We narrowed to 30,195 results for: REP
-
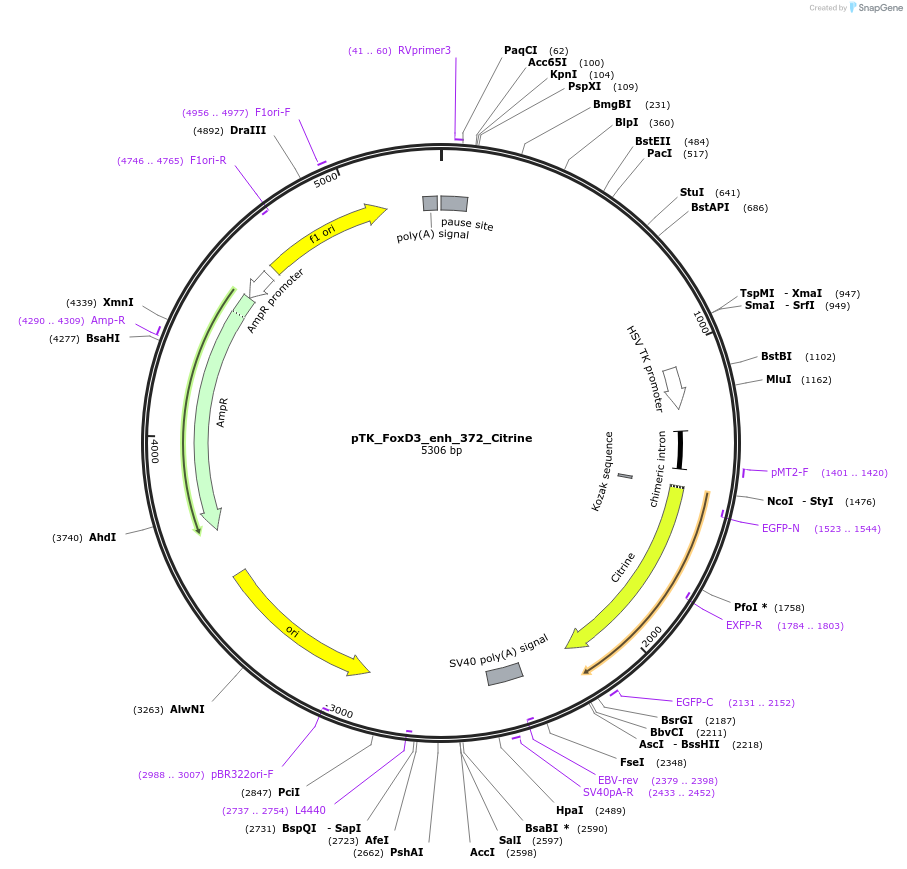
Plasmid#130583Purposeenhancer reporterDepositorInsertFoxD3_enh_372
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
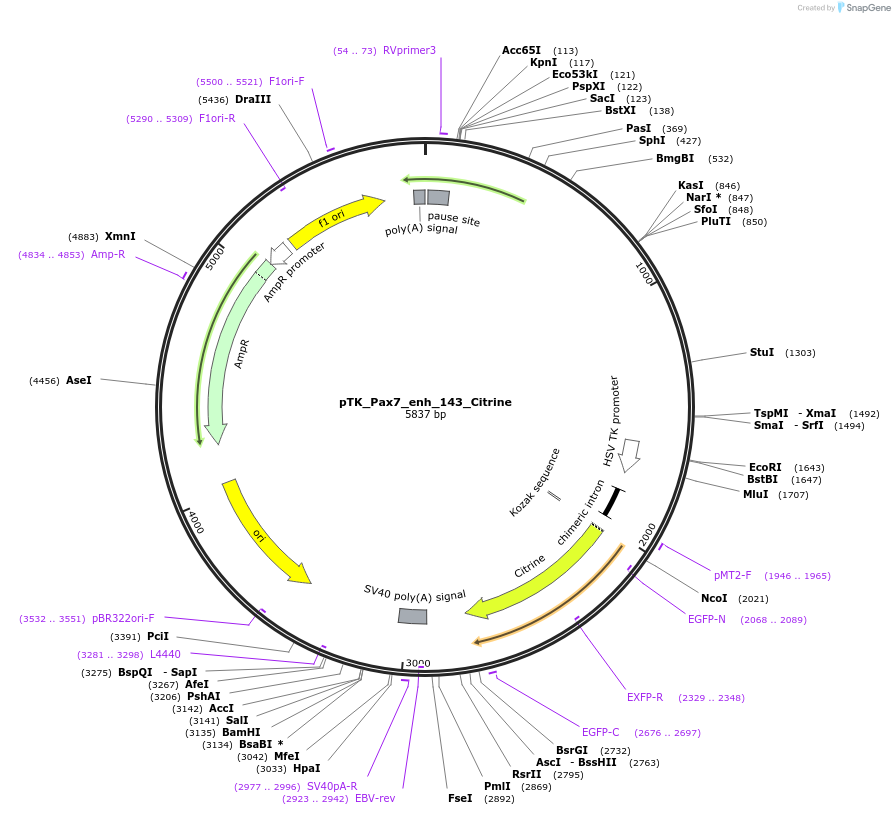
pTK_Pax7_enh_143_Citrine
Plasmid#130602Purposeenhancer reporterDepositorInsertPax7_enh_143
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
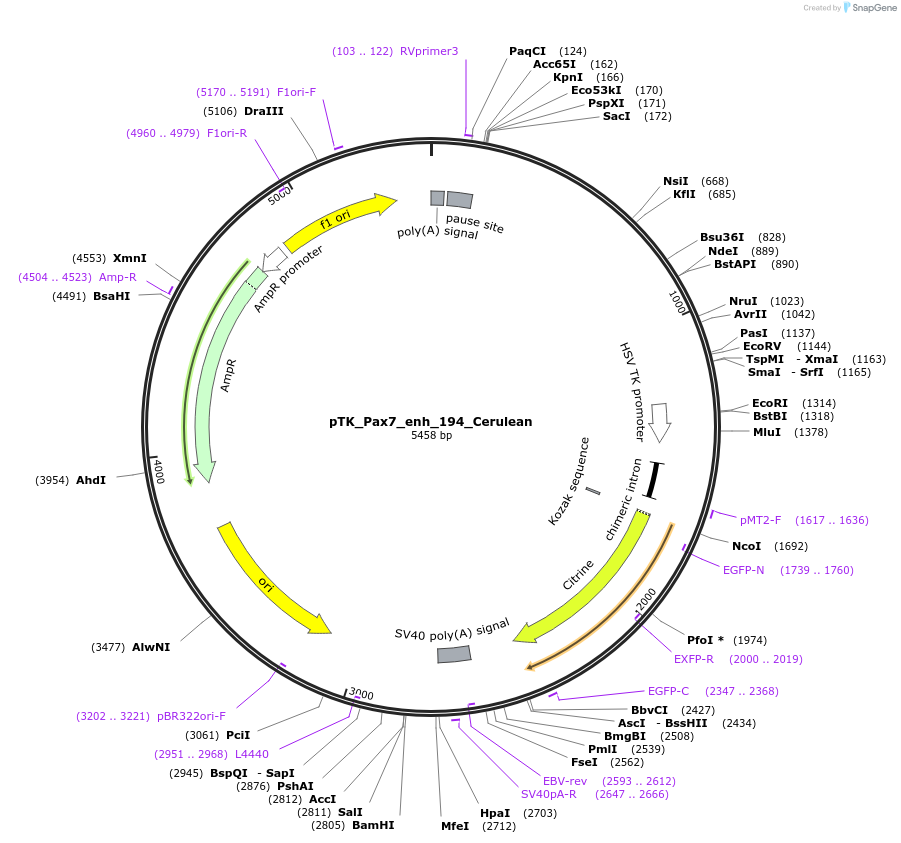
pTK_Pax7_enh_194_Cerulean
Plasmid#130603Purposeenhancer reporterDepositorInsertPax7_enh_194
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
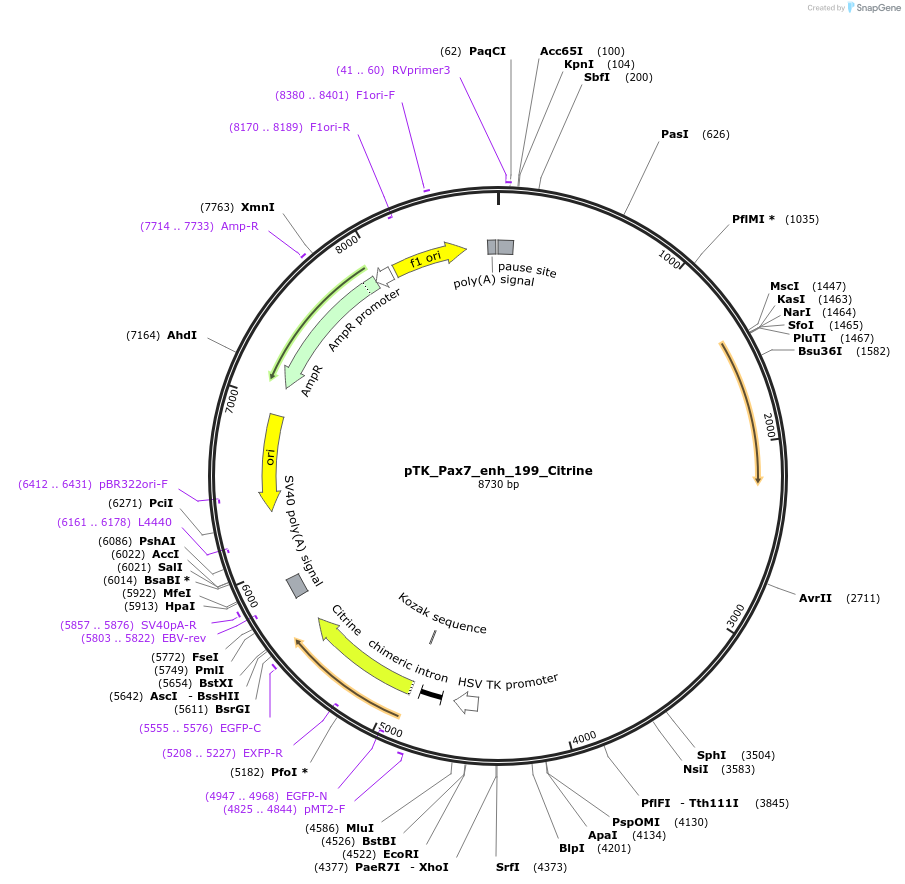
pTK_Pax7_enh_199_Citrine
Plasmid#130605Purposeenhancer reporterDepositorInsertPax7_enh_199
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
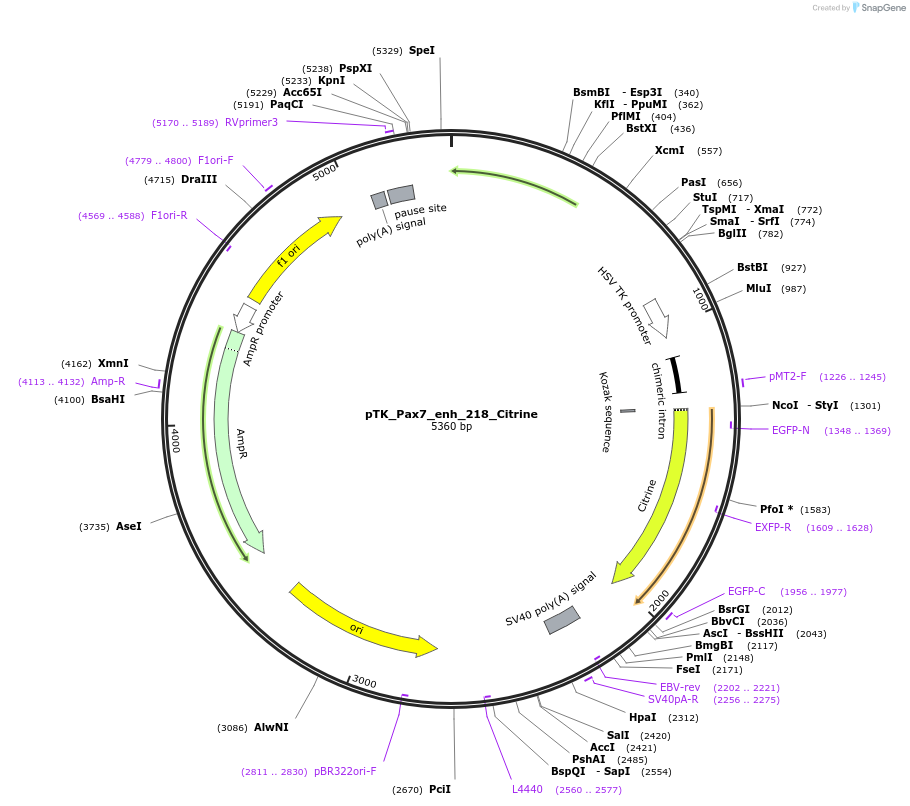
pTK_Pax7_enh_218_Citrine
Plasmid#130607Purposeenhancer reporterDepositorInsertPax7_enh_218
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
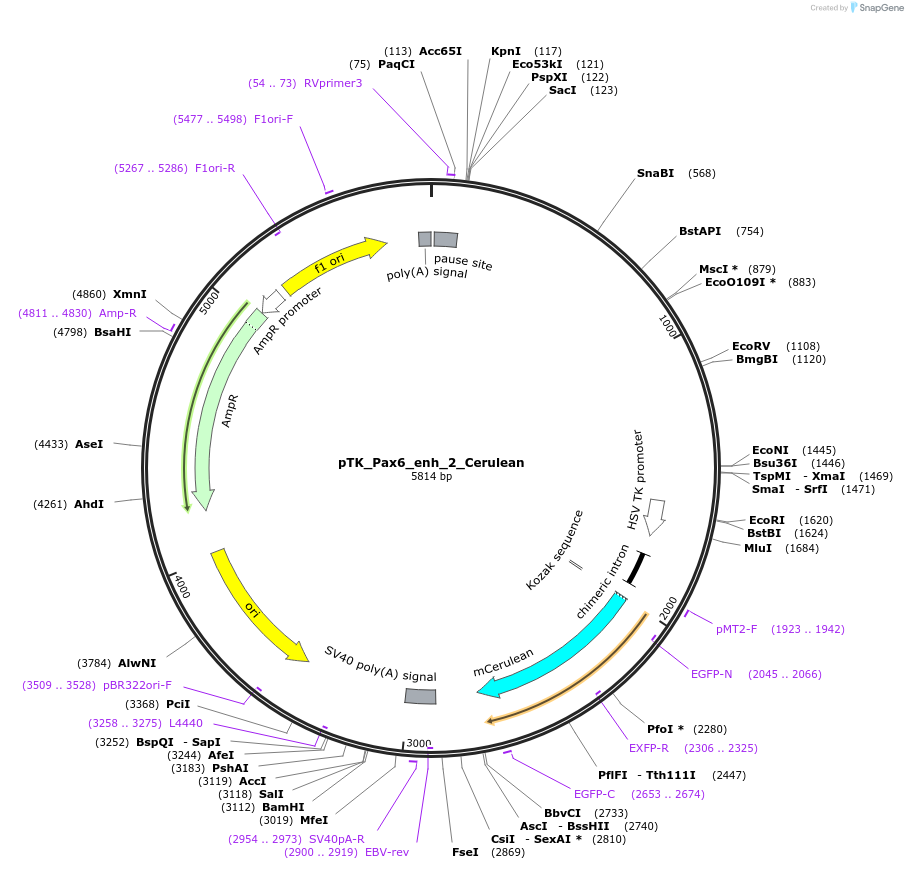
pTK_Pax6_enh_2_Cerulean
Plasmid#130620Purposeenhancer reporterDepositorInsertPax6_enh_2
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
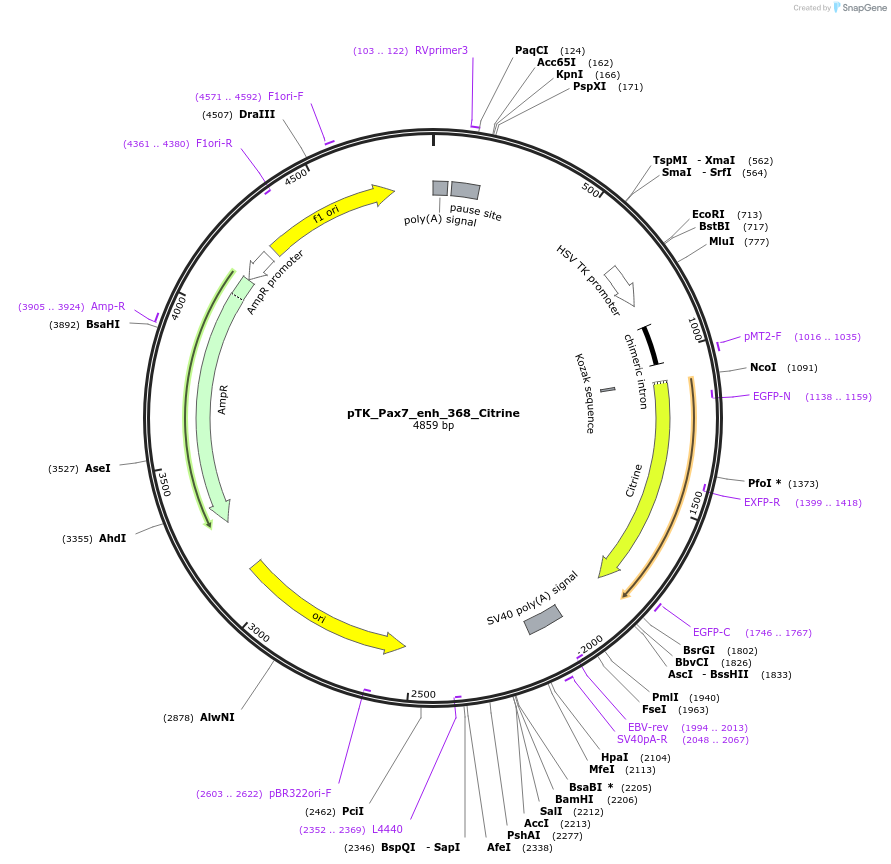
pTK_Pax7_enh_368_Citrine
Plasmid#130608Purposeenhancer reporterDepositorInsertPax7_enh_368
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceOct. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
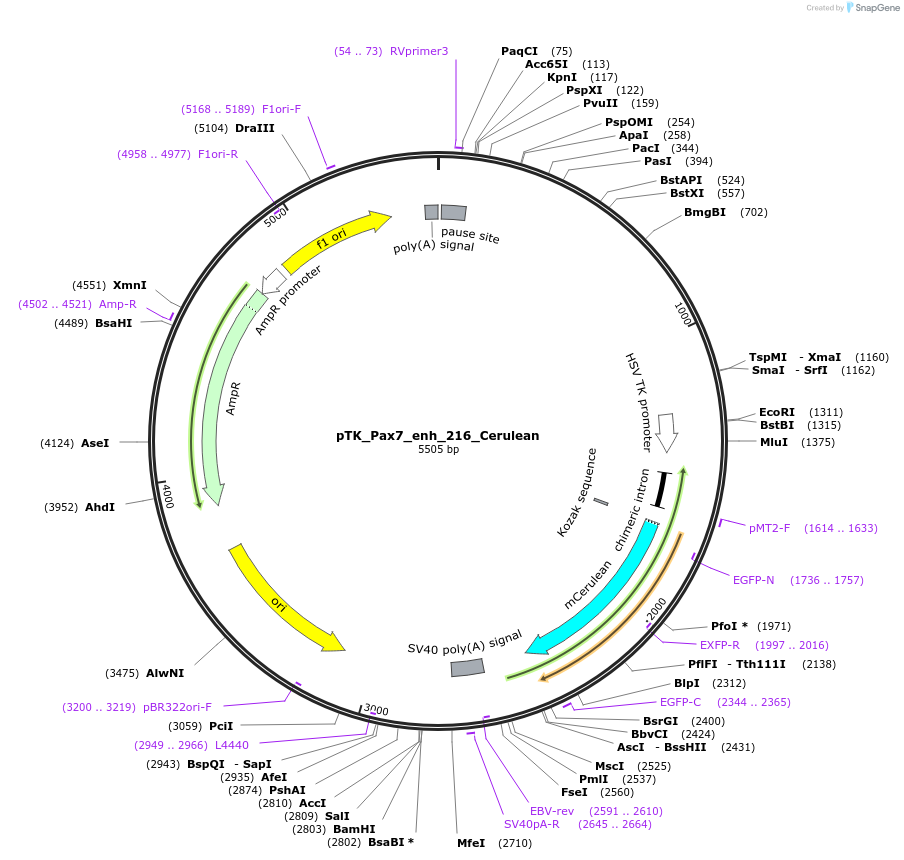
pTK_Pax7_enh_216_Cerulean
Plasmid#130606Purposeenhancer reporterDepositorInsertPax7_enh_216
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
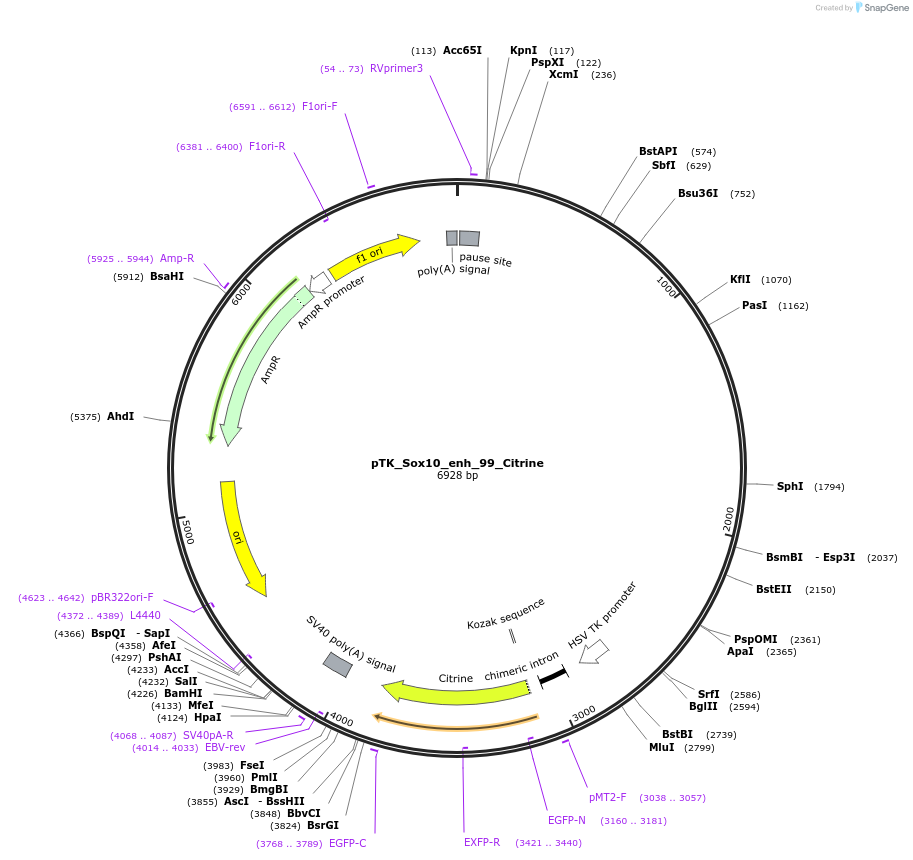
pTK_Sox10_enh_99_Citrine
Plasmid#130588Purposeenhancer reporterDepositorInsertSox10_enh_99
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
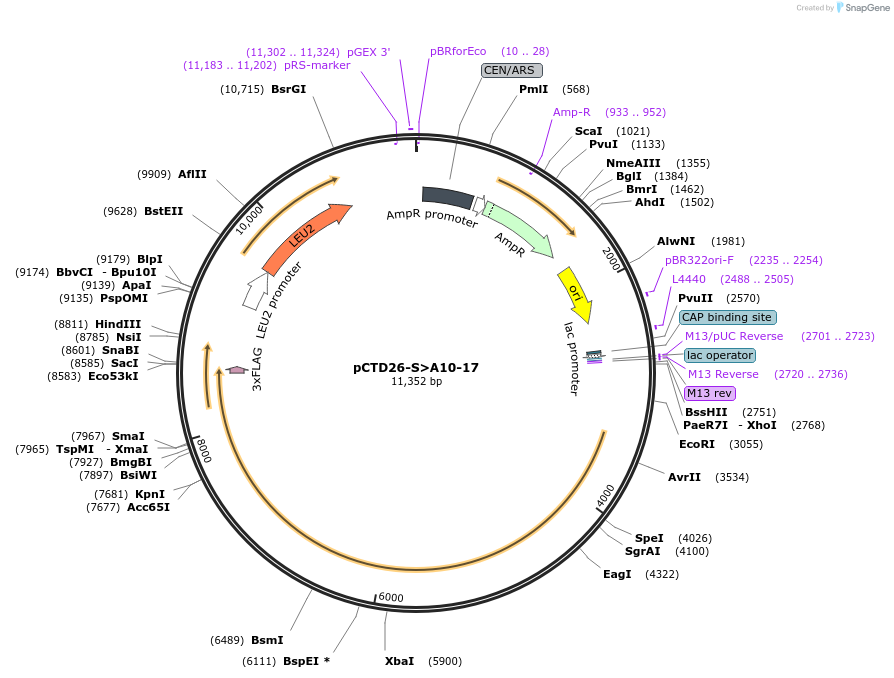
pCTD26-S>A10-17
Plasmid#112826PurposepSMF2 with all S to A mutations in only repeats 10-17.DepositorInsertRPB1 (RPO21 Budding Yeast)
ExpressionYeastMutationpSMF2 with all S to A mutations in only repeats 1…PromoterNative promoterAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
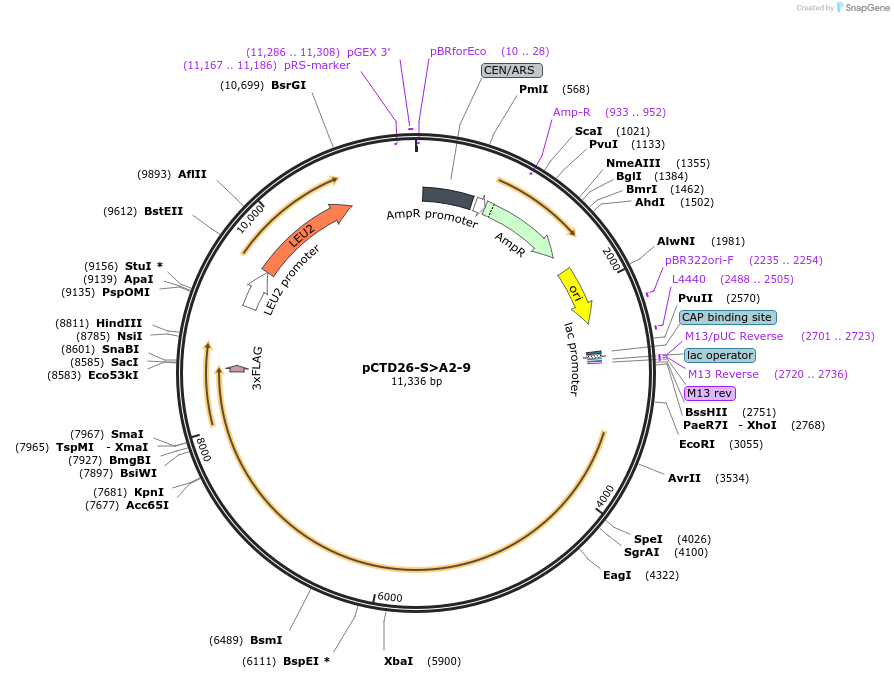
pCTD26-S>A2-9
Plasmid#112825PurposepSMF2 with all S to A mutations in only repeats 2-9.DepositorInsertRPB1 (RPO21 Budding Yeast)
ExpressionYeastMutationpSMF2 with all S to A mutations in only repeats 2…PromoterNative promoterAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
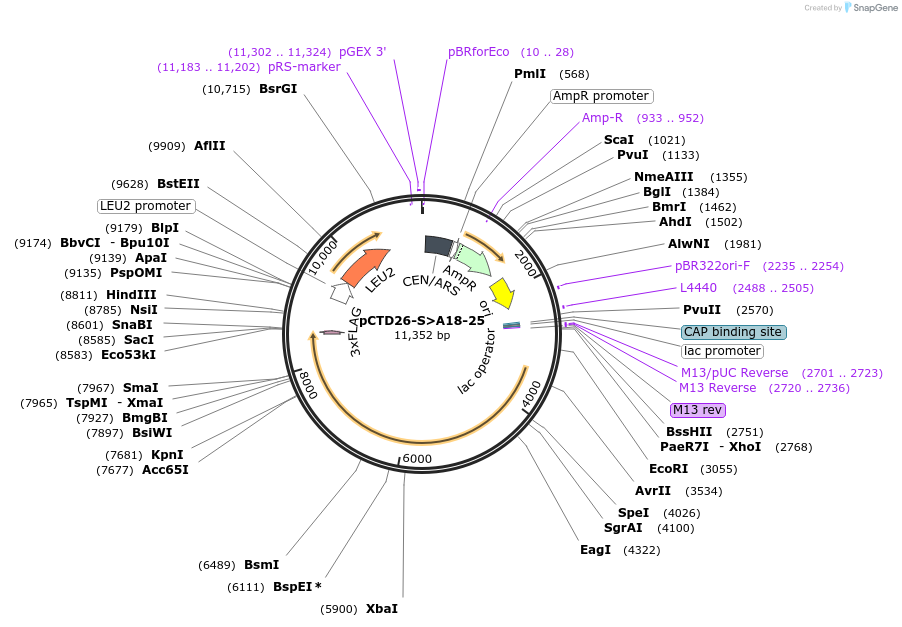
pCTD26-S>A18-25
Plasmid#112827PurposepSMF2 with all S to A mutations in only repeats 18-25.DepositorInsertRPB1 (RPO21 Budding Yeast)
ExpressionYeastMutationpSMF2 with all S to A mutations in only repeats 1…PromoterNative promoterAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
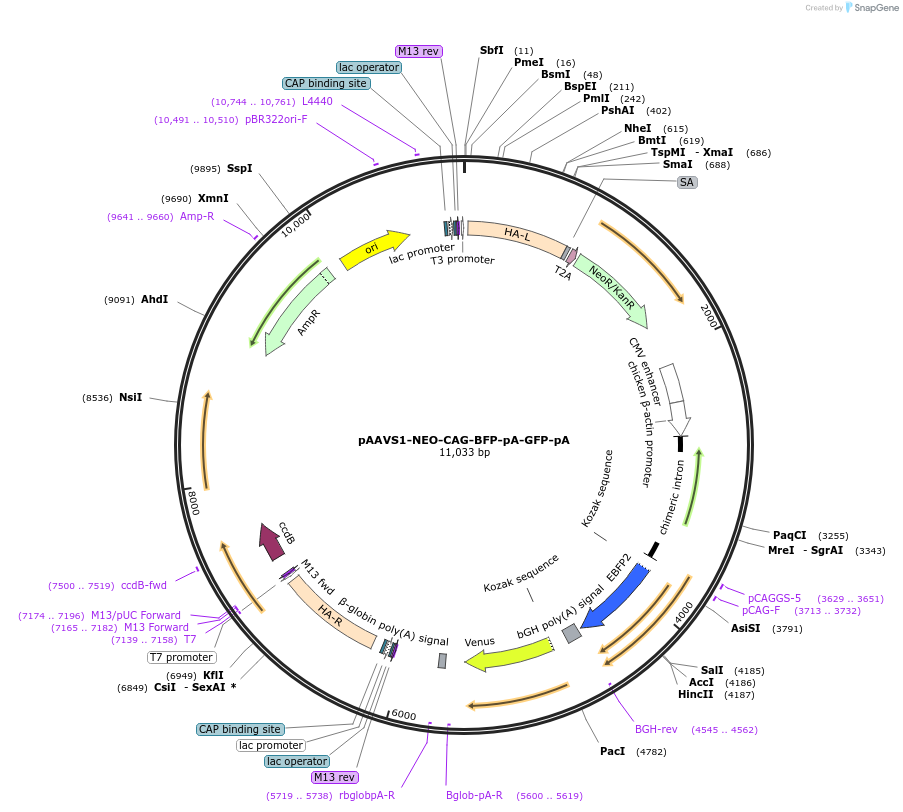
pAAVS1-NEO-CAG-BFP-pA-GFP-pA
Plasmid#149344PurposeVector for targeting the Replace reporter to the AAVS1 locusDepositorInsertBFP
ExpressionMammalianPromoterCAGAvailable SinceJan. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
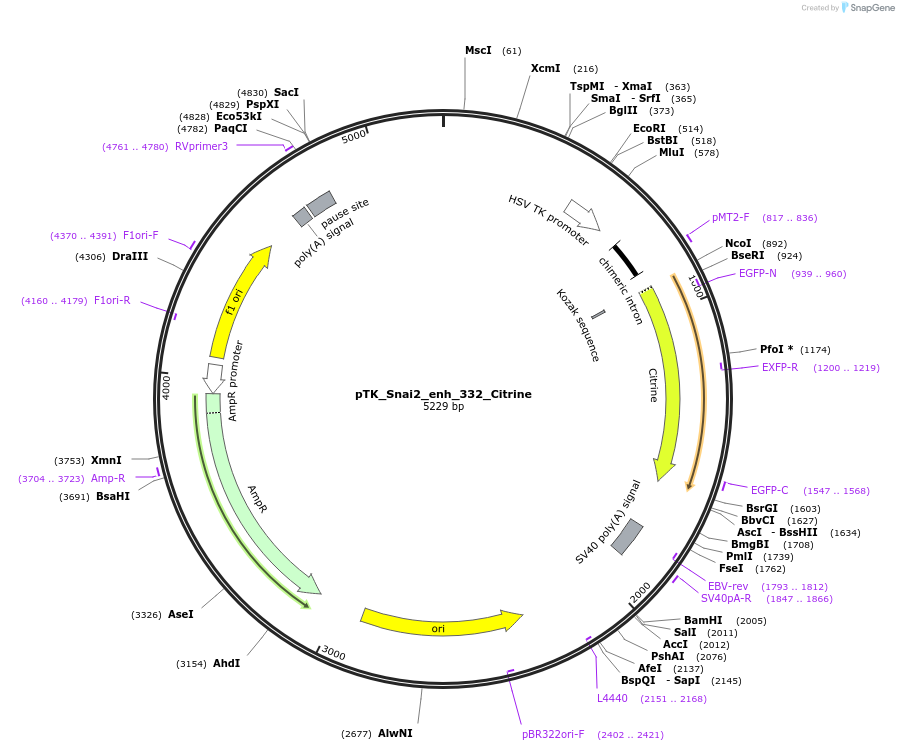
pTK_Snai2_enh_332_Citrine
Plasmid#130577Purposeenhancer reporterDepositorInsertSnai2_enh_332
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
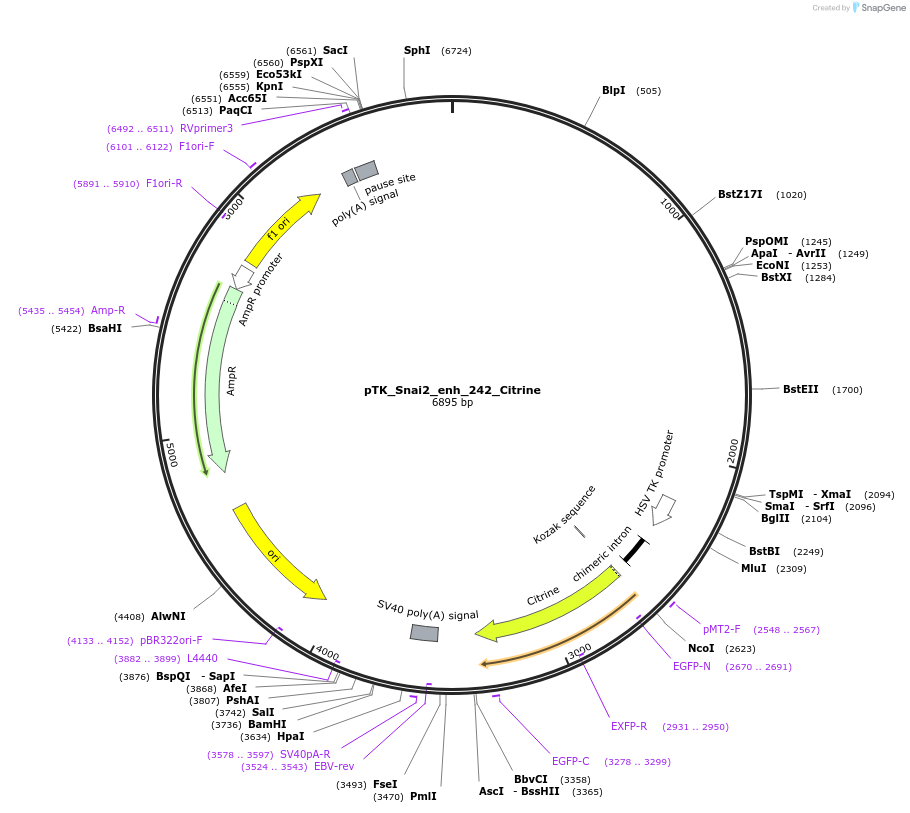
pTK_Snai2_enh_242_Citrine
Plasmid#130576Purposeenhancer reporterDepositorInsertSnai2_enh_242
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
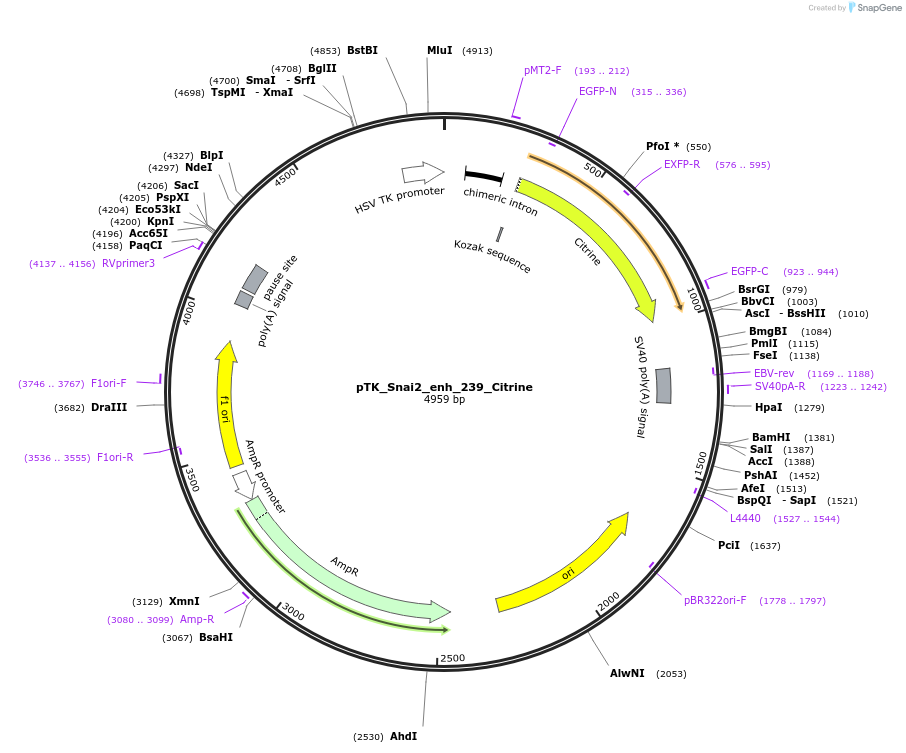
pTK_Snai2_enh_239_Citrine
Plasmid#130575Purposeenhancer reporterDepositorInsertSnai2_enh_239
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceNov. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
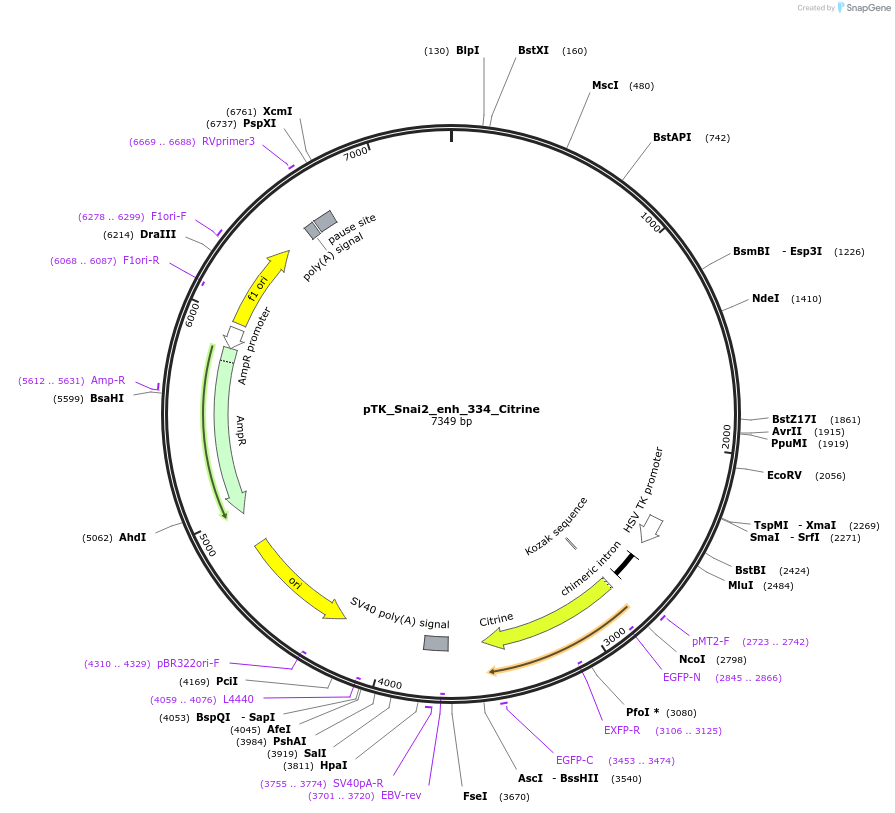
pTK_Snai2_enh_334_Citrine
Plasmid#130578Purposeenhancer reporterDepositorInsertSnai2_enh_334
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
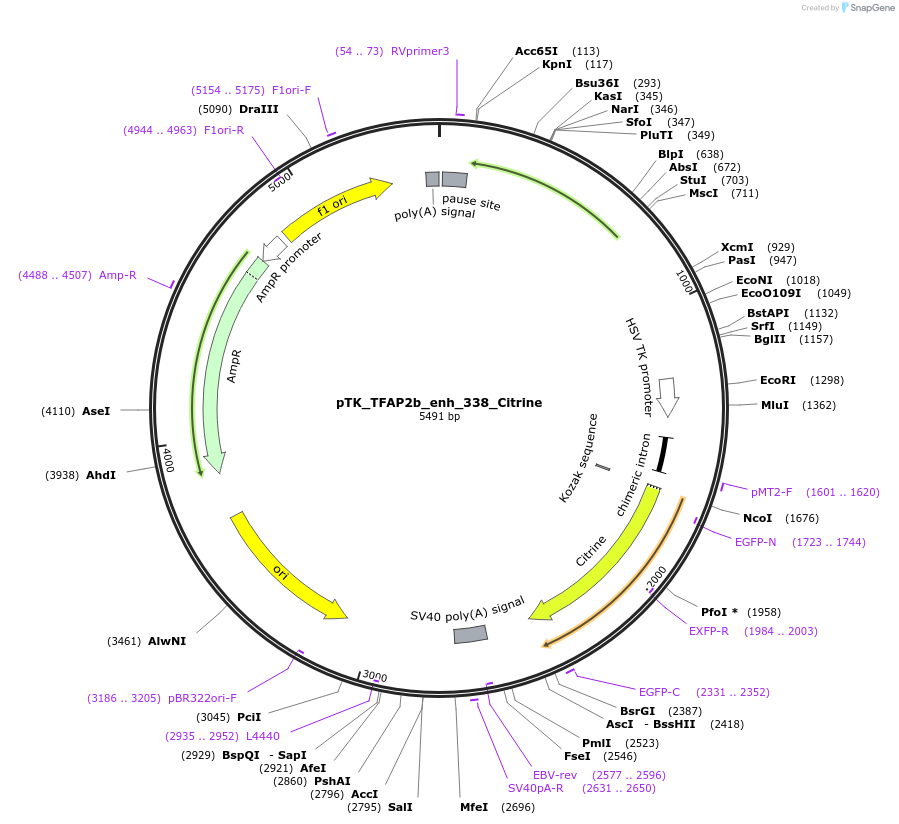
pTK_TFAP2b_enh_338_Citrine
Plasmid#130619Purposeenhancer reporterDepositorInsertTFAP2b_enh_338
UseEnhancer reporterPromoterthymidine kinase minimal promoterAvailable SinceOct. 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
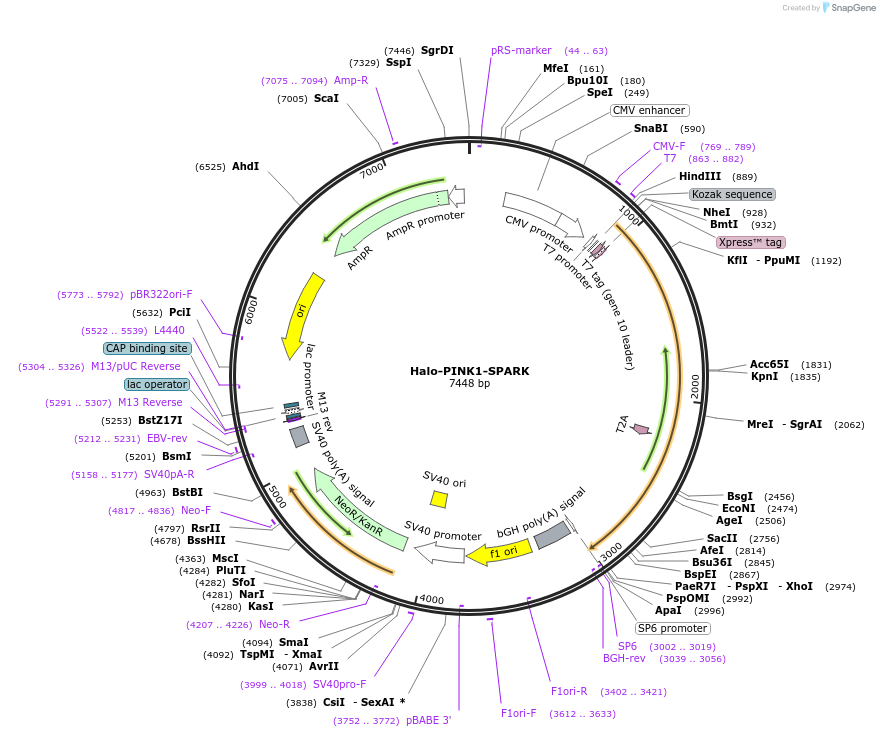
Halo-PINK1-SPARK
Plasmid#248088PurposeHaloTag labeled PINK1-SPARKDepositorInsertHalo-PINK1-SPARK
ExpressionMammalianPromoterCMVAvailable SinceDec. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
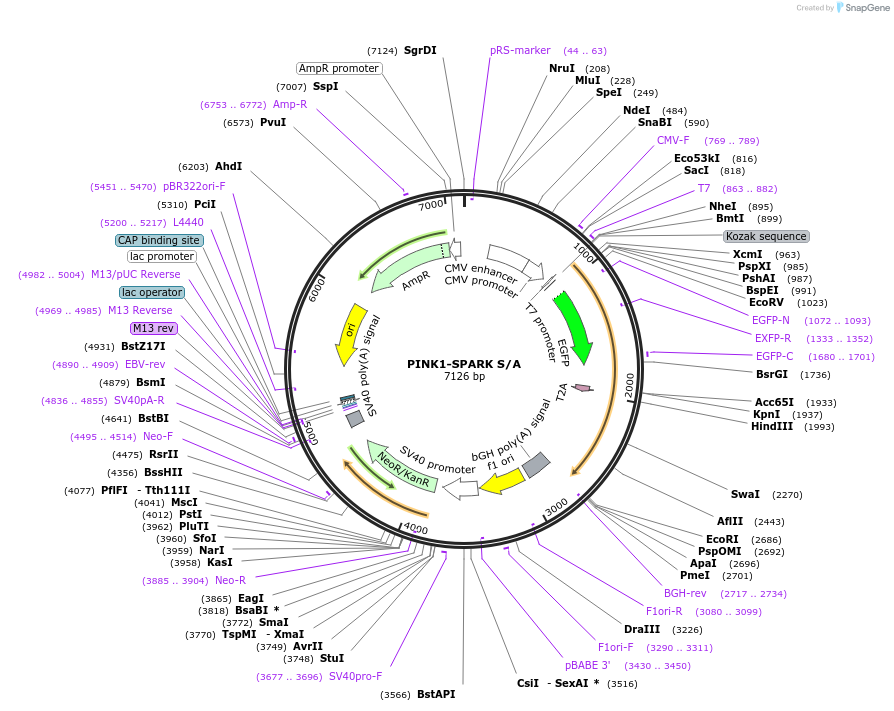
PINK1-SPARK S/A
Plasmid#248087PurposePINK1-SPARK control construct with PINK1 phosphosite mutatedDepositorInsertPINK1-SPARK S/A
ExpressionMammalianPromoterCMVAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only