We narrowed to 5,531 results for: PEP
-
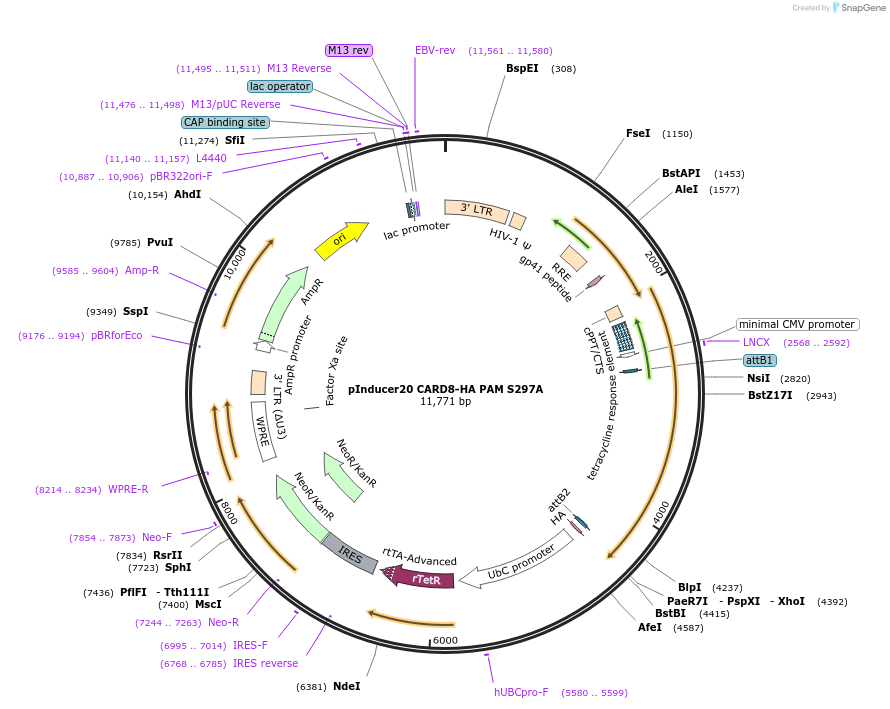
Plasmid#169983PurposeTet/Dox-inducible mammalian expression of CARD8 that is autoprocessing-deficient (S297A) with sgRNA binding site mutationsDepositorAvailable SinceMay 13, 2021AvailabilityAcademic Institutions and Nonprofits only
-
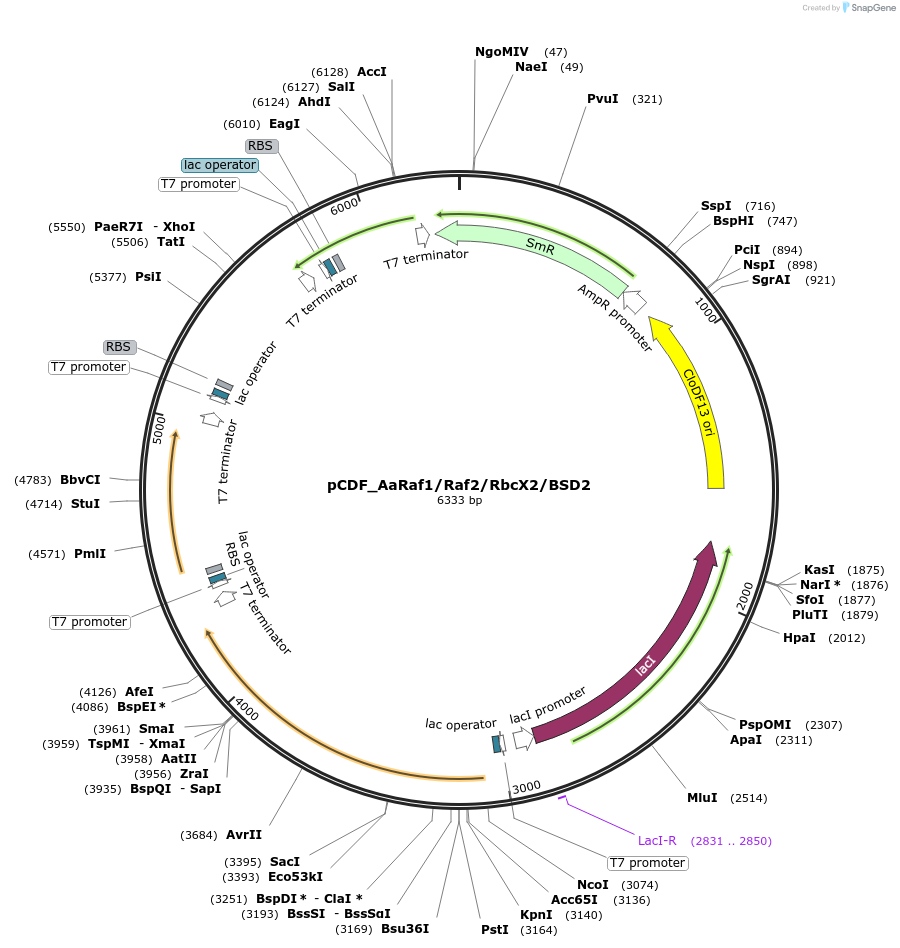
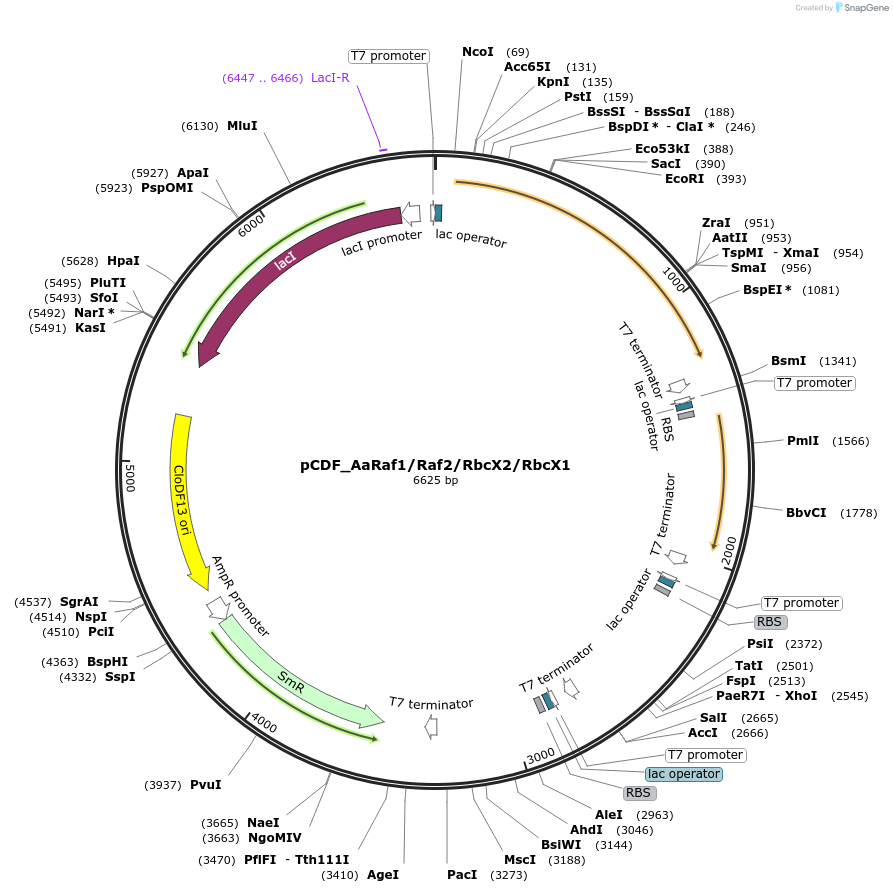
pCDF_AaRaf1/Raf2/RbcX2/BSD2
Plasmid#229507PurposeExpresses Anthoceros agrestis chaperones: Raf1,Raf2,RbcX2,BSD2DepositorInsertsRubisco accumulation factor 1
Rubisco accumulation factor 2
RbcX2
BSD2
ExpressionBacterialMutationN terminus chloroplast transit peptide truncationPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
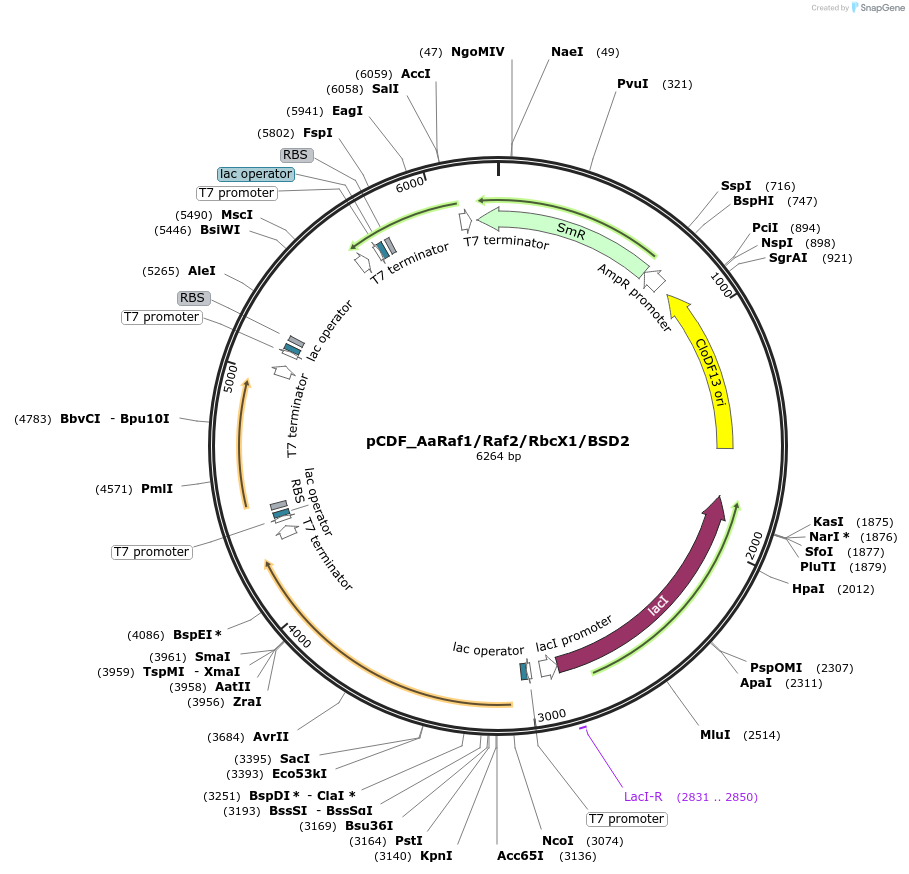
pCDF_AaRaf1/Raf2/RbcX1/BSD2
Plasmid#229508PurposeExpresses Anthoceros agrestis chaperones: Raf1,Raf2,RbcX1,BSD2DepositorInsertsRubisco accumulation factor 1
Rubisco accumulation factor 2
RbcX1
BSD2
ExpressionBacterialMutationN terminus chloroplast transit peptide truncationPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
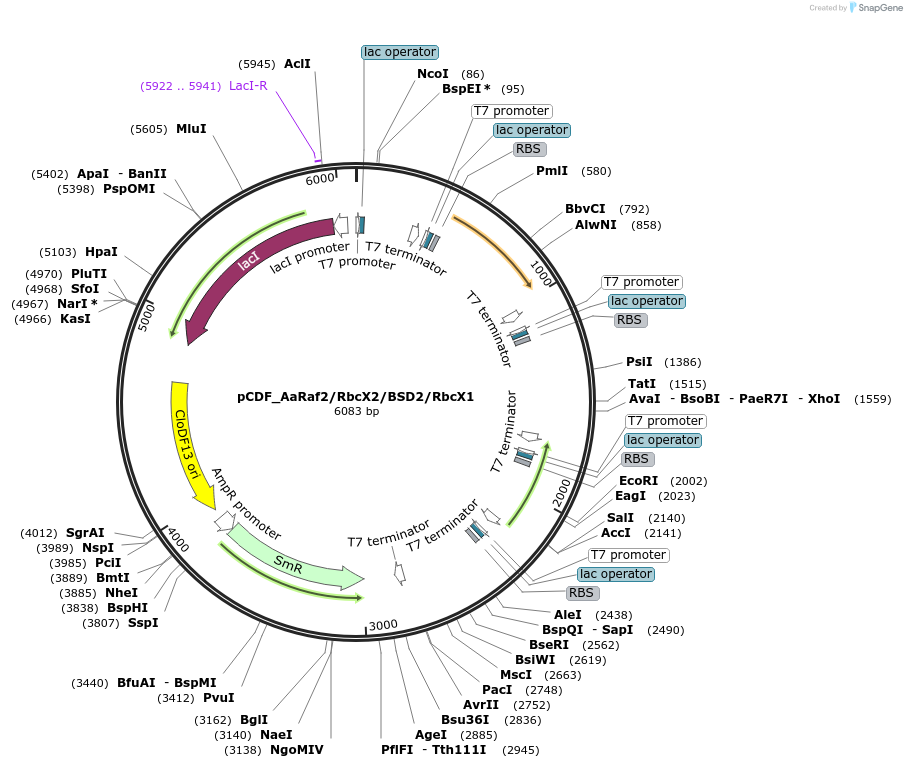
pCDF_AaRaf2/RbcX2/BSD2/RbcX1
Plasmid#229512PurposeExpresses Anthoceros agrestis chaperones: Raf2,RbcX2,BSD2,RbcX1DepositorInsertsRubisco accumulation factor 2
RbcX2
BSD2
RbcX1
ExpressionBacterialMutationN terminus chloroplast transit peptide truncationPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
pCDF_AaRaf1/Raf2/RbcX2/RbcX1
Plasmid#229514PurposeExpresses Anthoceros agrestis chaperones: Raf1,Raf2,RbcX2,RbcX1DepositorInsertsRubisco accumulation factor 1
Rubisco accumulation factor 2
RbcX2
RbcX1
ExpressionBacterialMutationN terminus chloroplast transit peptide truncationPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
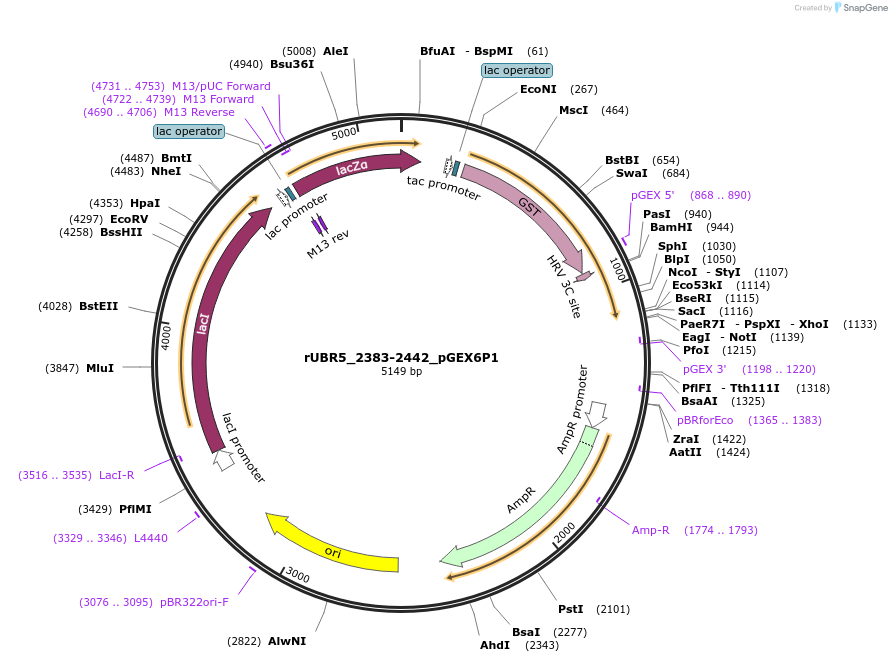
rUBR5_2383-2442_pGEX6P1
Plasmid#209791PurposeBacterial expression of UBR5 Mlle domainDepositorInsertMlle domain of UBR5 (Ubr5 Rat)
TagsGSTExpressionBacterialMutationaa 2383-2442 onlyPromotertacAvailable SinceNov. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
pInducer20 CARD8-HA PAM L368G
Plasmid#169985PurposeTet/Dox-inducible mammalian expression of CARD8 with a site III mutation (L368G) with sgRNA binding site mutationsDepositorAvailable SinceMay 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
pInducer20 CARD8-HA PAM R394E
Plasmid#169987PurposeTet/Dox-inducible mammalian expression of CARD8 with a site III mutation (R394E) with sgRNA binding site mutationsDepositorAvailable SinceMay 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
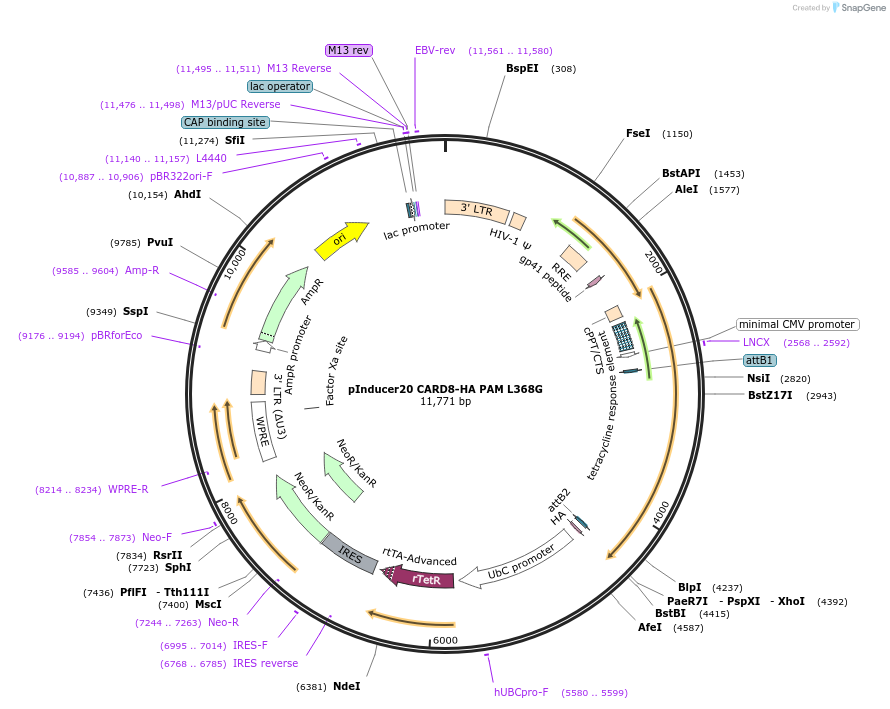
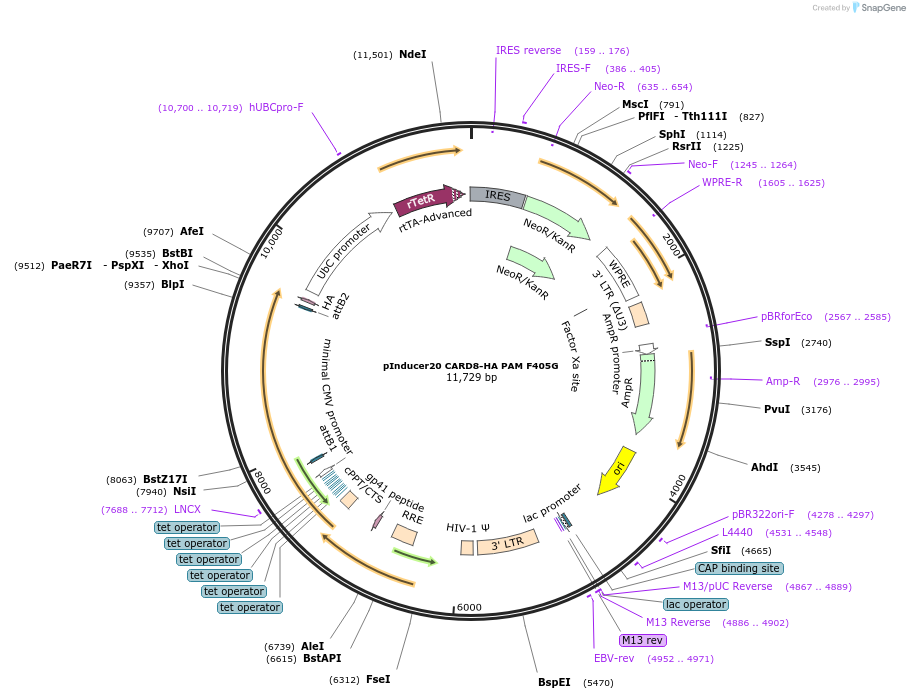
pInducer20 CARD8-HA PAM F405G
Plasmid#169988PurposeTet/Dox-inducible mammalian expression of CARD8 with a site III mutation (F405G) with sgRNA binding site mutationsDepositorAvailable SinceMay 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
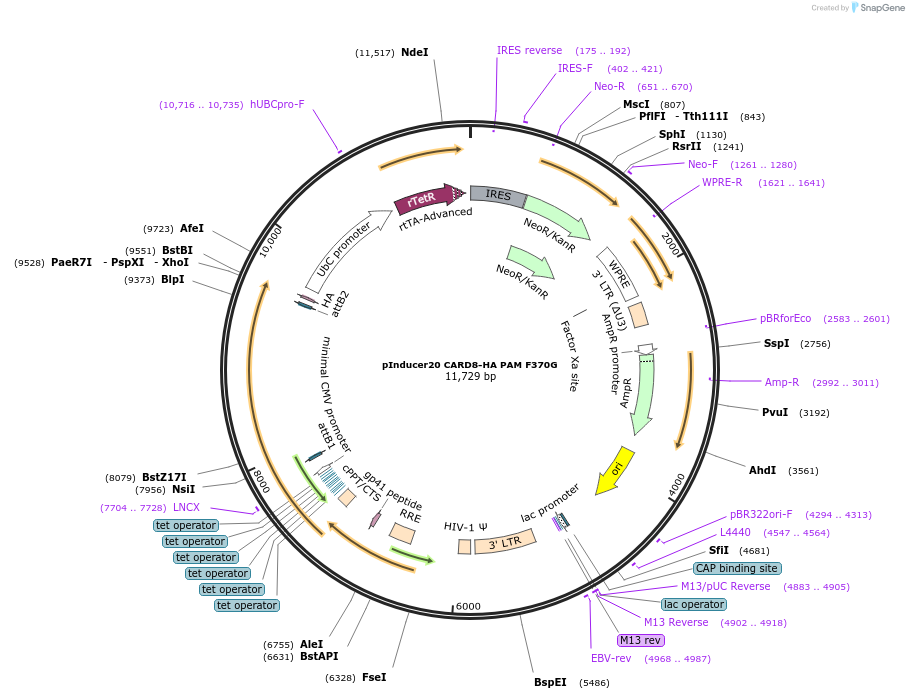
pInducer20 CARD8-HA PAM F370G
Plasmid#169986PurposeTet/Dox-inducible mammalian expression of CARD8 with a site III mutation (F370G) with sgRNA binding site mutationsDepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
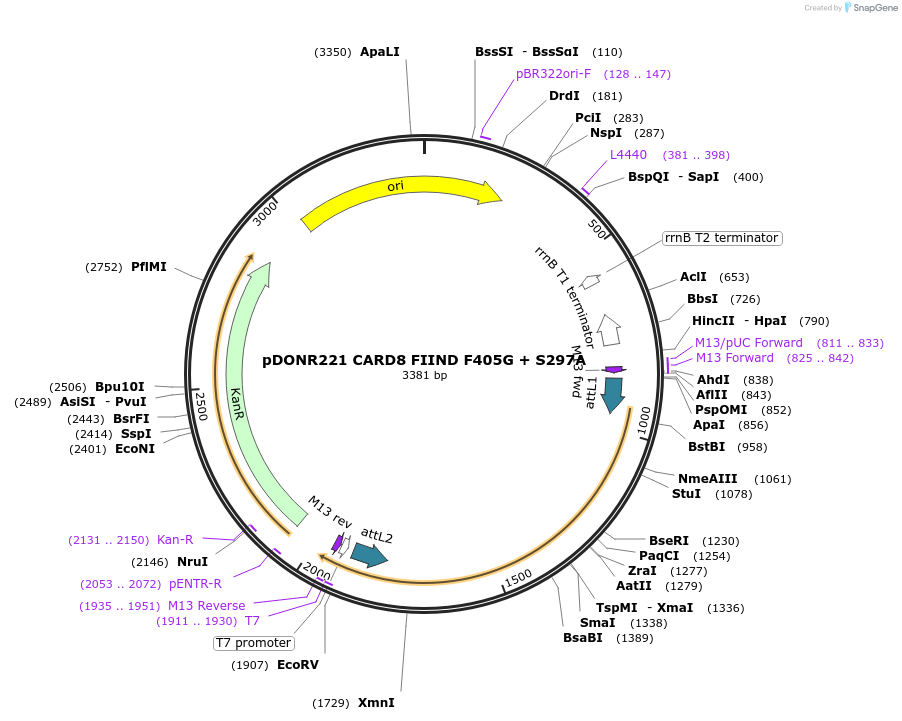
pDONR221 CARD8 FIIND F405G + S297A
Plasmid#169974PurposepDONR221 for Gateway shuttling. Contains the autoprocessing-deficient CARD8 FIIND (L162M-P434, no stop codon) with an interface III mutation (F405G)DepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
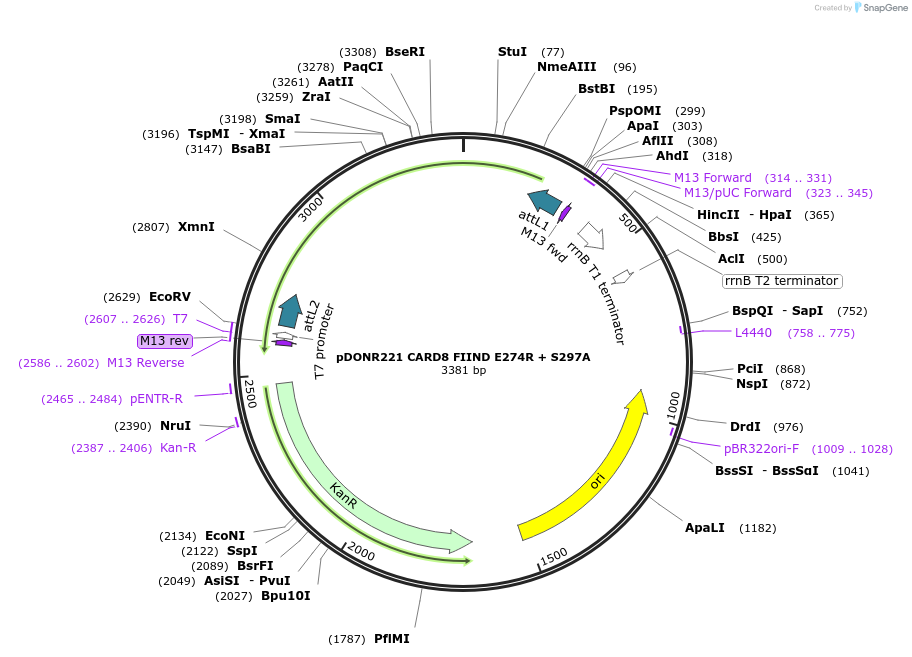
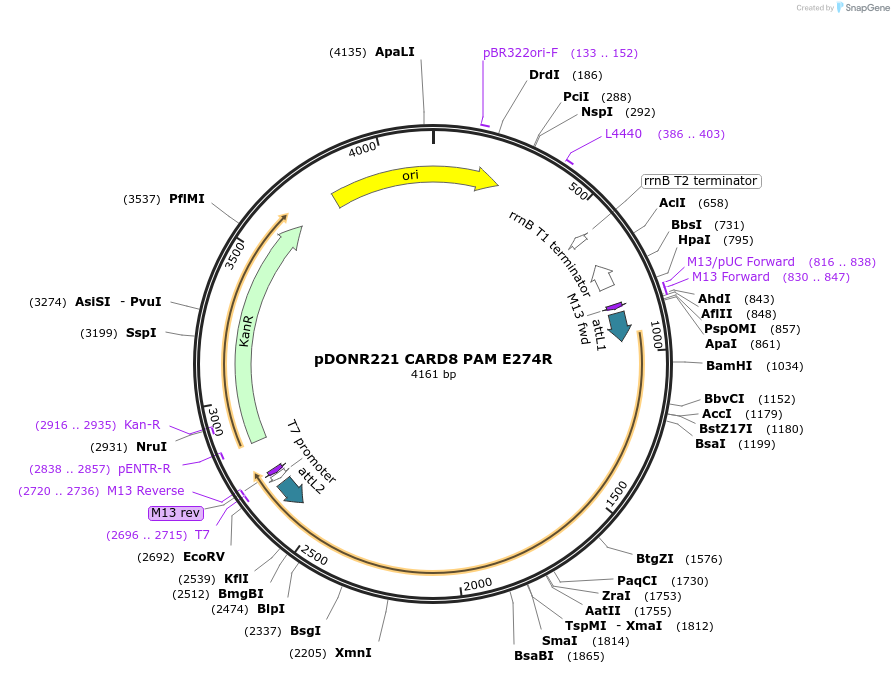
pDONR221 CARD8 FIIND E274R + S297A
Plasmid#169968PurposepDONR221 for Gateway shuttling. Contains the CARD8 autoprocessing-deficient FIIND (L162M-P434, no stop codon) that cannot bind DPP9 at site I (E274R)DepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
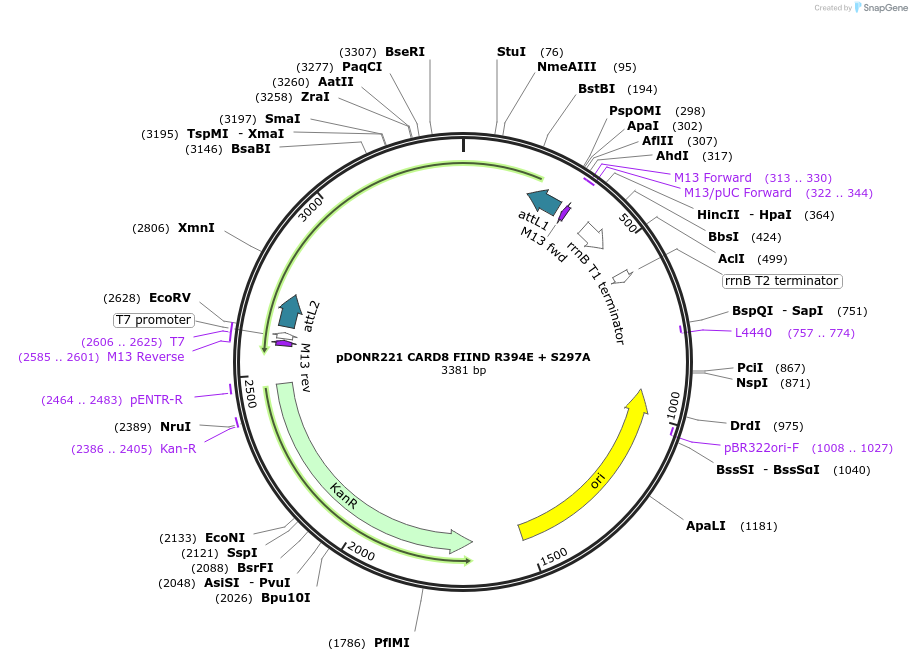
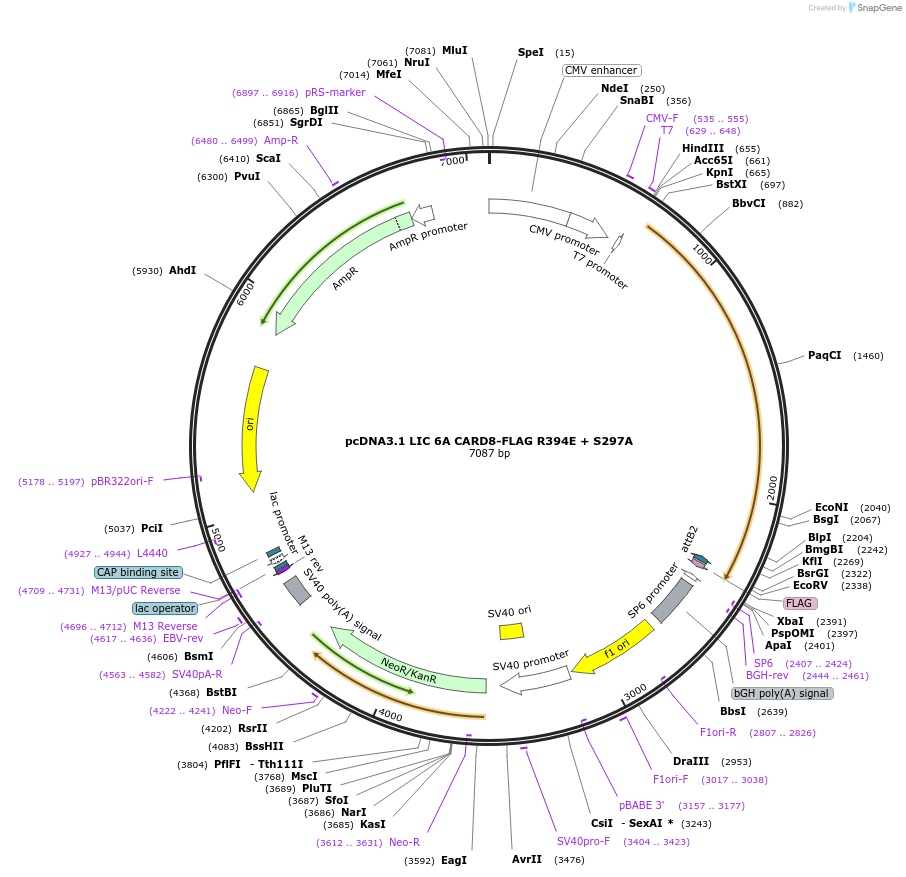
pDONR221 CARD8 FIIND R394E + S297A
Plasmid#169972PurposepDONR221 for Gateway shuttling. Contains the autoprocessing-deficient CARD8 FIIND (L162M-P434, no stop codon) with an interface III mutation (R394E)DepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
pDONR221 CARD8 PAM E274R
Plasmid#169977PurposepDONR221 for Gateway shuttling. Contains CARD8 that cannot bind DPP9 due to a mutation at site I (E274R) with sgRNA binding site mutationsDepositorAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 LIC 6A CARD8-FLAG R394E + S297A
Plasmid#169955PurposeMammalian expression of CARD8-FLAG with a site III mutation (R394E) with an additional autoprocessing mutation (S297A)DepositorAvailable SinceMay 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
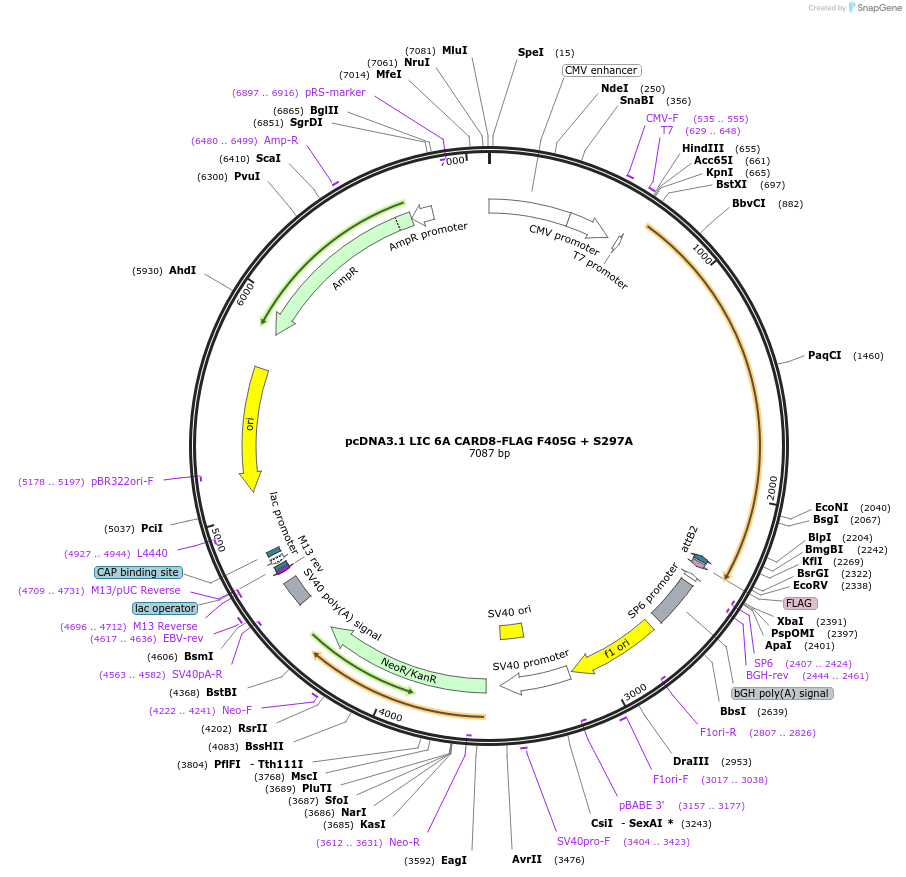
pcDNA3.1 LIC 6A CARD8-FLAG F405G + S297A
Plasmid#169957PurposeMammalian expression of CARD8-FLAG with a site III mutation (F405G) with an additional autoprocessing mutation (S297A)DepositorAvailable SinceMay 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
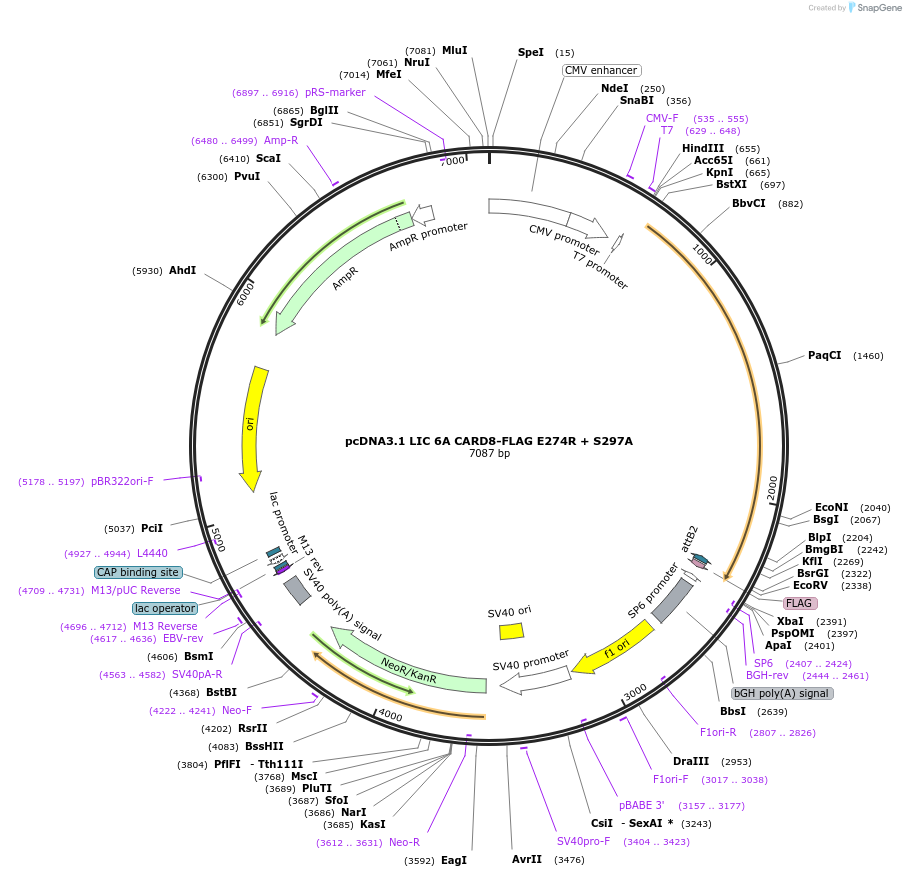
pcDNA3.1 LIC 6A CARD8-FLAG E274R + S297A
Plasmid#169951PurposeMammalian expression of CARD8-FLAG that cannot bind DPP9 due to a mutation at site I (E274R) with an additional autoprocessing mutation (S297A)DepositorAvailable SinceMay 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
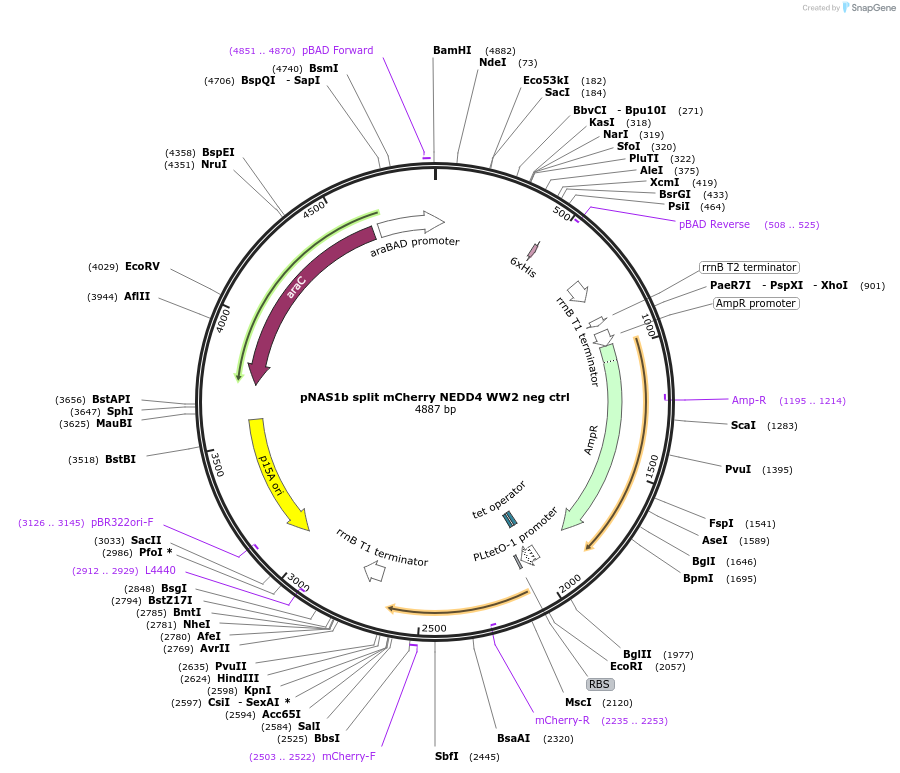
pNAS1b split mCherry NEDD4 WW2 neg ctrl
Plasmid#112030PurposeMode #2 phosphosite negative control (split mCherry, using NEDD4 WW2 binding domain paired with negative control peptide)DepositorInsertNEDD4 WW2 domain fused to C-terminal split mCherry, neg ctrl interacting peptide fused to N-terminal split mCherry
TagsSplit mCherryExpressionBacterialAvailable SinceAug. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
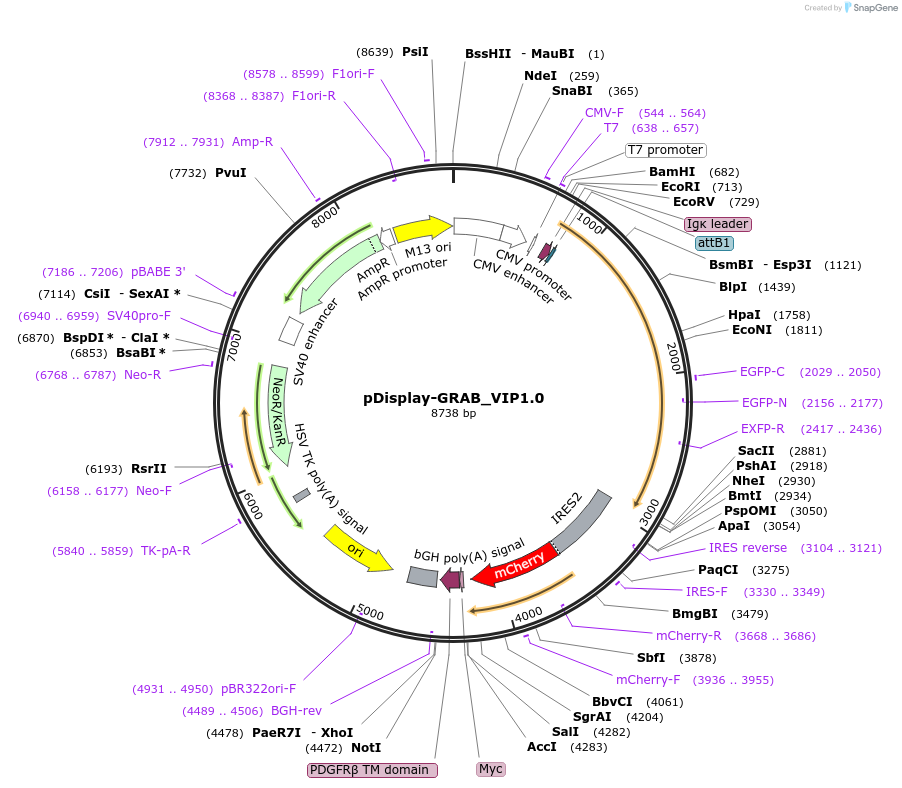
pDisplay-GRAB_VIP1.0
Plasmid#208682PurposeExpresses the genetically-encoded fluorescent vasoactive intestinal peptide (VIP) sensor GRAB_VIP1.0 in mammalian cellsDepositorInsertGPCR activation based vasoactive intestinal peptide (VIP) sensor GRAB_VIP1.0
ExpressionMammalianPromoterCMVAvailable SinceJan. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
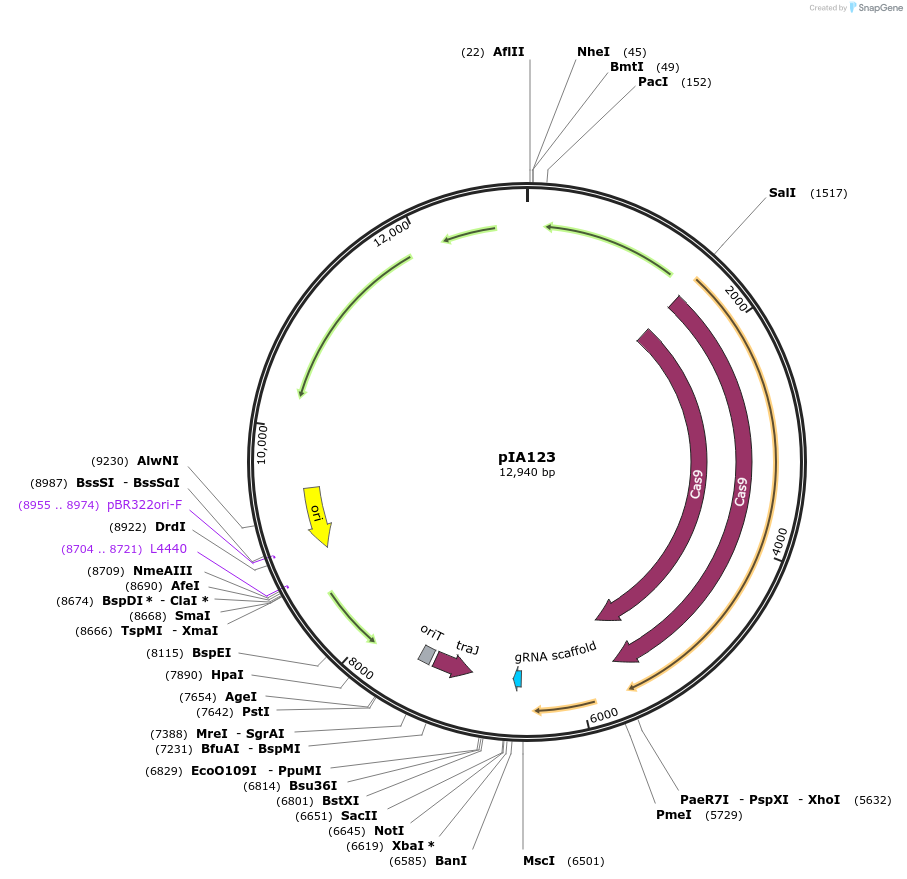
pIA123
Plasmid#234038PurposeE. coli-C. difficile shuttle vector for CRISPR editing in C. difficile. Pxyl::Cas9-opt Pgdh::sgRNA-cdr_0985. PstI site to add DNA for homology directed repair. MscI and NotI for replacement of sgRNA.DepositorInsertsCas9
sgRNA
UseCRISPRExpressionBacterialPromoterPgdh and PxylAvailable SinceMarch 19, 2025AvailabilityAcademic Institutions and Nonprofits only