We narrowed to 9,316 results for: sacs
-
Plasmid#133656PurposeIntegration of PEX3-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast peroxisome.DepositorAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only
-
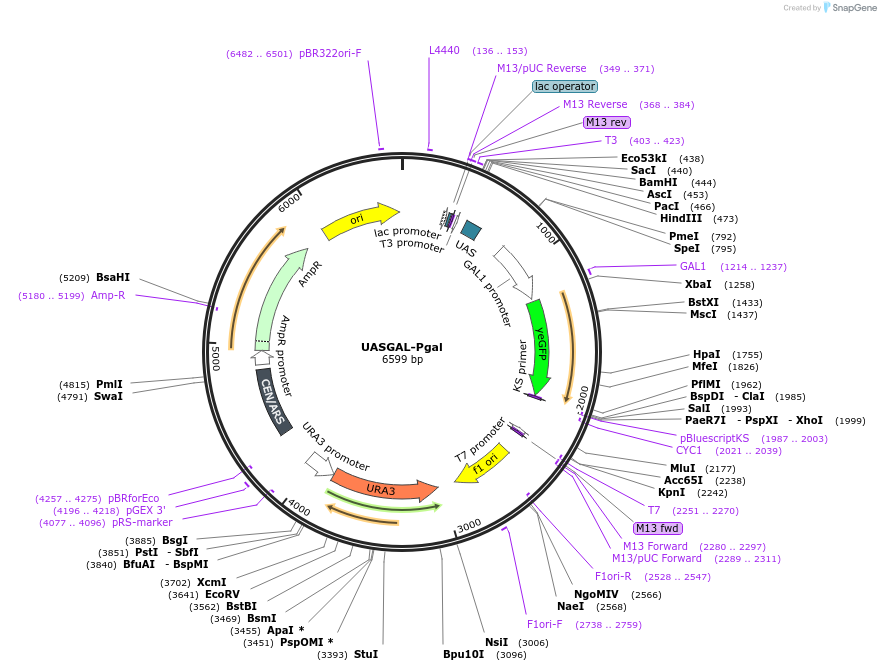
UASGAL-Pgal
Plasmid#60338PurposeContains a promoter of activating sequence derived from native galactose promoter coupled with full length native galactose promoterDepositorInsertpromoter
UseSynthetic Biology; Yeast synthetic hybrid promote…ExpressionYeastAvailable SinceNov. 6, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
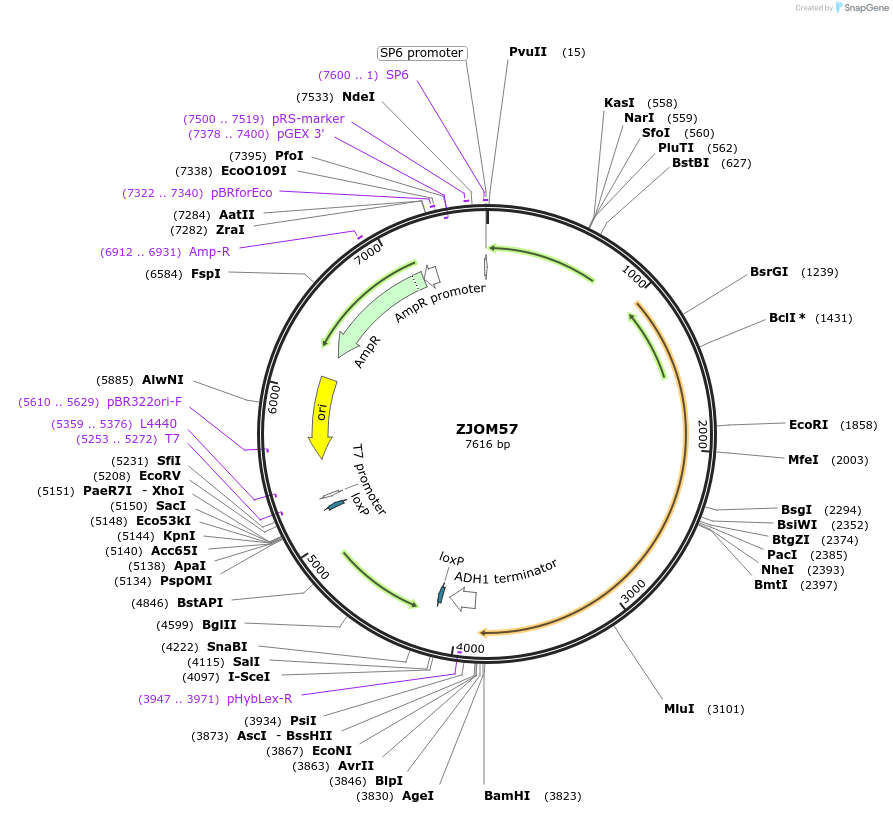
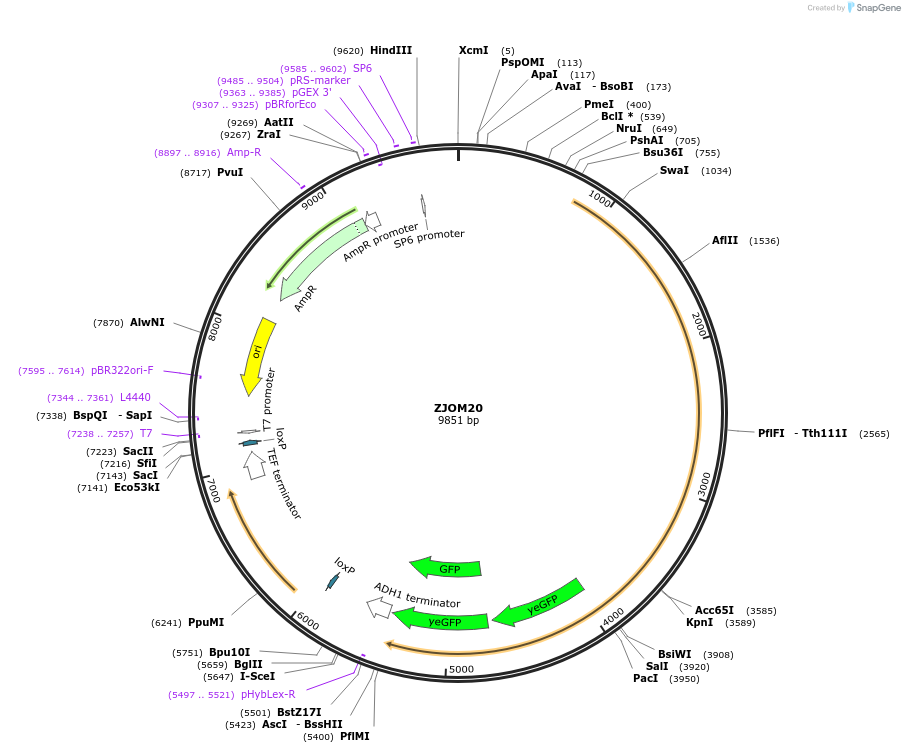
ZJOM20
Plasmid#133644PurposeIntegration of PEX1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast peroxisome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
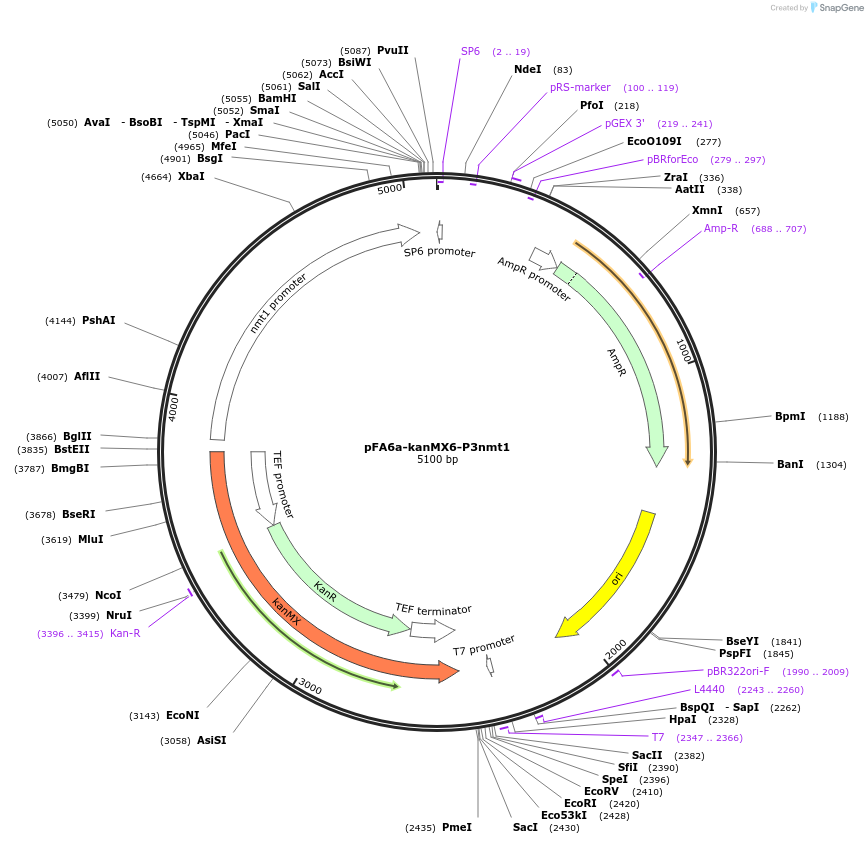
pFA6a-kanMX6-P3nmt1
Plasmid#39280DepositorInsertnmt1 promoter (nmt1 Fission Yeast)
UseYeast genomic targetingTagsA. gossypii translation elongation factor 1a gene…Mutation-1163 to +6 of nmt1 locusAvailable SinceAug. 14, 2012AvailabilityAcademic Institutions and Nonprofits only -
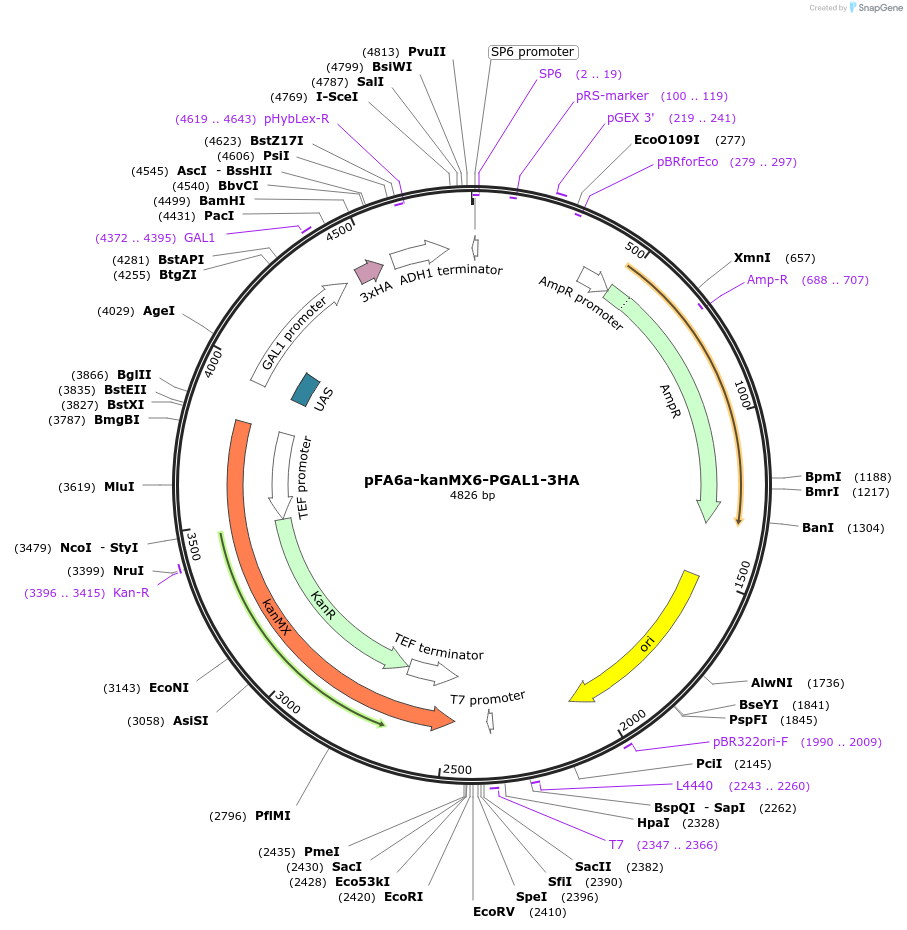
pFA6a-kanMX6-PGAL1-3HA
Plasmid#41608DepositorInsertGAL1 promoter (GAL1 Budding Yeast)
UseYeast genomic targetingTags3xHA, A. gossypii translation elongation factor 1…Available SinceDec. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
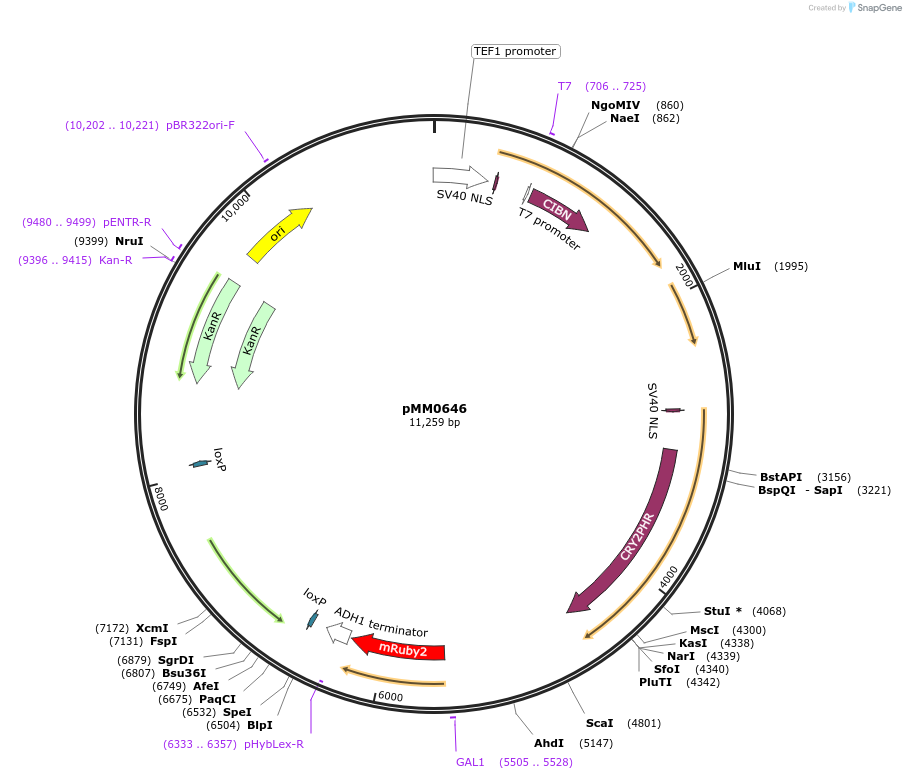
pMM0646
Plasmid#129007PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2 with loxable KIURA3 markerDepositorInsertKanR-ColE1 URA 5'-pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZCRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-loxP-KIURA3-loxP-URA3 3'
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
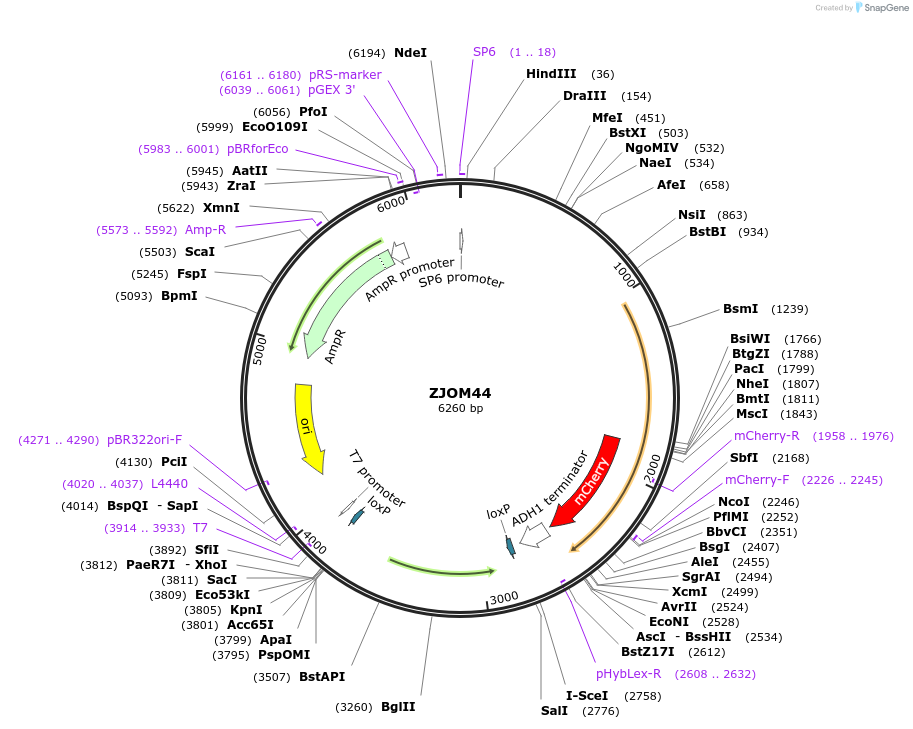
ZJOM44
Plasmid#133653PurposeIntegration of SNF7-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
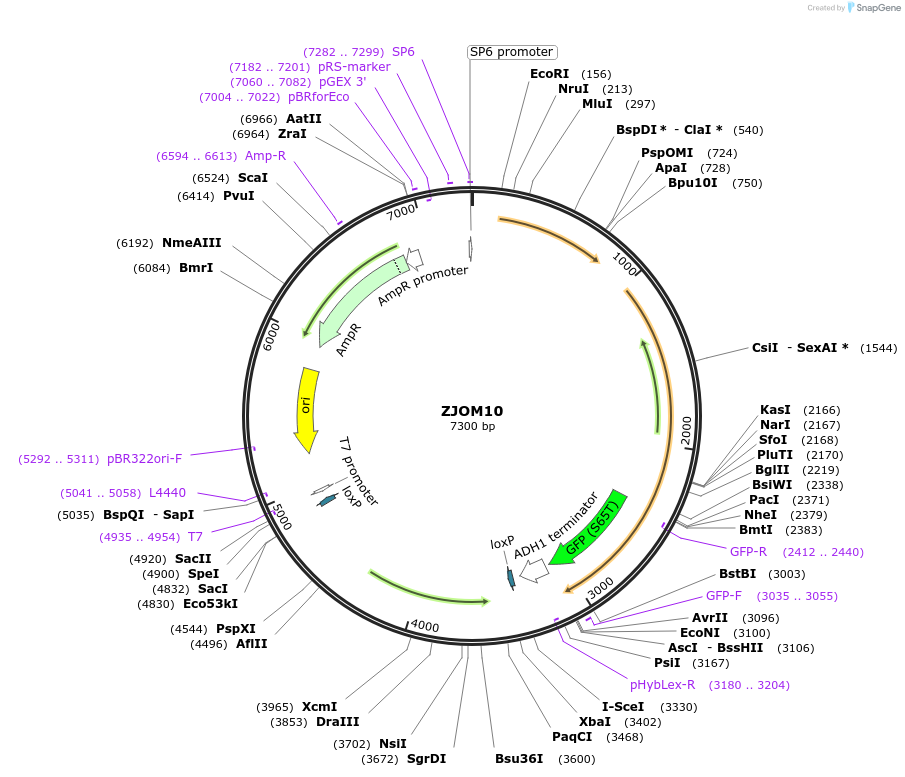
ZJOM10
Plasmid#133639PurposeIntegration of VPS4-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM5
Plasmid#133635PurposeIntegration of CHS5-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
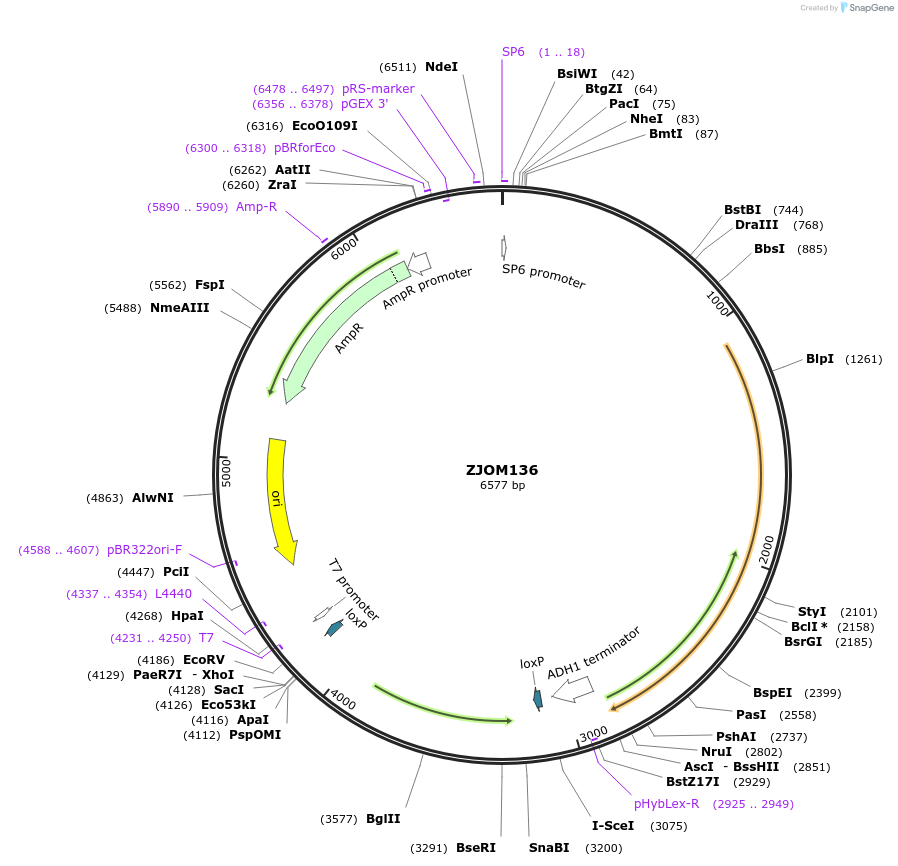
ZJOM136
Plasmid#133660PurposeIntegration of ELO3-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
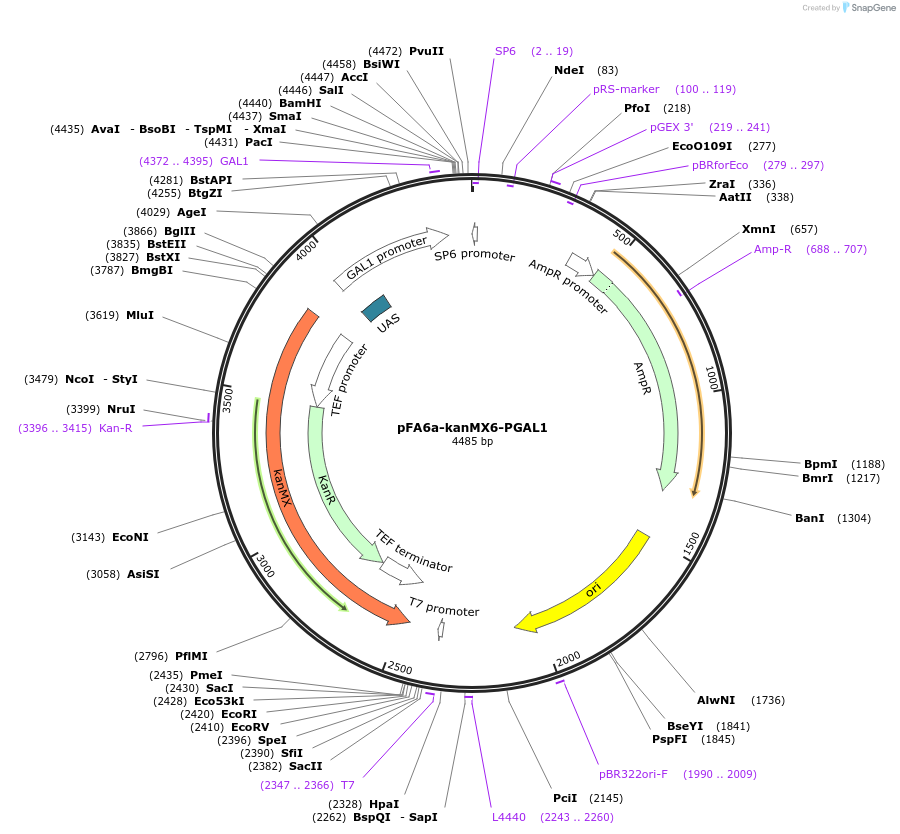
pFA6a-kanMX6-PGAL1
Plasmid#41605DepositorInsertGAL1 promoter (GAL1 Budding Yeast)
UseYeast genomic targetingTagsA. gossypii translation elongation factor 1a gene…Available SinceDec. 10, 2012AvailabilityAcademic Institutions and Nonprofits only -
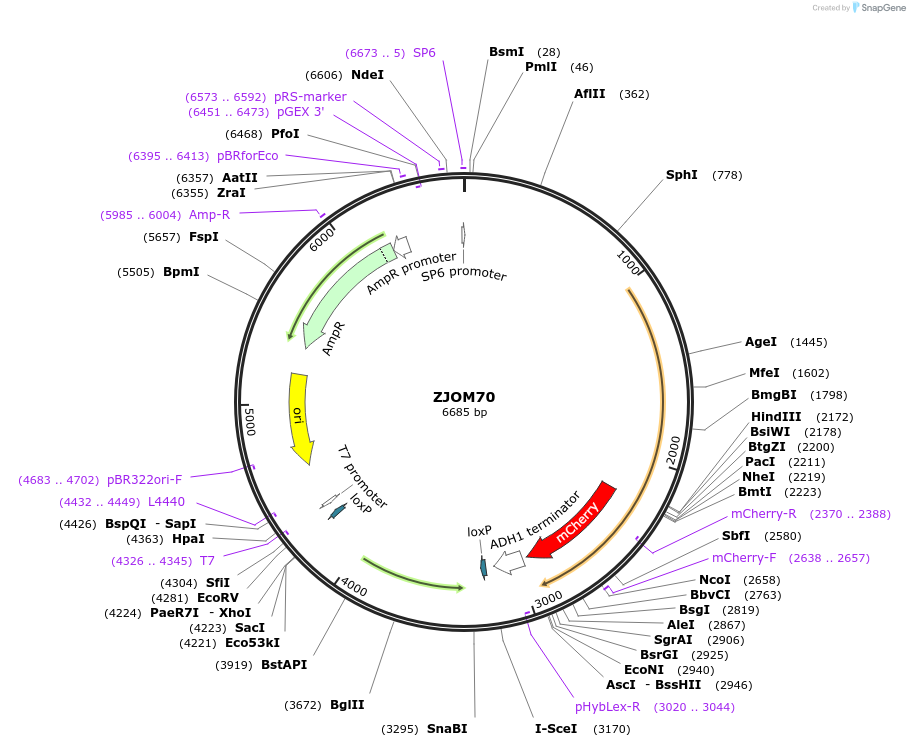
ZJOM70
Plasmid#133658PurposeIntegration of ERG6-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
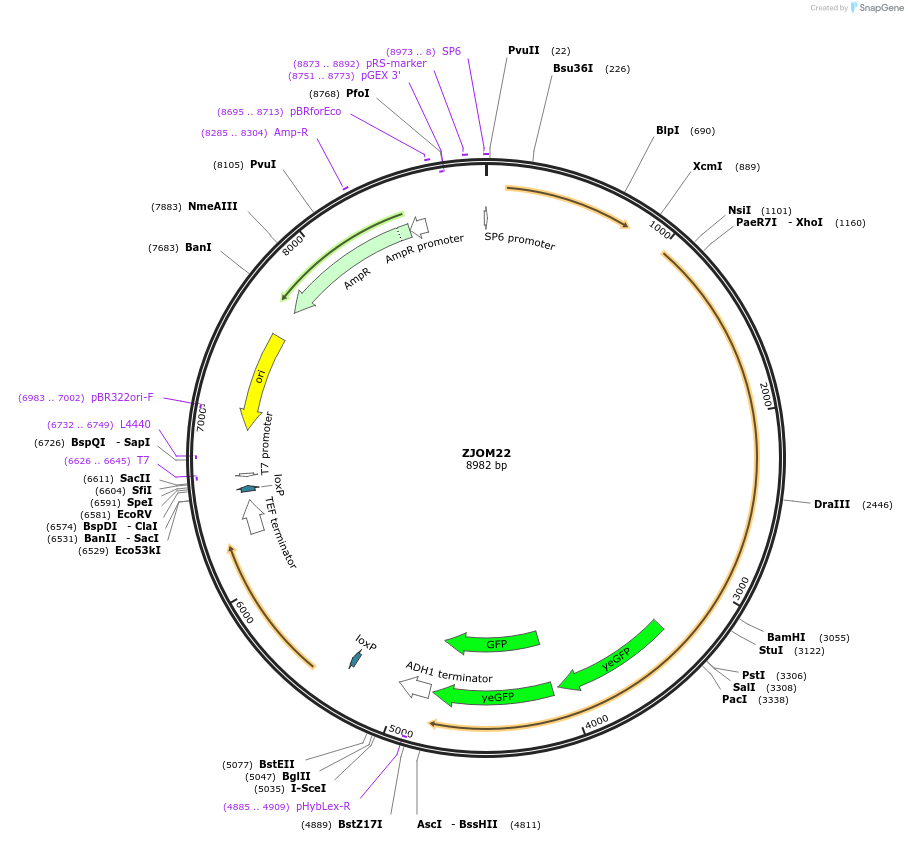
ZJOM22
Plasmid#133629PurposeIntegration of EMC1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
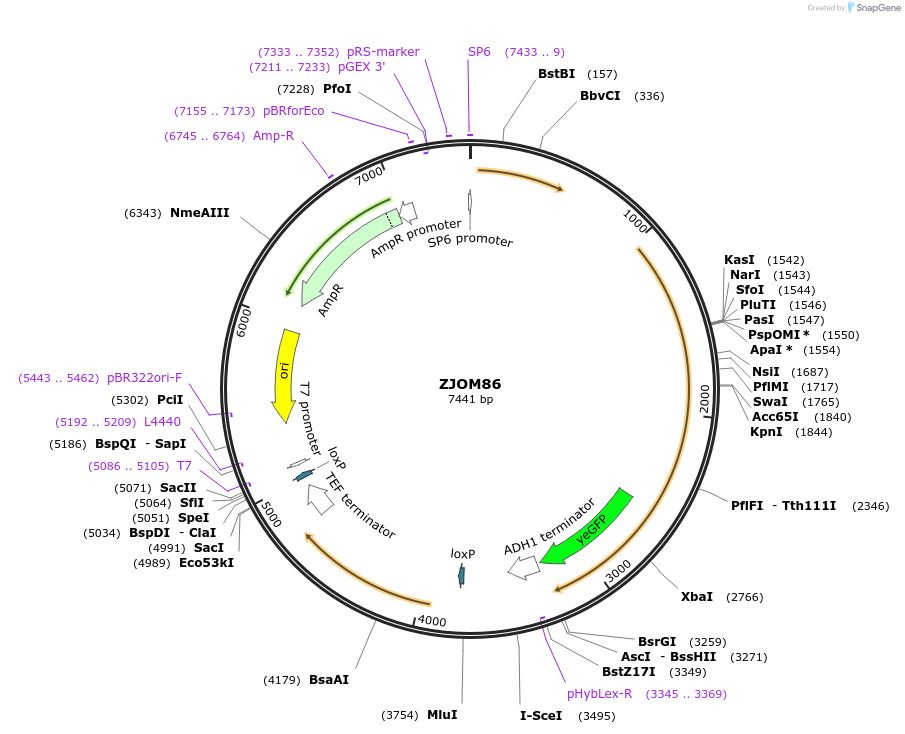
ZJOM86
Plasmid#133634PurposeIntegration of ANP1-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast early golgi.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
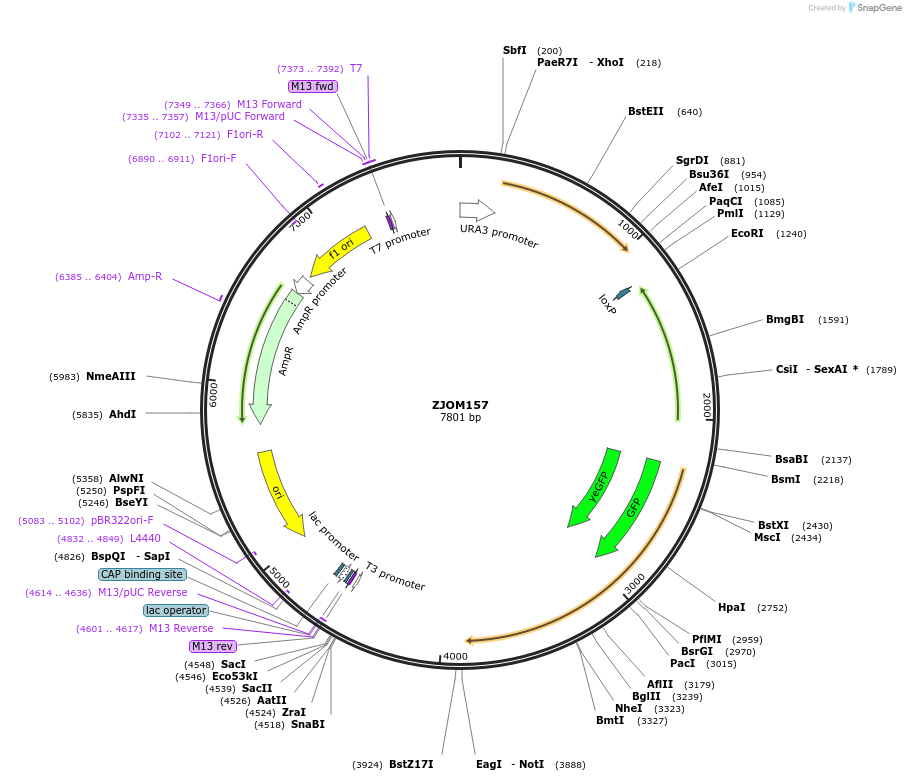
ZJOM157
Plasmid#133640PurposeIntegration of GFP-PEP12 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis).Marker for yeast late endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
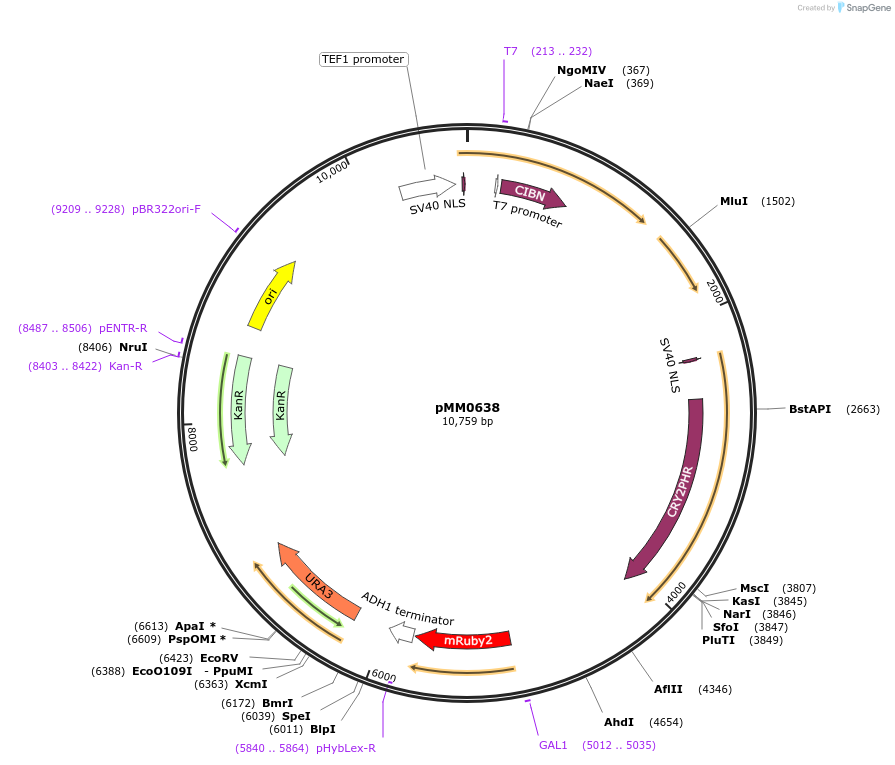
pMM0638
Plasmid#129005PurposeIntegration vector for optogenetic machinery (pSTRONG-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2DepositorInsert5' URA3 -pTEF1-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-URA3 URA3 3'- KanR-ColE1
ExpressionYeastAvailable SinceOct. 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
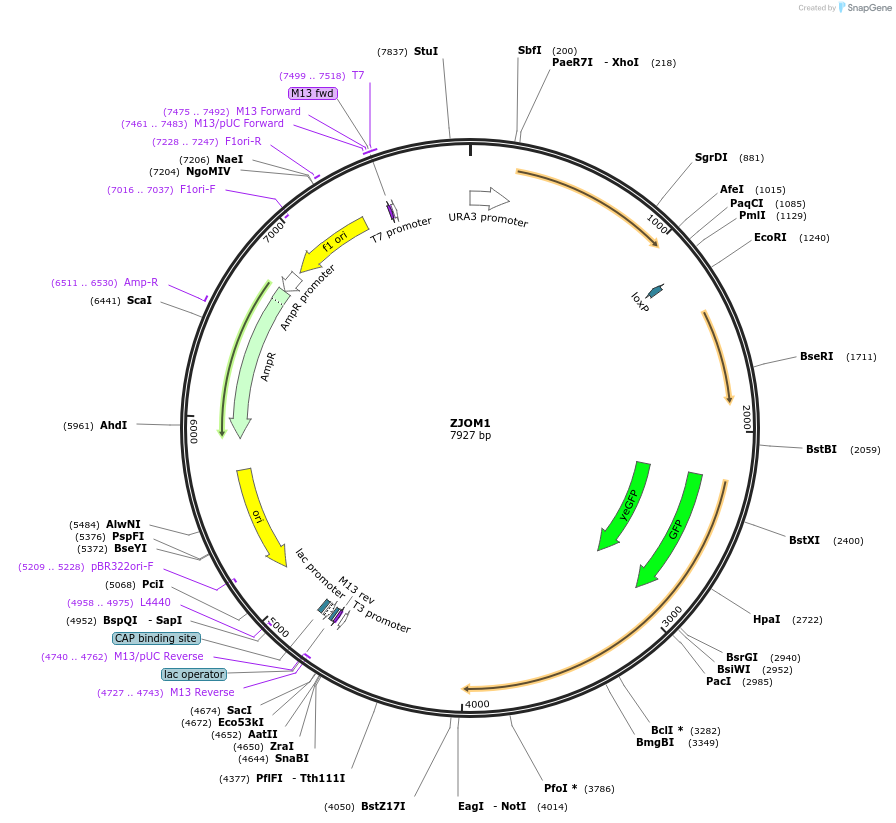
ZJOM1
Plasmid#133632PurposeIntegration of GFP-SED5 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
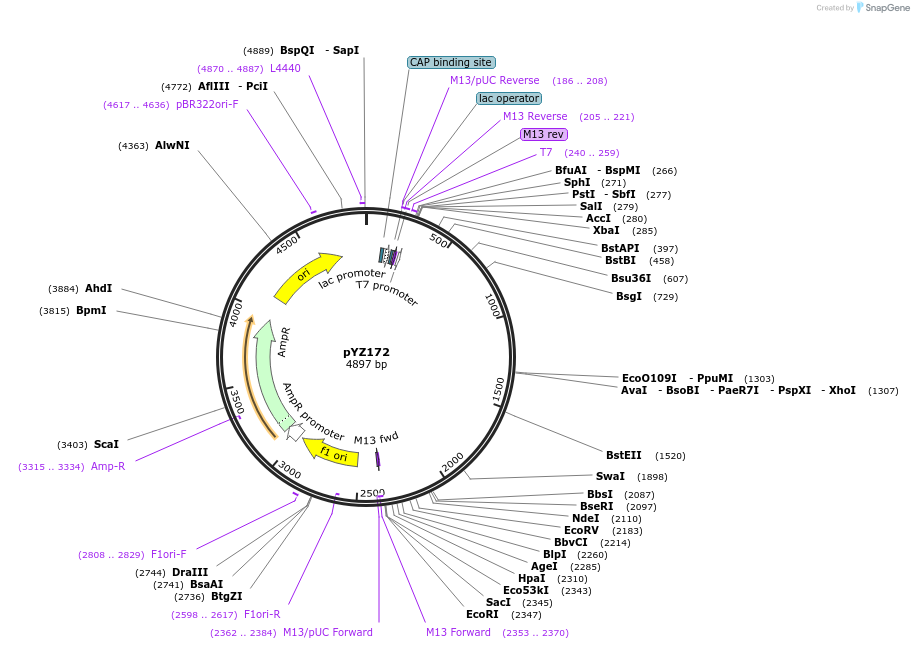
pYZ172
Plasmid#98409PurposeDonor DNA plasmid to introduce lys9 deletion in S. pombe. Linearization with Not1 digestion.DepositorInsertsupstream homologous region to delete lys9 in genome by recombination
downstream homologous region to delete lys9 in genome by recombination
UseCRISPR and Synthetic BiologyAvailable SinceFeb. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
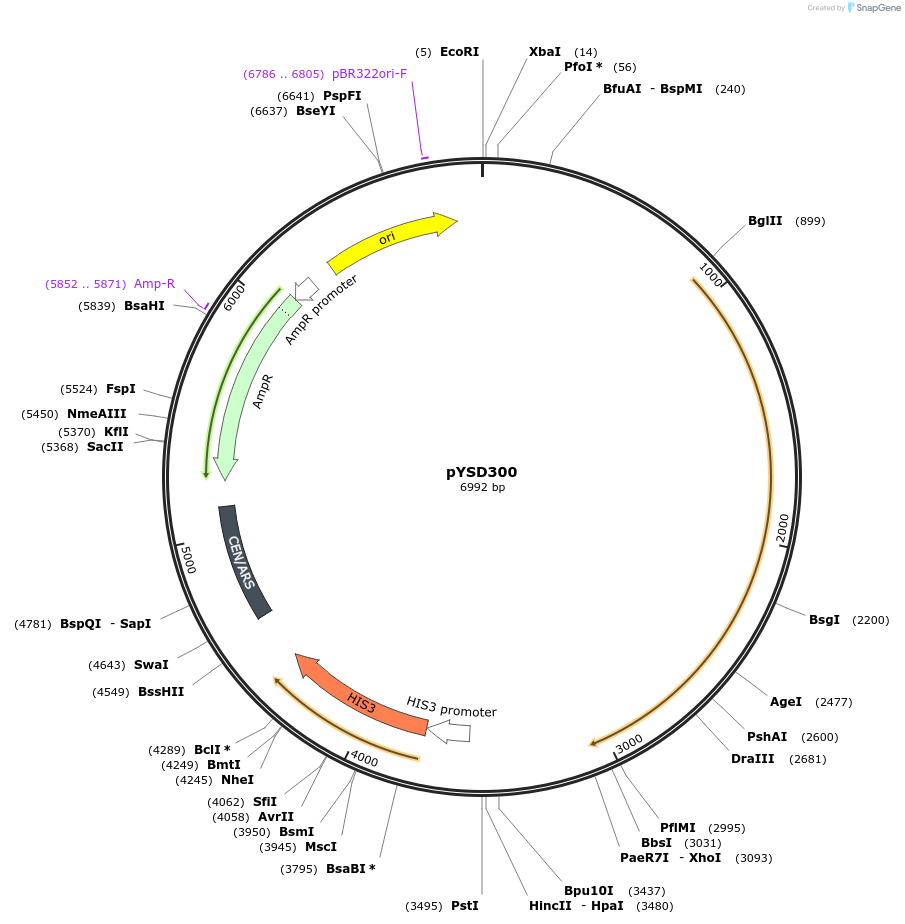
pYSD300
Plasmid#212930PurposeEncodes the S. cerevisiae Aga1 protein (Part3) with pGBK1 promoter, tENO2 terminator, HIS3 selectable marker and CEN/ARS yeast origin.DepositorInsertAGA1 (AGA1 Budding Yeast)
ExpressionYeastAvailable SinceFeb. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
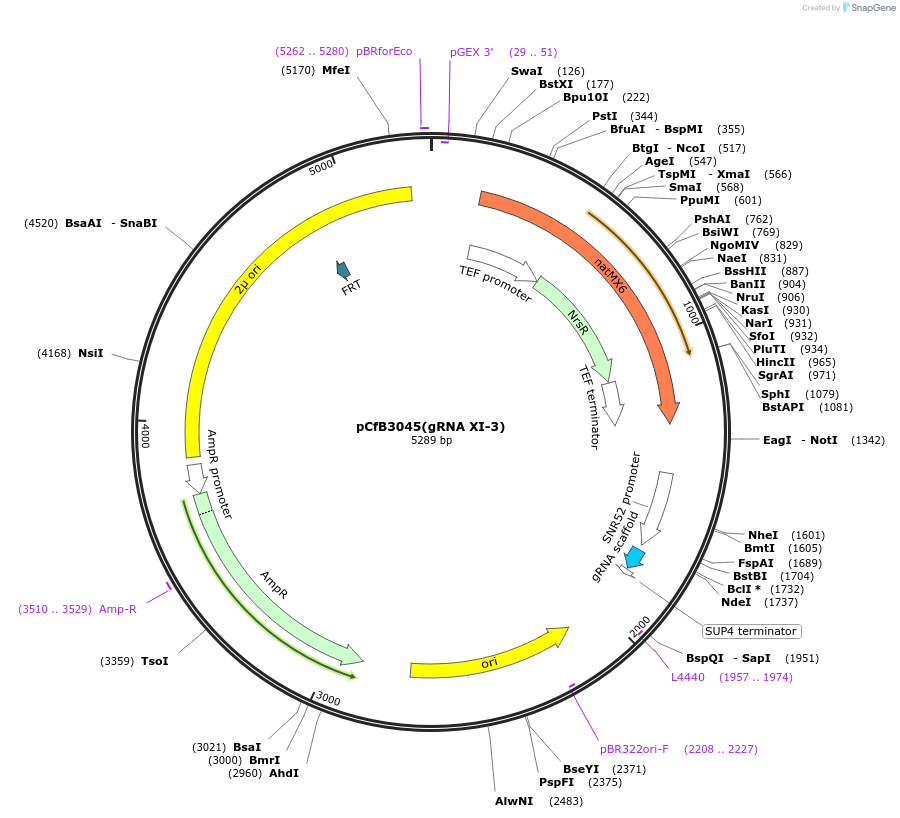
pCfB3045(gRNA XI-3)
Plasmid#73287PurposeEasyClone-MarkerFree guiding RNA vector to direct Cas9 to cut at site XI-3DepositorInsertguiding RNA
UseCRISPR; GrnaExpressionYeastAvailable SinceNov. 23, 2016AvailabilityAcademic Institutions and Nonprofits only