We narrowed to 5,133 results for: codon optimized
-
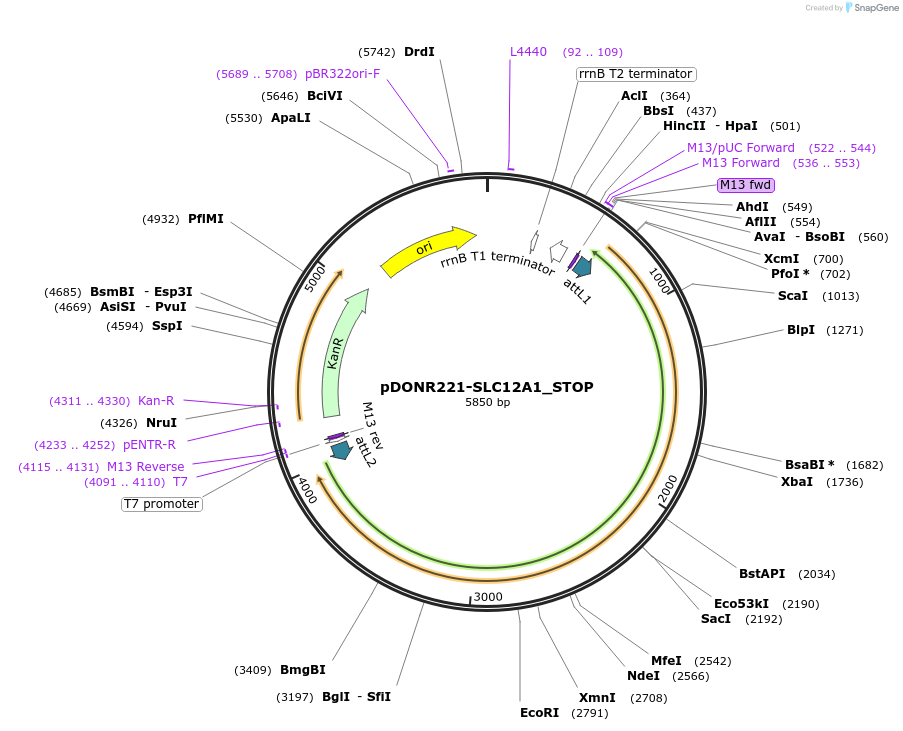
Plasmid#161062PurposeGateway entry clone with codon-optimized ORF sequence for subcloning into custom expression vectors. Contains a STOP codon at the end of the codon-optimized ORF sequence.DepositorInsertSLC12A1 (SLC12A1 Human)
ExpressionMammalianAvailable SinceJune 21, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
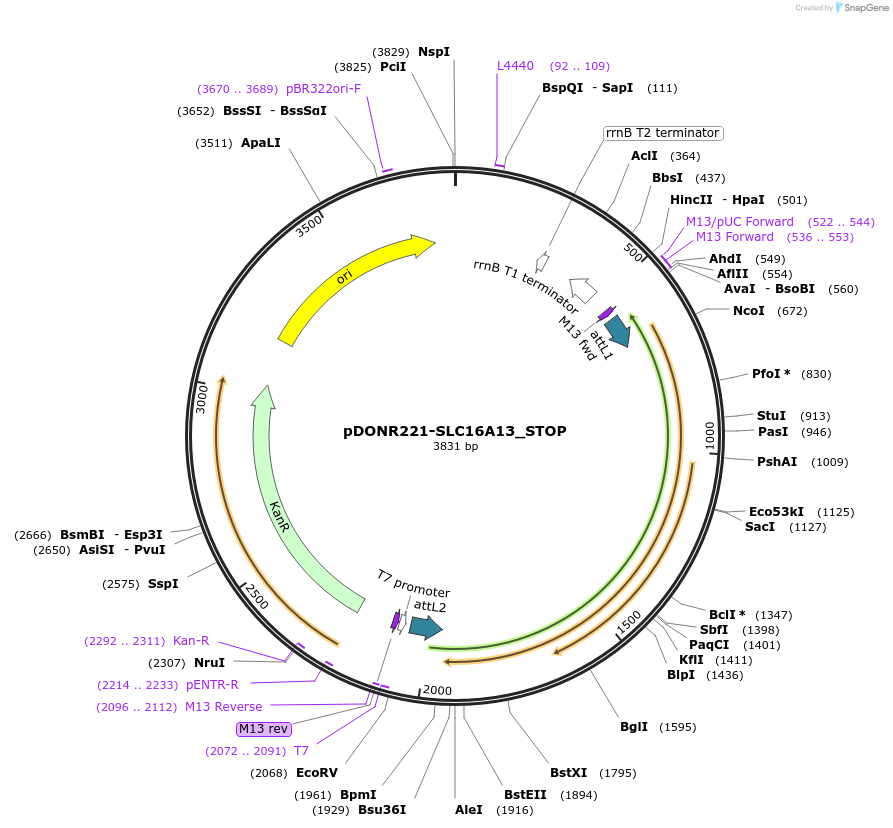
pDONR221-SLC16A13_STOP
Plasmid#161041PurposeGateway entry clone with codon-optimized ORF sequence for subcloning into custom expression vectors. Contains a STOP codon at the end of the codon-optimized ORF sequence.DepositorInsertSLC16A13 (SLC16A13 Human)
ExpressionMammalianAvailable SinceJune 21, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
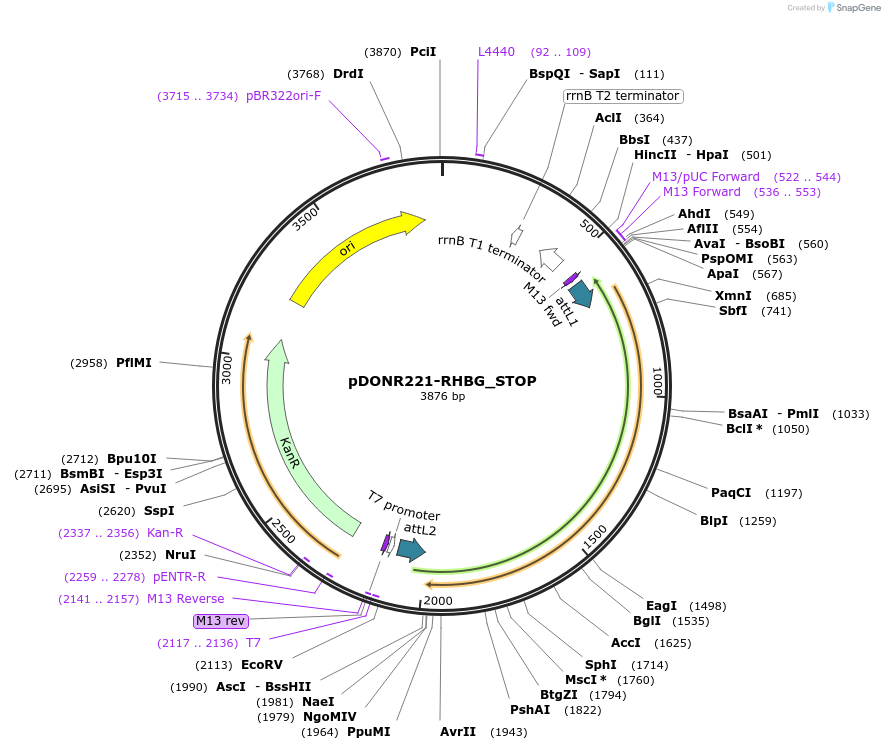
pDONR221-RHBG_STOP
Plasmid#161049PurposeGateway entry clone with codon-optimized ORF sequence for subcloning into custom expression vectors. Contains a STOP codon at the end of the codon-optimized ORF sequence.DepositorInsertRHBG (RHBG Human)
ExpressionMammalianAvailable SinceJune 21, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
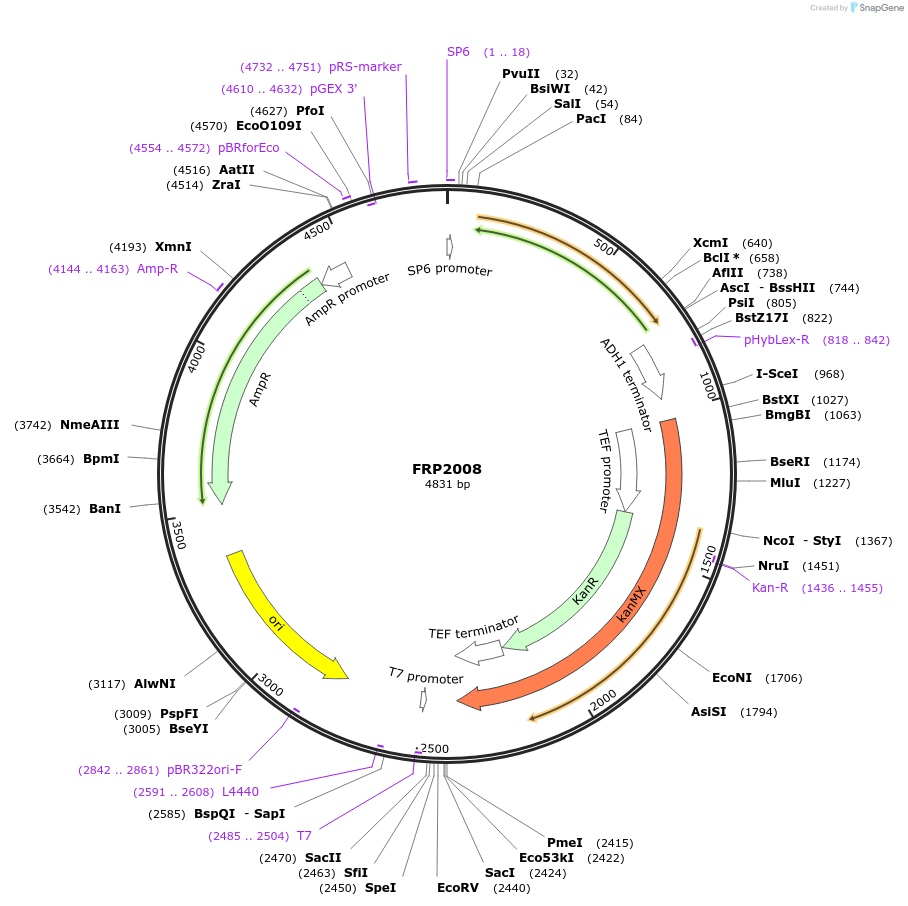
FRP2008
Plasmid#159312PurposeYeast tagging vector to create gene fusion with yeast codon optimized mKOkappa under KanMX selectionDepositorTypeEmpty backboneExpressionYeastMutationV2delAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
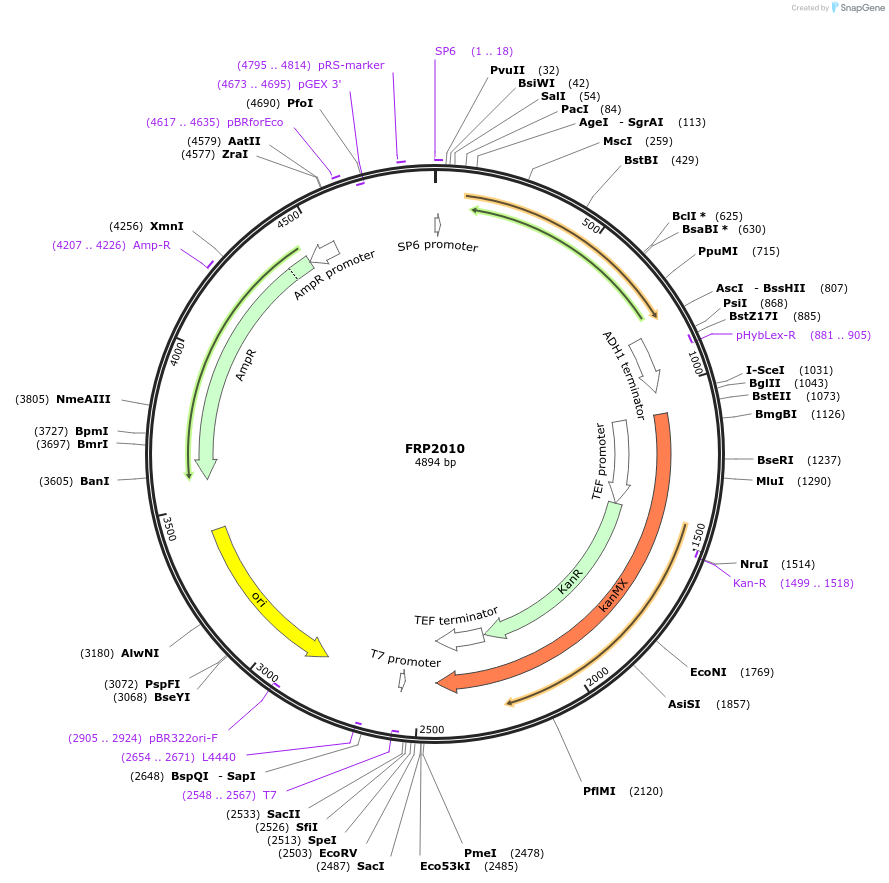
FRP2010
Plasmid#159313PurposeYeast tagging vector to create gene fusion with yeast codon optimized tSapphire under KanMX selectionDepositorTypeEmpty backboneExpressionYeastMutationV2del, L231HAvailable SinceSept. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
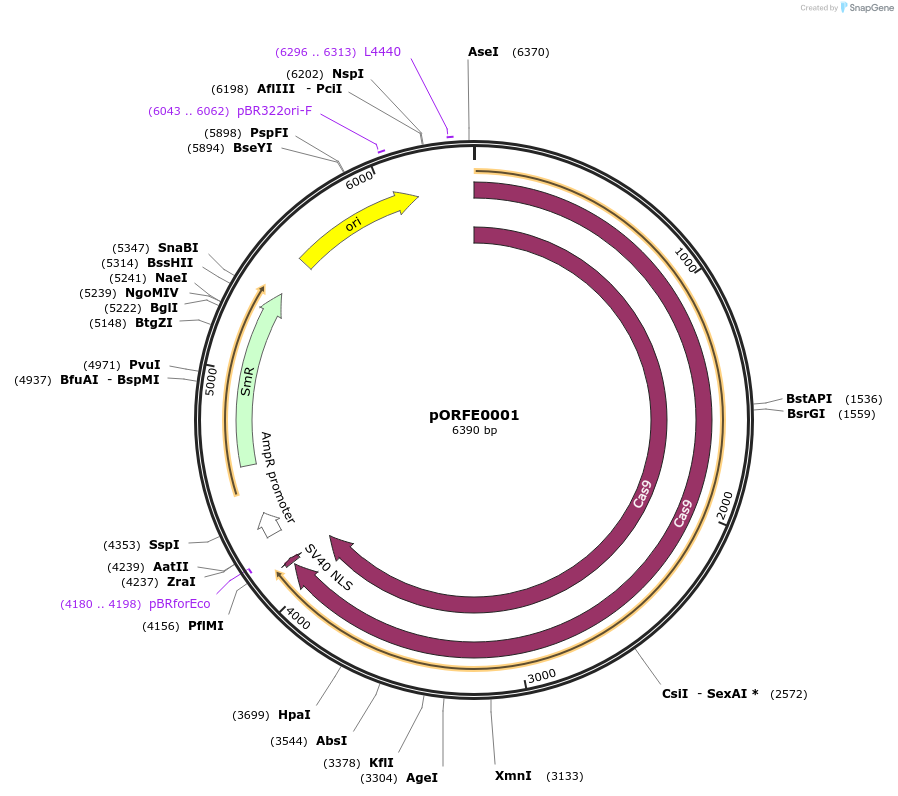
pORFE0001
Plasmid#112070PurposeAtCas9 module CDS1DepositorInsertAtCAS9
UseCRISPRExpressionPlantMutationPlant codon optimized Cas9 S. pyogenesAvailable SinceAug. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
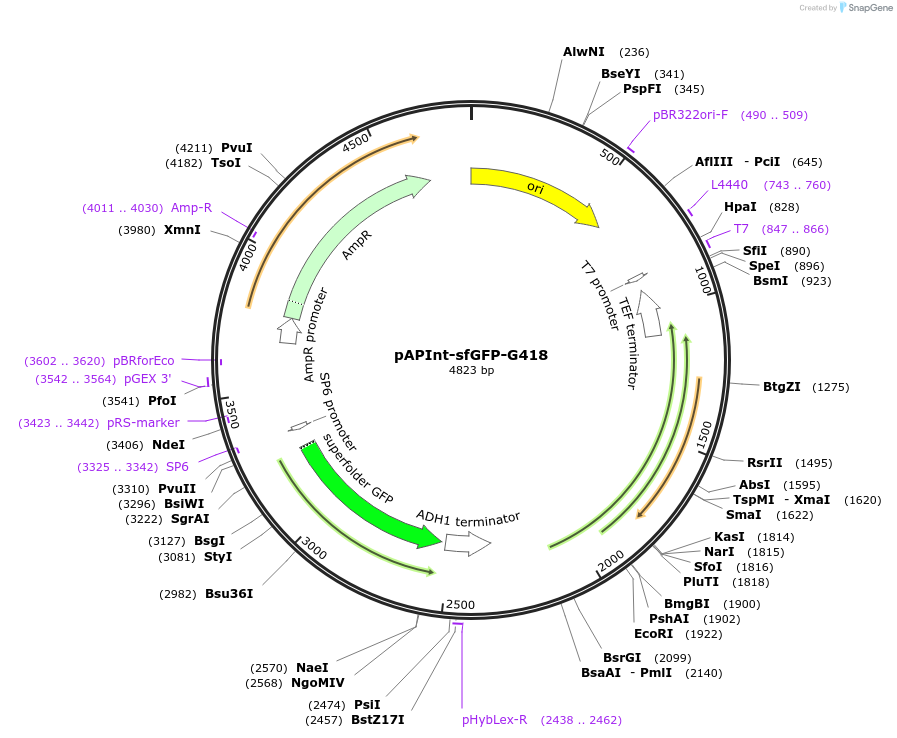
pAPInt-sfGFP-G418
Plasmid#236481PurposePlasmid for deleting or C-terminally tagging endogenous genes with sfGFP and selection on G418.DepositorInsertsfGFP
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
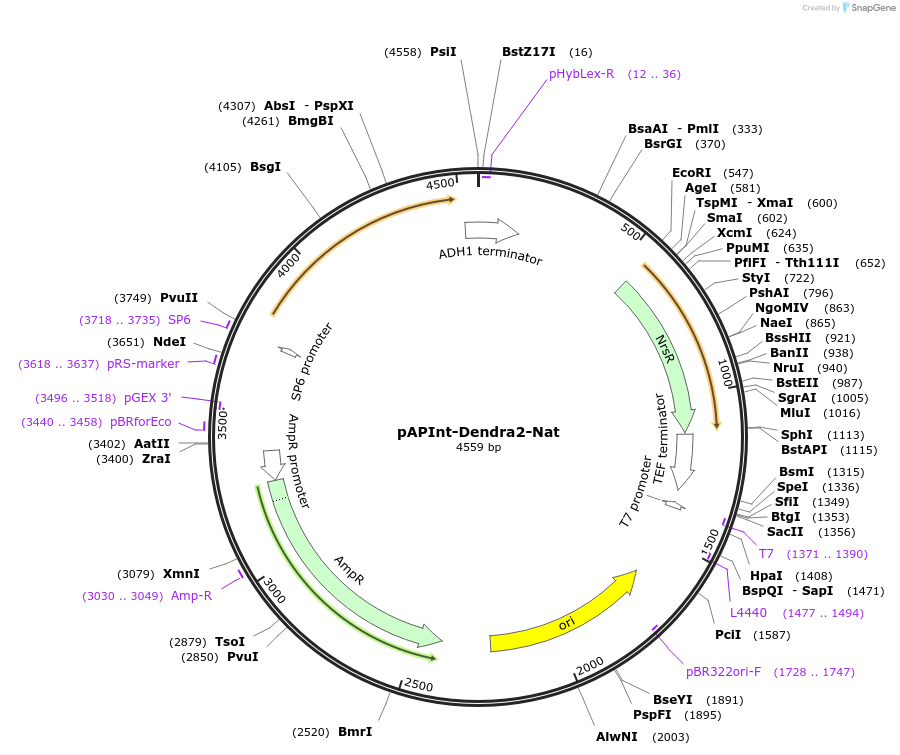
pAPInt-Dendra2-Nat
Plasmid#236494PurposePlasmid for deleting or C-terminally tagging endogenous genes with Dendra2 and selection on Nat.DepositorInsertDendra2
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
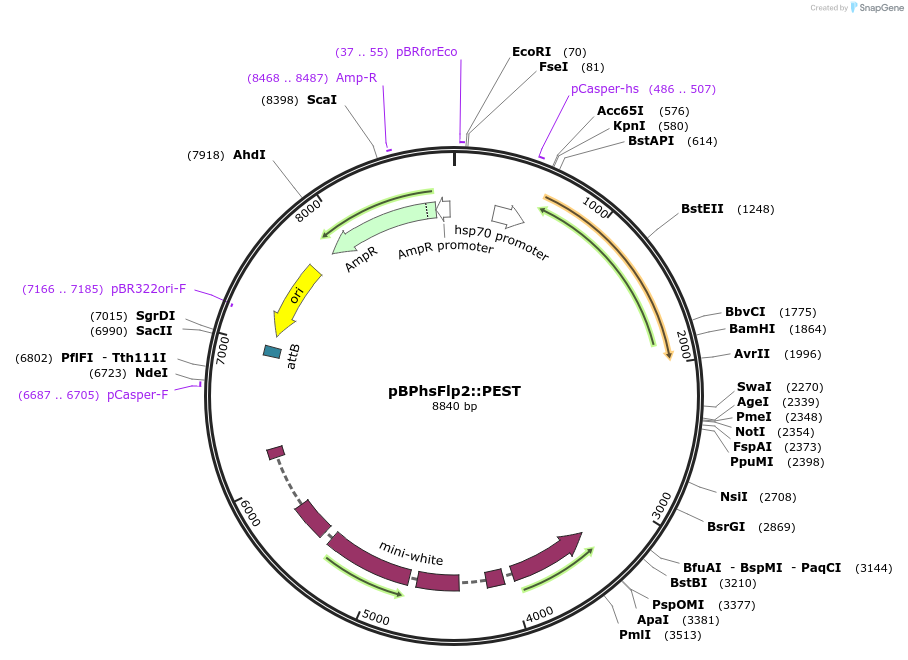
pBPhsFlp2::PEST
Plasmid#63171PurposeDrosophila codon-optimized Flp2::mODC-PEST fusion gene driven from Drosophila Hsp70Bb Heat Shock PromoterDepositorInserthsFlp2::PEST
ExpressionInsectAvailable SinceMay 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
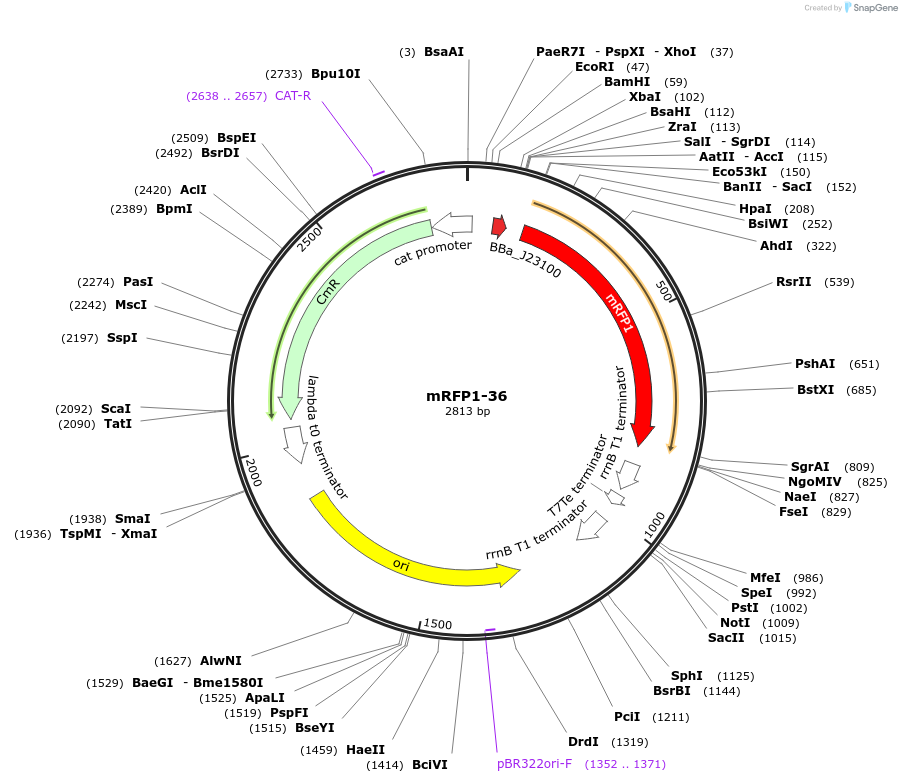
mRFP1-36
Plasmid#63825PurposeA member of an RBS library for mRFP1 with different translation ratesDepositorInsertmRFP1
ExpressionBacterialMutationCodon optimized for E.coliPromoterJ23100Available SinceMay 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
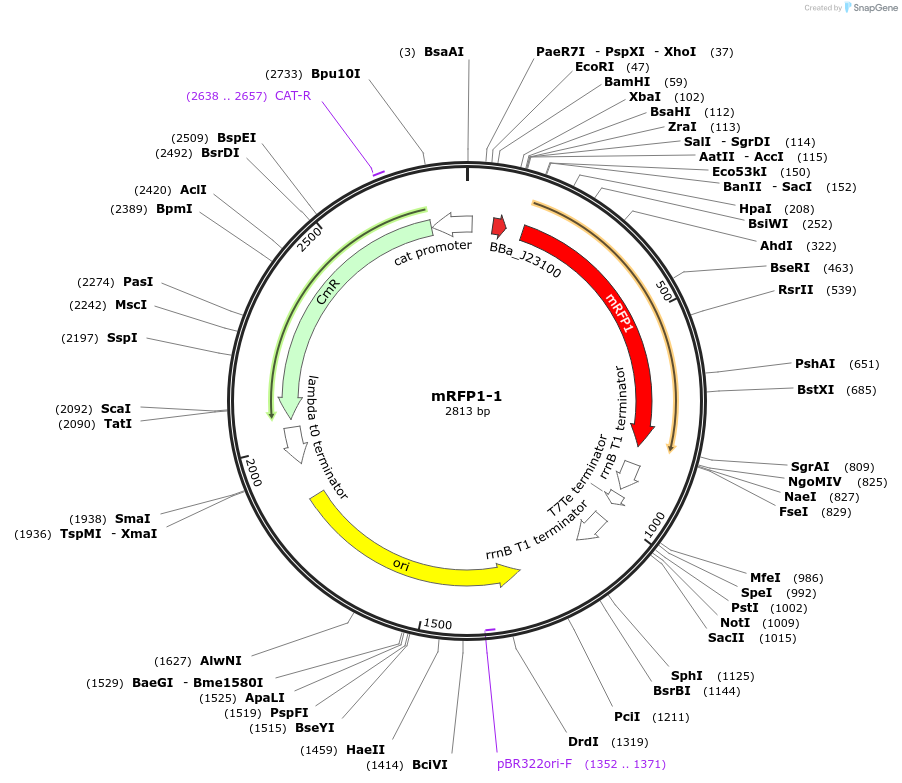
mRFP1-1
Plasmid#63816PurposeA member of an RBS library for mRFP1 with different translation ratesDepositorInsertmRFP1
ExpressionBacterialMutationCodon optimized for E.coliPromoterJ23100Available SinceMay 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
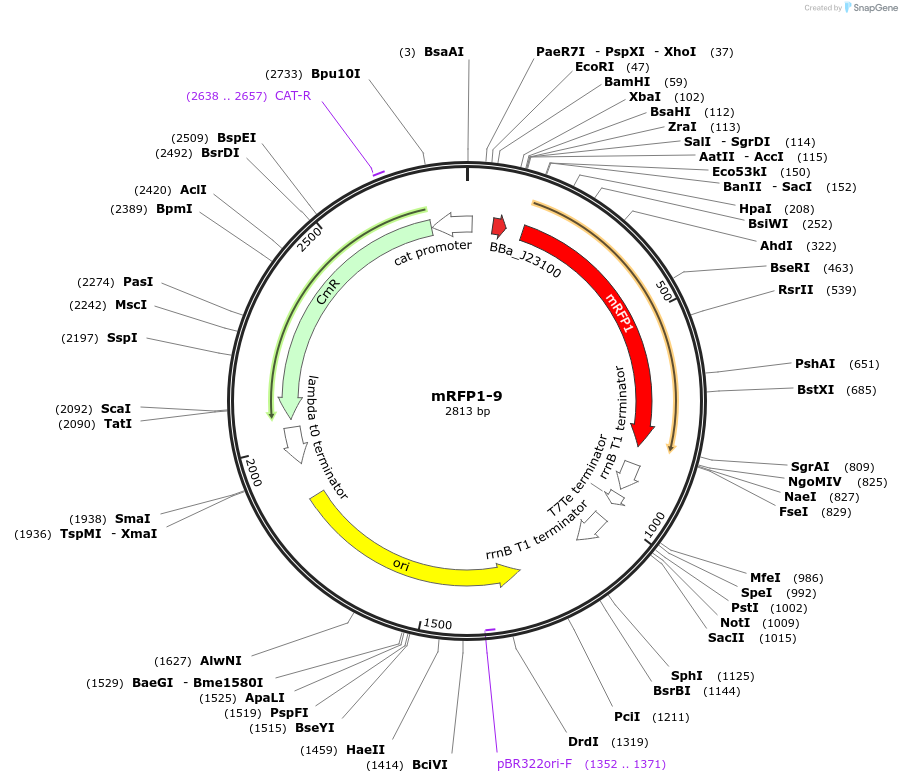
mRFP1-9
Plasmid#63818PurposeA member of an RBS library for mRFP1 with different translation ratesDepositorInsertmRFP1
ExpressionBacterialMutationCodon optimized for E.coliPromoterJ23100Available SinceMay 15, 2015AvailabilityAcademic Institutions and Nonprofits only -
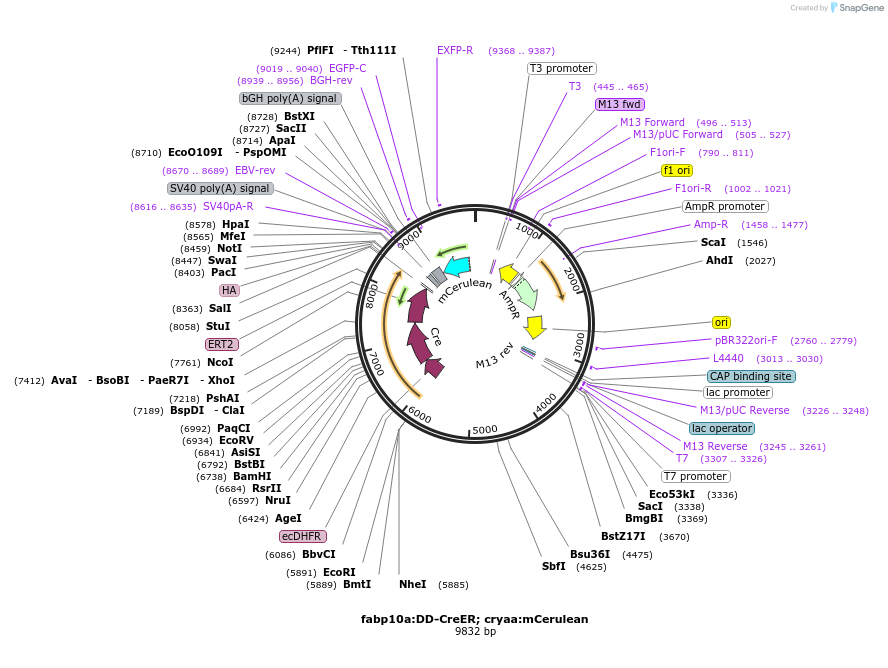
fabp10a:DD-CreER; cryaa:mCerulean
Plasmid#230076PurposeExpression of destabilized CreER in zebrafish hepatocytes.DepositorInsertltDHFR(DD)
UseZebrafish expressionTagsCreER and HAMutationamino acids mutations: R12H, N23S, G67S, V78A, E1…Promoterfabp10aAvailable SinceMay 30, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
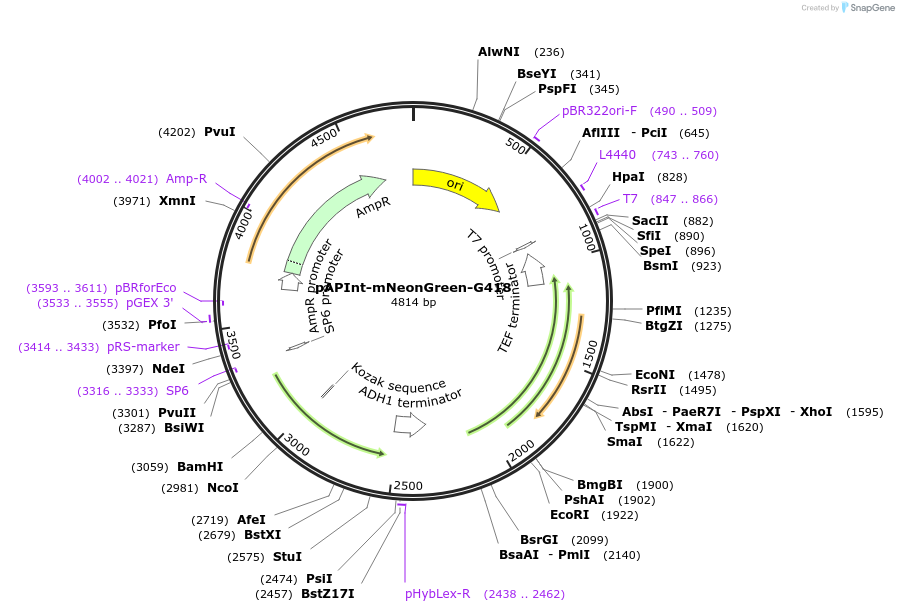
pAPInt-mNeonGreen-G418
Plasmid#236484PurposePlasmid for deleting or C-terminally tagging endogenous genes with mNeonGreen and selection on G418.DepositorInsertmNeonGreen
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
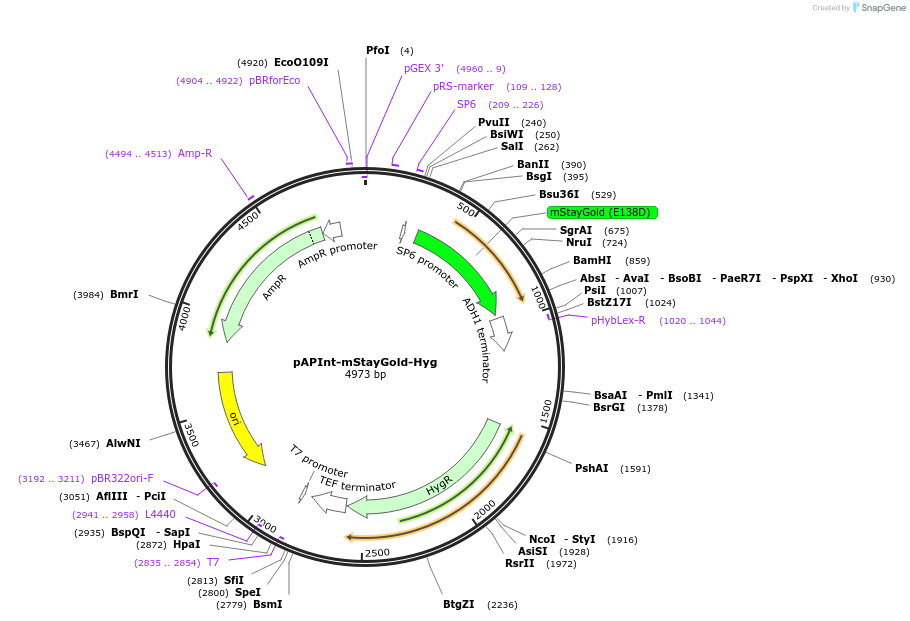
pAPInt-mStayGold-Hyg
Plasmid#236486PurposePlasmid for deleting or C-terminally tagging endogenous genes with mStayGold and selection on Hyg.DepositorInsertmStayGold
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
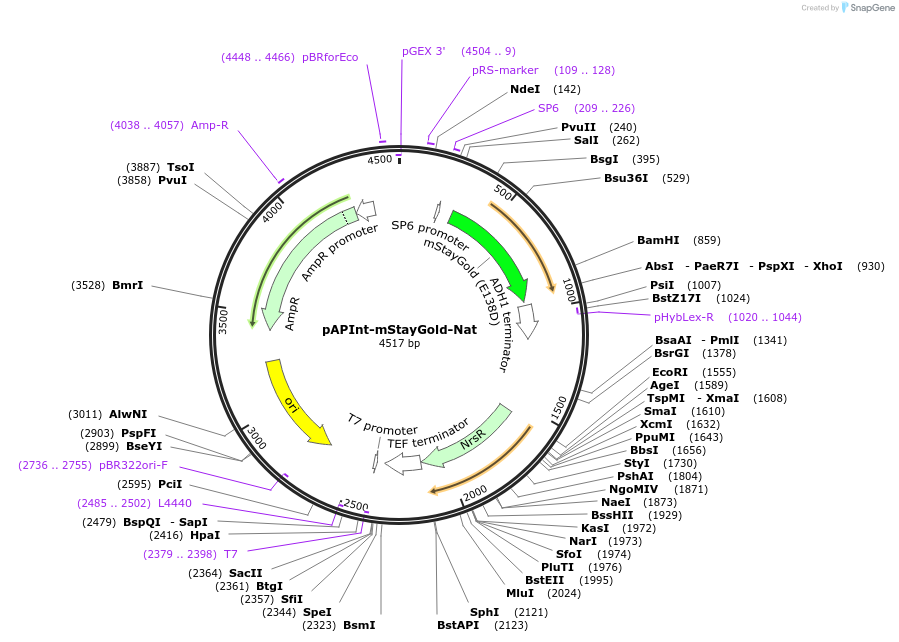
pAPInt-mStayGold-Nat
Plasmid#236485PurposePlasmid for deleting or C-terminally tagging endogenous genes with mStayGold and selection on Nat.DepositorInsertmStayGold
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
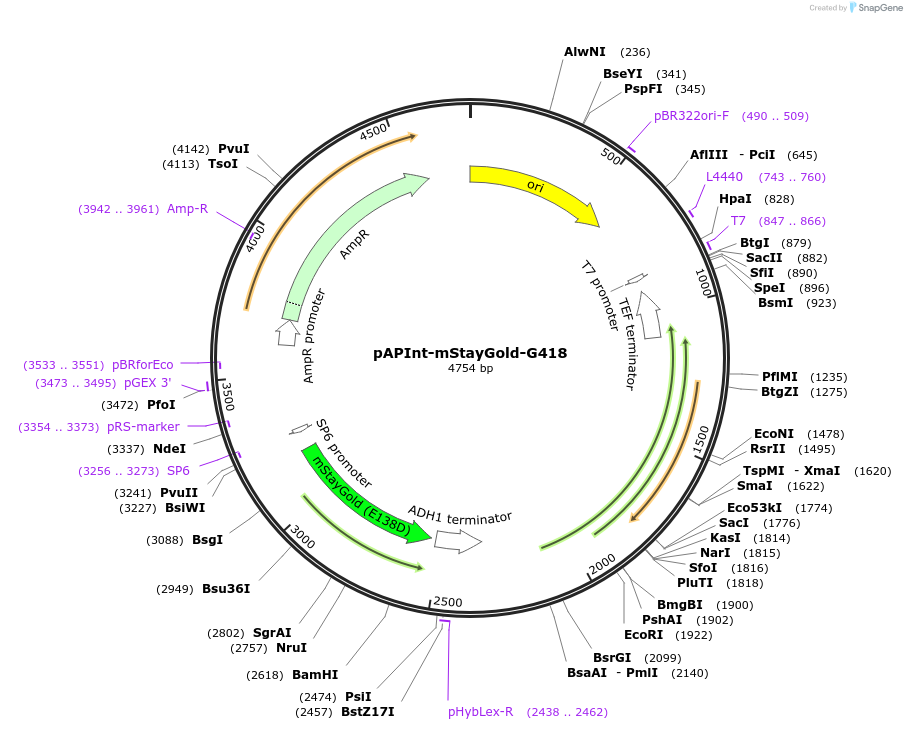
pAPInt-mStayGold-G418
Plasmid#236487PurposePlasmid for deleting or C-terminally tagging endogenous genes with mStayGold and selection on G418.DepositorInsertmStayGold
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
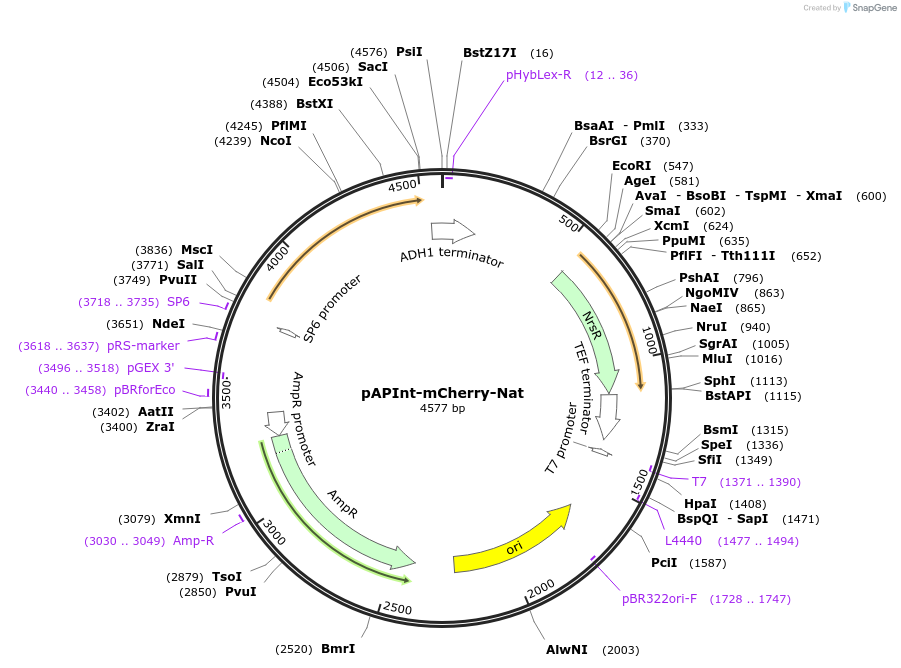
pAPInt-mCherry-Nat
Plasmid#236488PurposePlasmid for deleting or C-terminally tagging endogenous genes with mCherry and selection on Nat.DepositorInsertmCherry
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
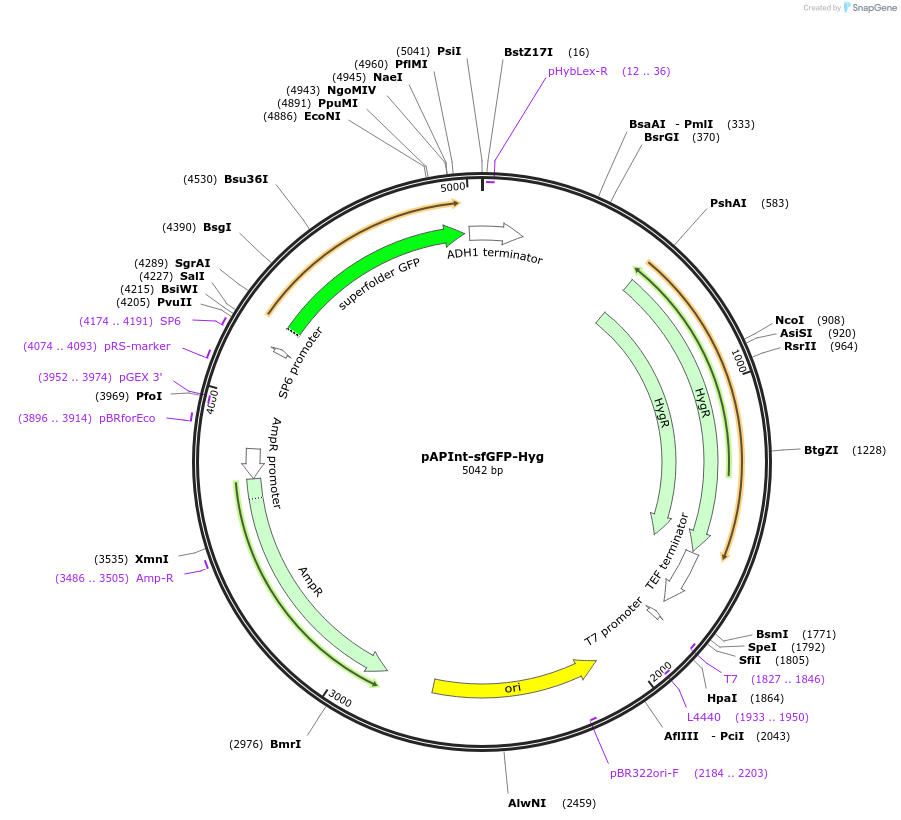
pAPInt-sfGFP-Hyg
Plasmid#236480PurposePlasmid for deleting or C-terminally tagging endogenous genes with sfGFP and selection on Hyg.DepositorInsertsfGFP
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
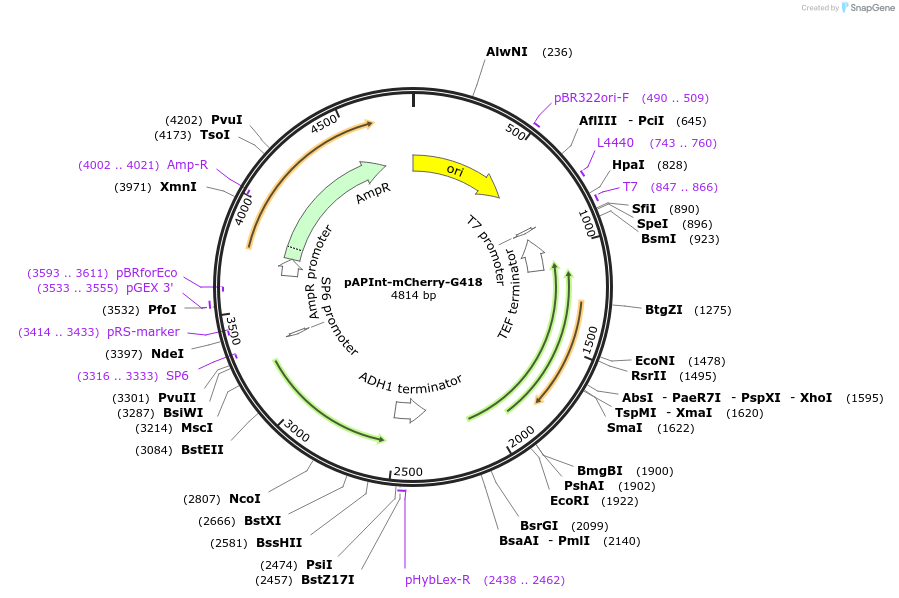
pAPInt-mCherry-G418
Plasmid#236490PurposePlasmid for deleting or C-terminally tagging endogenous genes with mCherry and selection on G418.DepositorInsertmCherry
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only