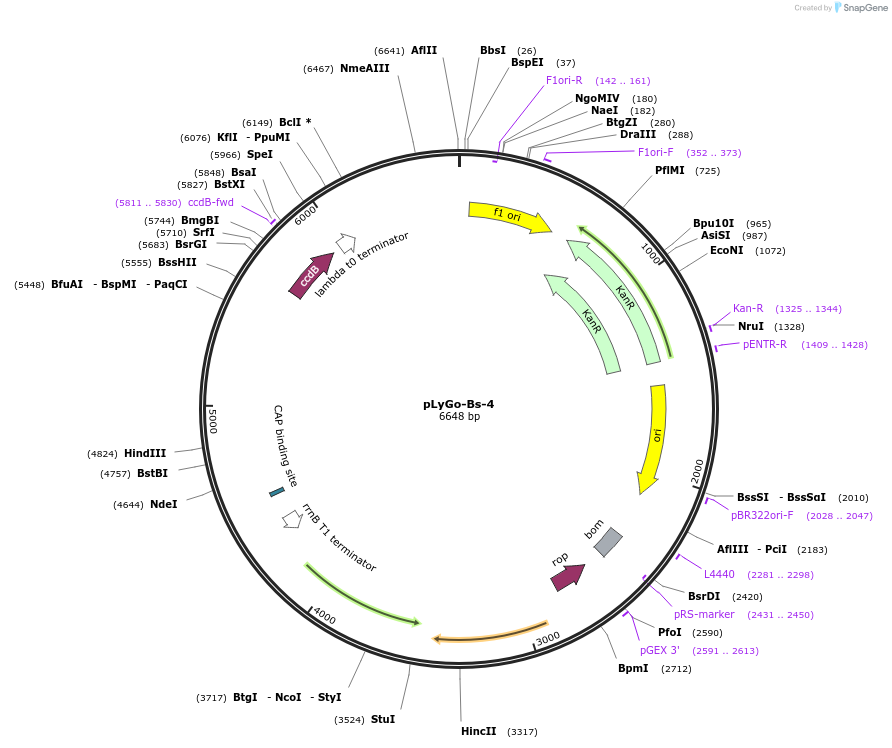
pLyGo-Bs-4
Plasmid#163142PurposepLyGo cloning vector for a sequence of interest (LPMO) in B. subtilis. Vector encoding the BatLPMO10 signal peptide and the LyGo cassette (SapI-ccdB-SapI)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialPromoterPamyL-PamyQ-PcryIIIA-cryIIIAstab49Available SinceJuly 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
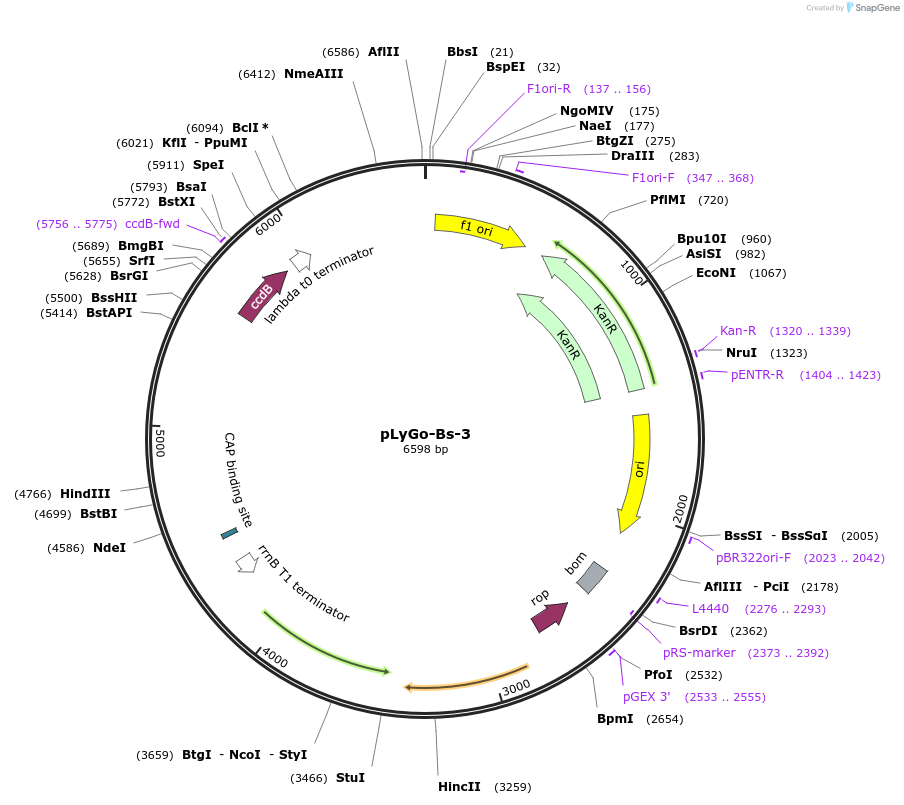
pLyGo-Bs-3
Plasmid#163141PurposepLyGo cloning vector for a sequence of interest (LPMO) in B. subtilis. Vector encoding the AprE signal peptide and the LyGo cassette (SapI-ccdB-SapI)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialPromoterPamyL-PamyQ-PcryIIIA-cryIIIAstab49Available SinceMay 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
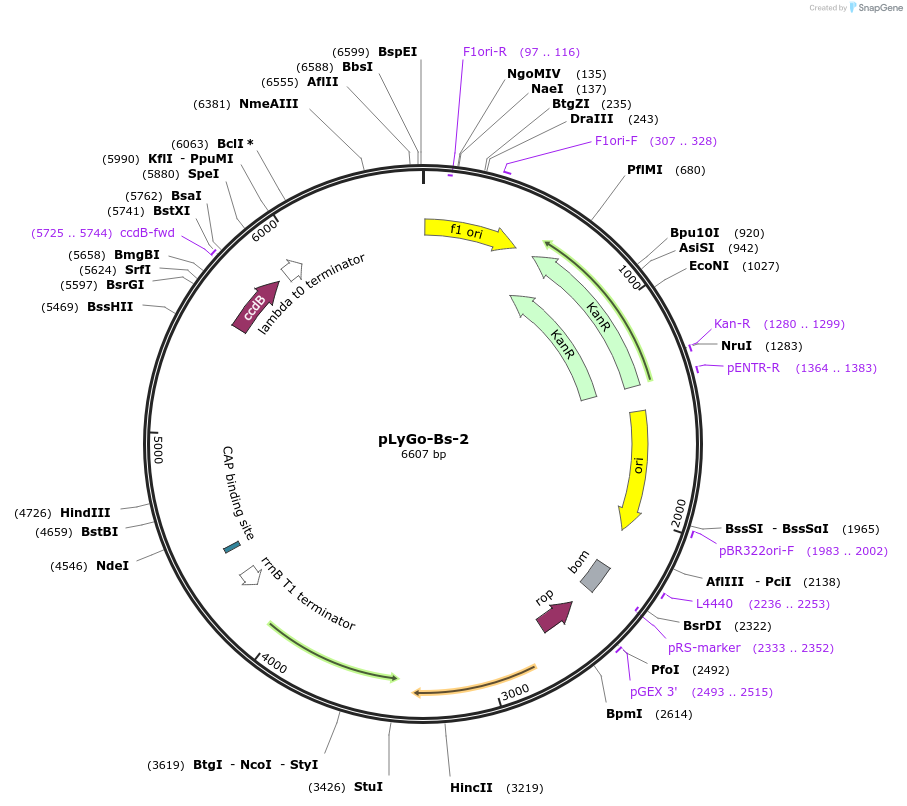
pLyGo-Bs-2
Plasmid#163140PurposepLyGo cloning vector for a sequence of interest (LPMO) in Bacillus subtilis. Vector encoding the AmyQ signal peptide and the LyGo cassette (SapI-ccdB-SapI)DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialPromoterPamyL-PamyQ-PcryIIIA-cryIIIAstab49Available SinceMay 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
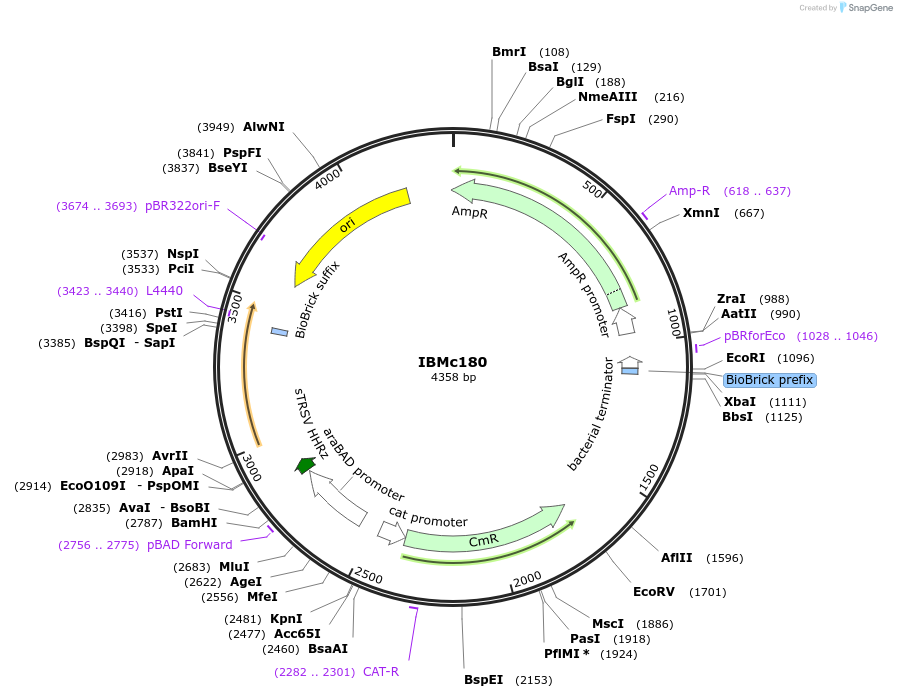
IBMc180
Plasmid#161942PurposeCarries Golden Gate substitution insert for acVHH-assisted bisection mappingDepositorInsertpSB1A3-BbsI-10aa-acVHH-T-cat-PBAD(SapI)-RiboJ-BCD22-acVHH-10aa-SapI
ExpressionBacterialPromoterPBADAvailable SinceMarch 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
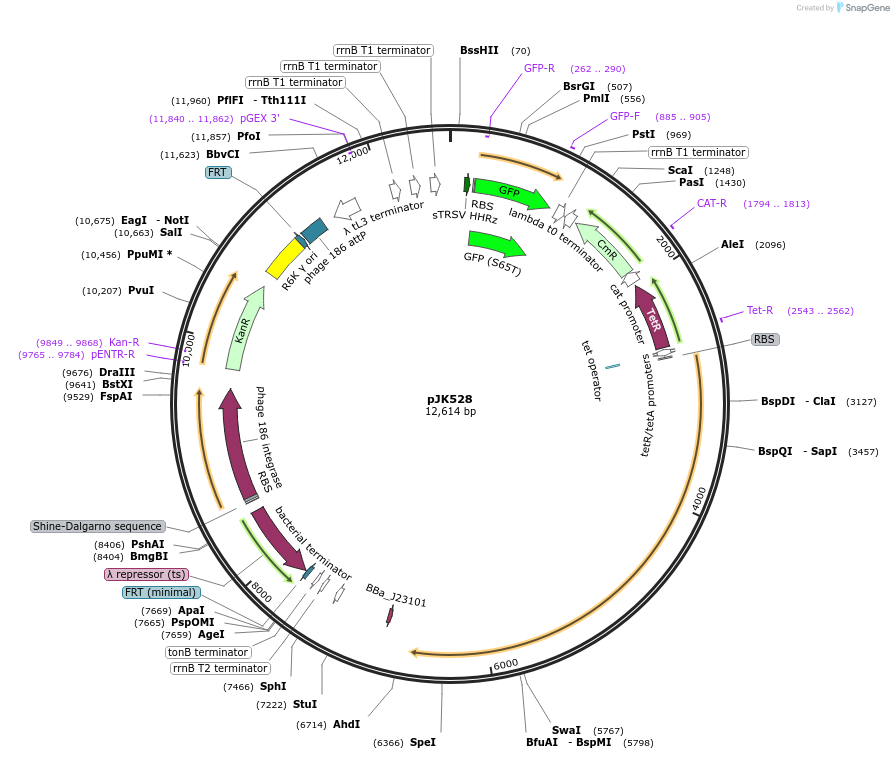
pJK528
Plasmid#155388PurposedCas12a oscillator, reverse crRNAs + PA4-mVenus. (Integration of 511; pOSIP)DepositorInsertsdCas12a (F. novicida)
PA4-mVenus
dCas12a oscillator crRNAs
UseCRISPRExpressionBacterialMutationD917A (nuclease-deactivating)Available SinceFeb. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
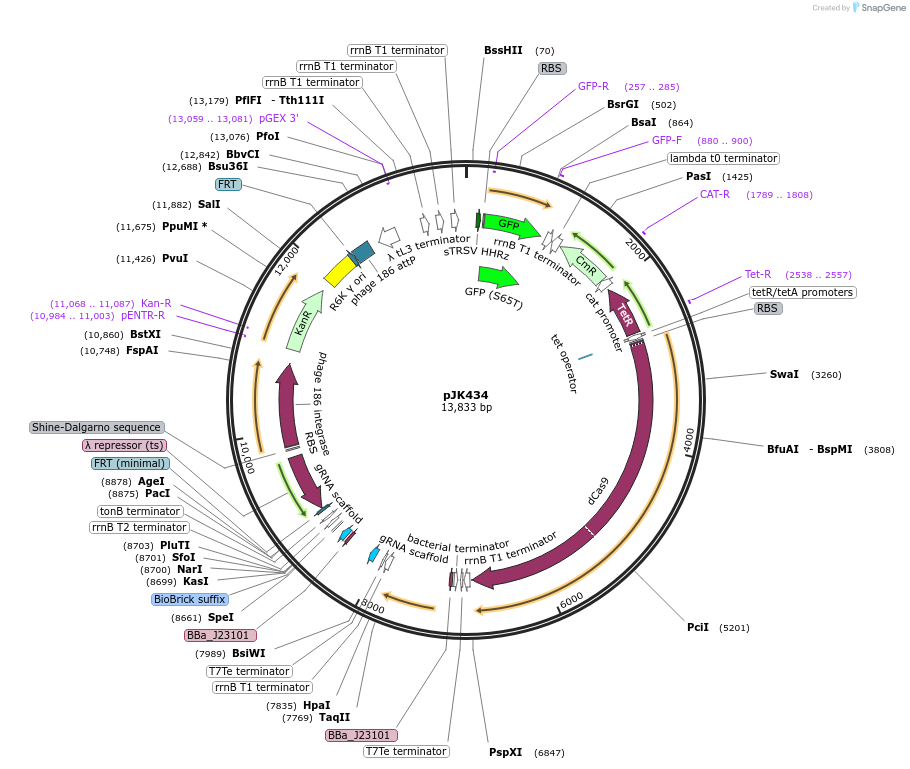
pJK434
Plasmid#155378PurposePphlF_A4NT, 1x cascade + [dCas9 + PA4-mVenus]. (Integration of 429; pOSIP)DepositorInsertsdCas9 (bacteria)
PA4-mVenus
PphlF_A4NT in dCas9 sgRNA oscillator v1
UseCRISPRExpressionBacterialMutationD10A H840A (catalytically inactive)Available SinceFeb. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
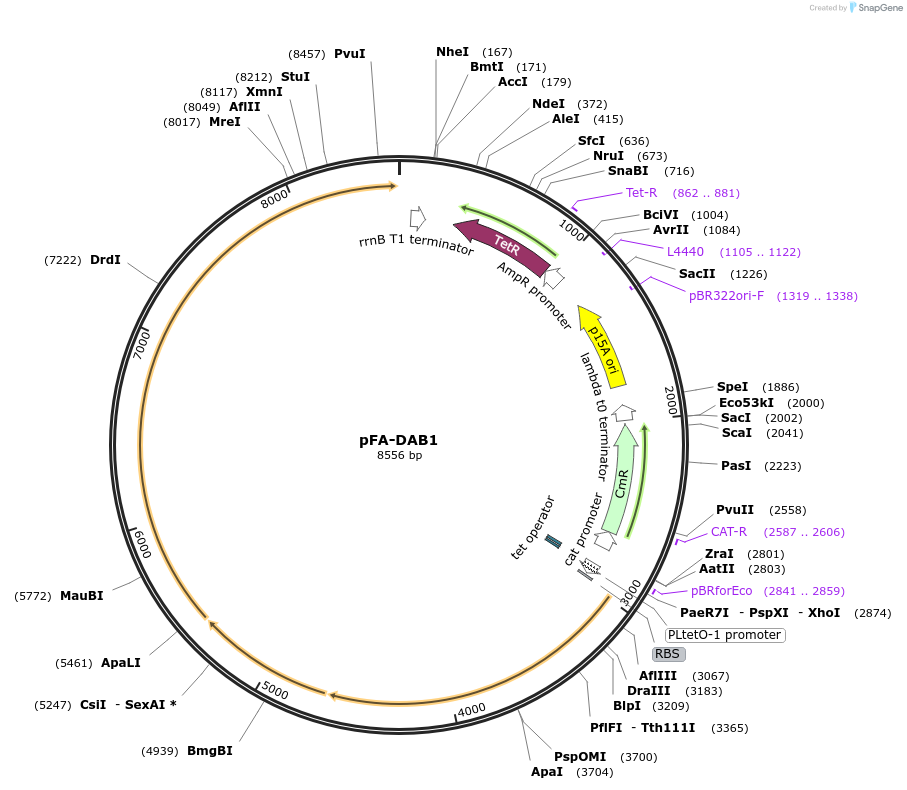
pFA-DAB1
Plasmid#162691PurposeExpresses DabAB1 inorganic carbon transport complexDepositorInsertDabAB1 inorganic carbon transport complex
ExpressionBacterialPromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
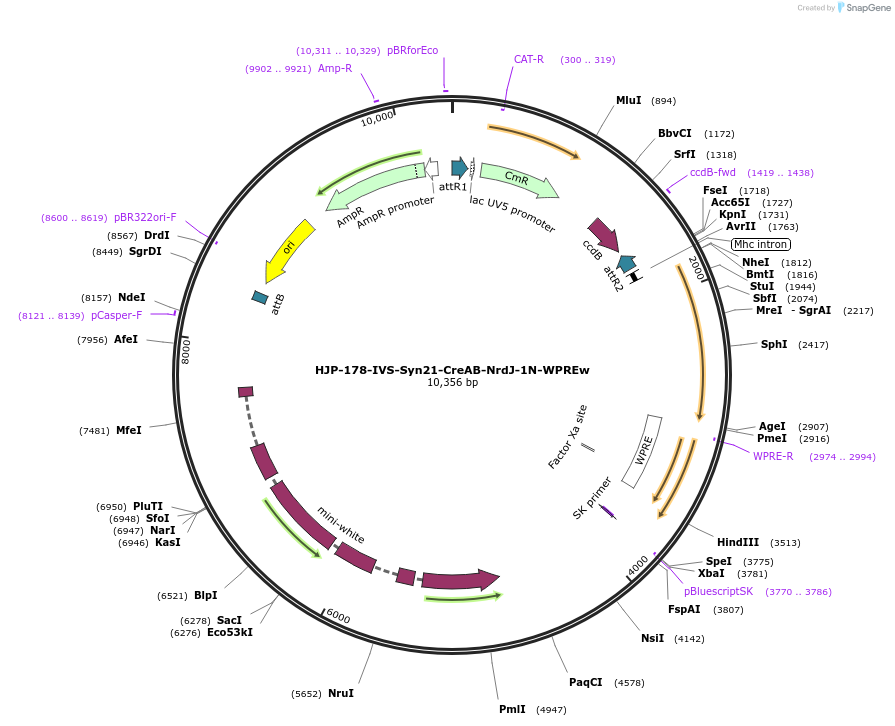
HJP-178-IVS-Syn21-CreAB-NrdJ-1N-WPREw
Plasmid#162473PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreAB
UseCre/LoxAvailable SinceJan. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
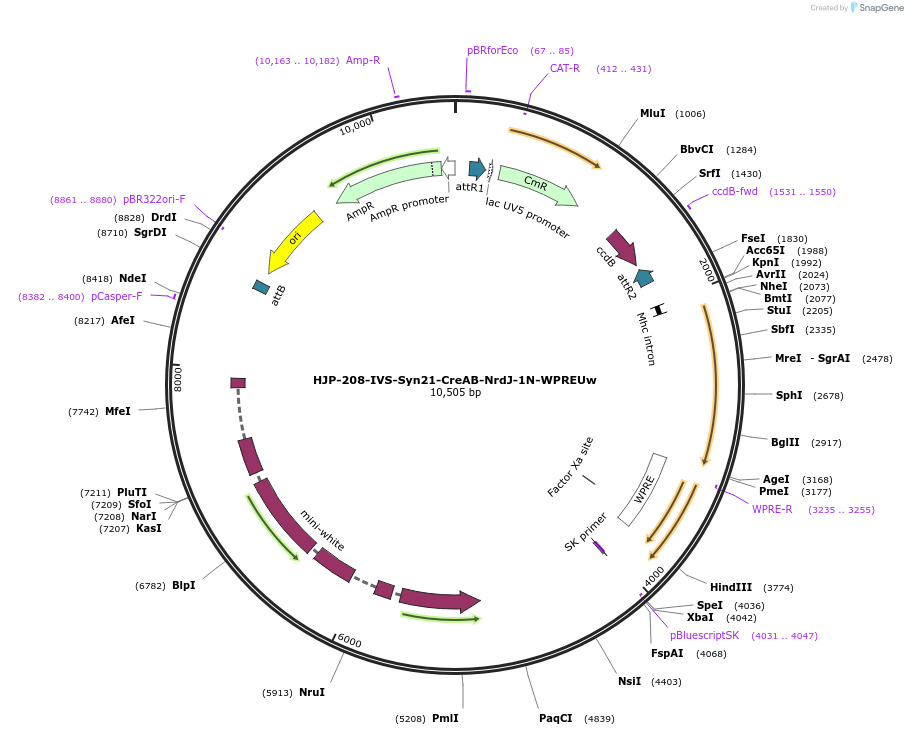
HJP-208-IVS-Syn21-CreAB-NrdJ-1N-WPREUw
Plasmid#162478PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreAB
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
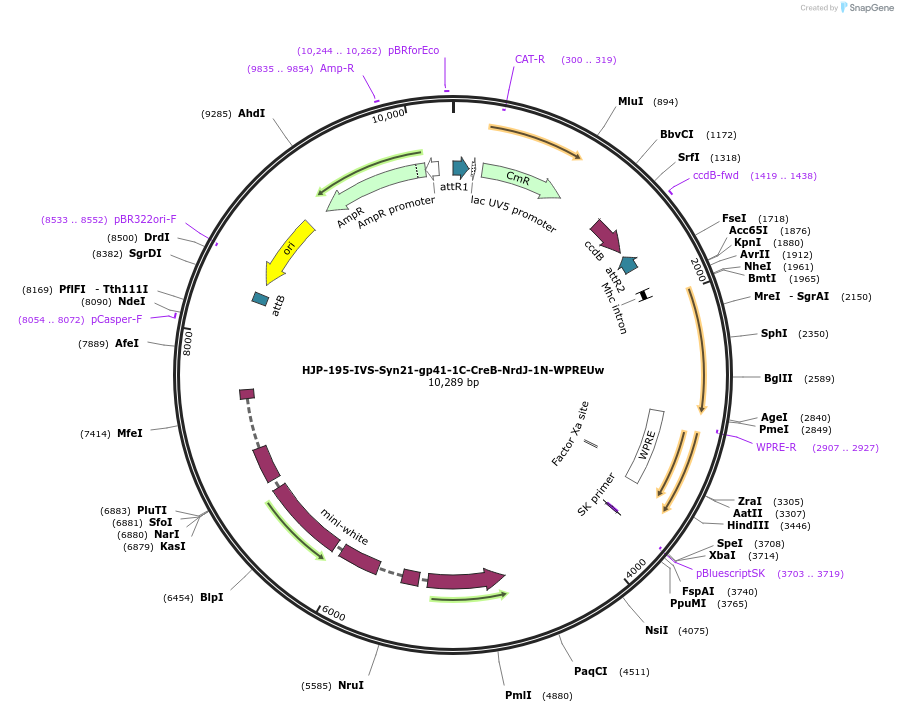
HJP-195-IVS-Syn21-gp41-1C-CreB-NrdJ-1N-WPREUw
Plasmid#162476PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreB
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
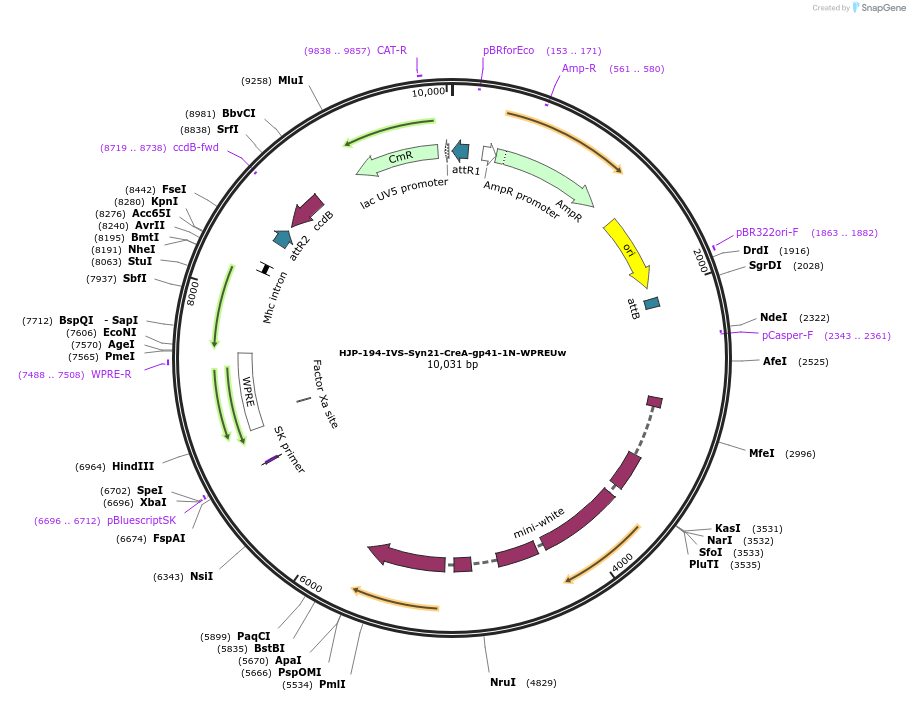
HJP-194-IVS-Syn21-CreA-gp41-1N-WPREUw
Plasmid#162475PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreA
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
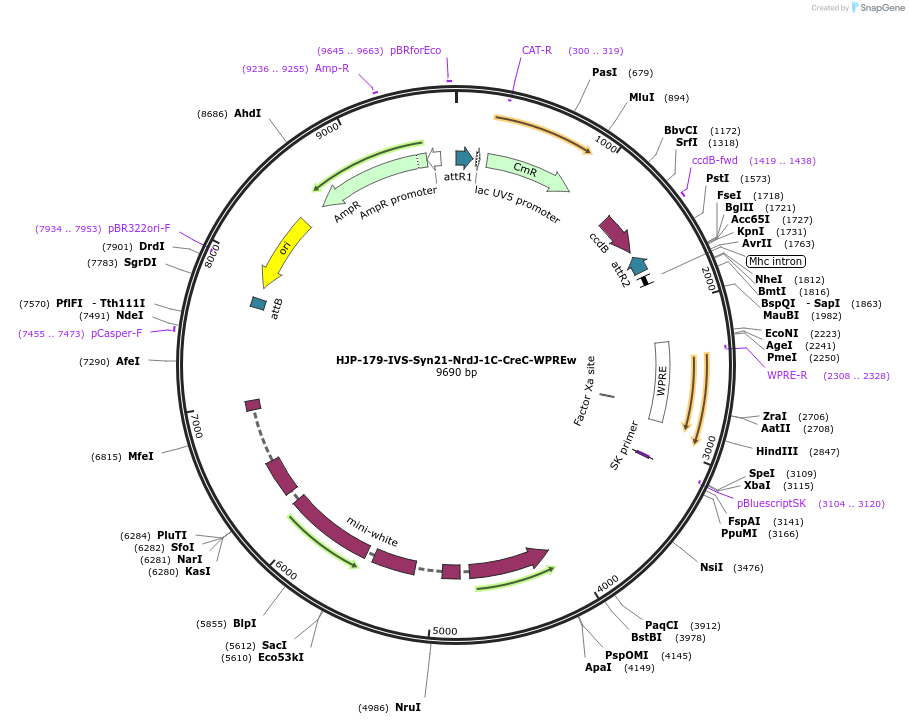
HJP-179-IVS-Syn21-NrdJ-1C-CreC-WPREw
Plasmid#162472PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreC
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
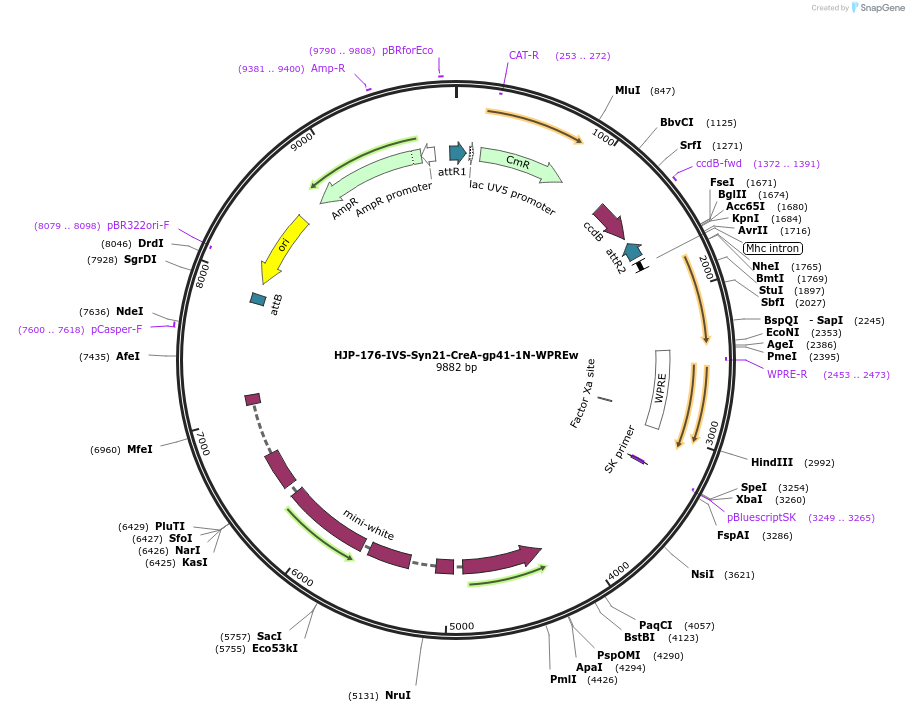
HJP-176-IVS-Syn21-CreA-gp41-1N-WPREw
Plasmid#162470PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreA
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
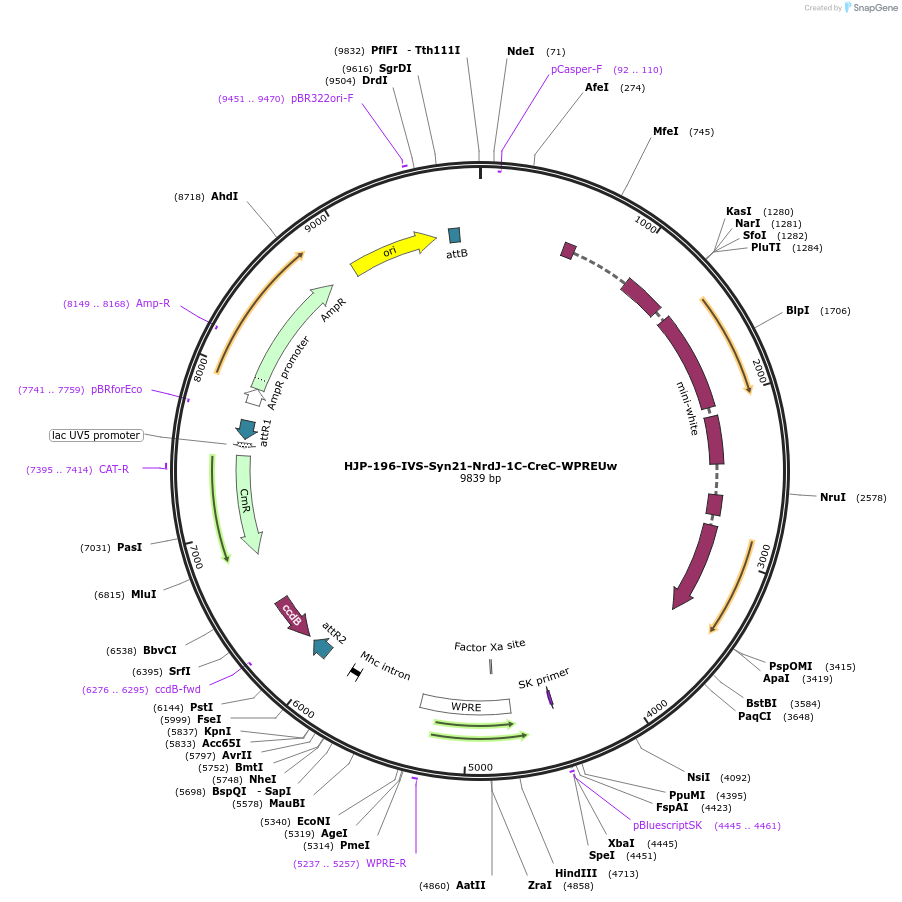
HJP-196-IVS-Syn21-NrdJ-1C-CreC-WPREUw
Plasmid#162477PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreC
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
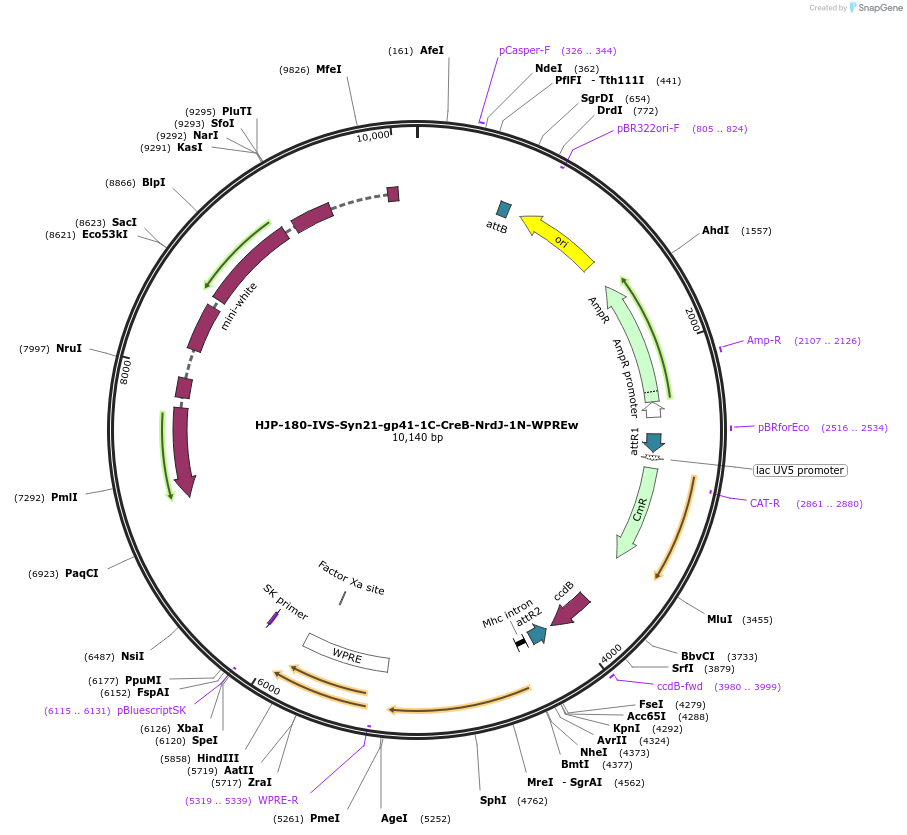
HJP-180-IVS-Syn21-gp41-1C-CreB-NrdJ-1N-WPREw
Plasmid#162471PurposeThis insert, when expressed in the same cell with other split Cre component, forms Cre recombinase through protein trans-splicing using split intein.DepositorInsertCreB
UseCre/LoxAvailable SinceDec. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
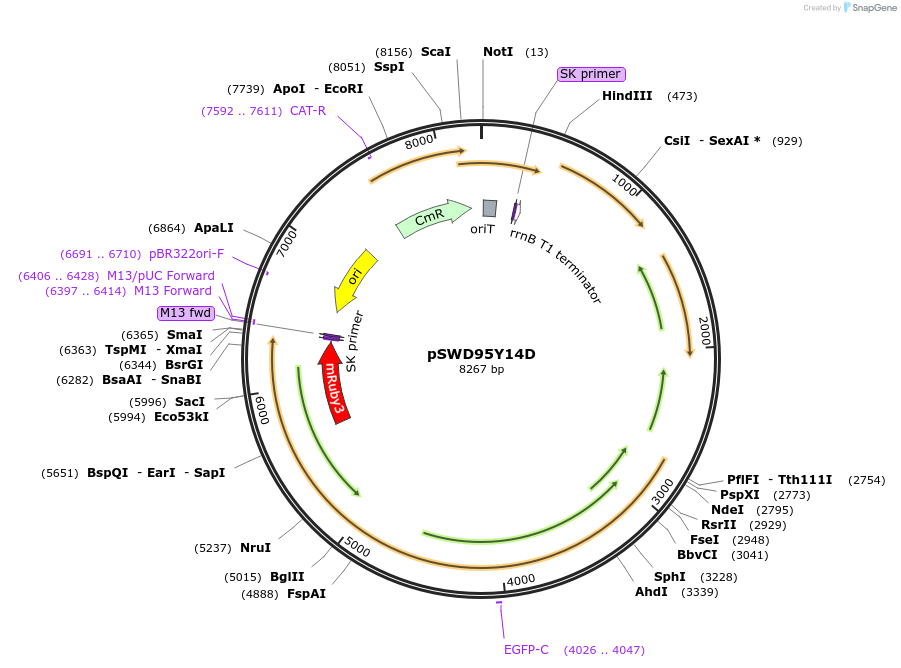
pSWD95Y14D
Plasmid#159098PurposeCckA FRET Sensor with cyclic-di-GMP point mutantDepositorInsertCckA 1-182, Linker SNVTRHRSATE, mClover3, CckA 183-691(Y514D), Linker SNVTRHRSAT, mRuby3
ExpressionBacterialMutationMutated Y514 to D514PromoterXyloseAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
pSWD95D293A
Plasmid#159099PurposeCckA FRET Sensor with DITE motif mutantDepositorInsertCckA 1-182, Linker SNVTRHRSATE, mClover3, CckA 183-691 (D293A), Linker SNVTRHRSAT, mRuby3
ExpressionBacterialMutationCckA D293 mutated to A293 and V199I on mRuby3 (P…PromoterXyloseAvailable SinceNov. 17, 2020AvailabilityAcademic Institutions and Nonprofits only