We narrowed to 5,675 results for: KRAS
-
Plasmid#240111PurposeA lentiviral (blue) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by synthtic JeT promoter.DepositorInsertmurine Nr1d1 promoter+Electra2-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded JeT PromoterAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
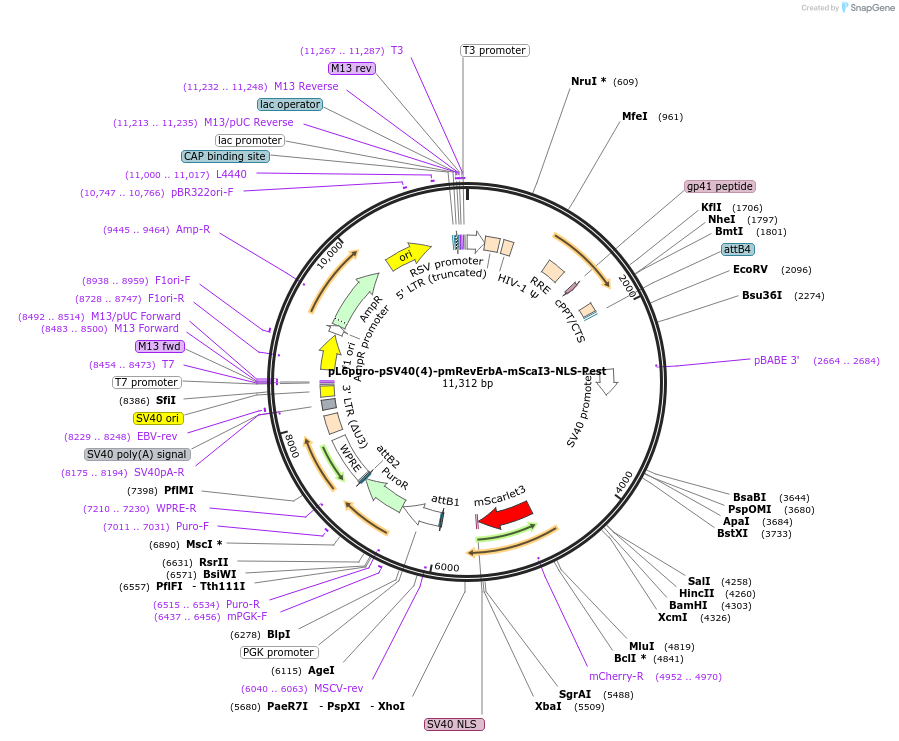
pL6puro-pSV40(4)-pmRevErbA-mScaI3-NLS-Pest
Plasmid#240118PurposeA lentiviral (red) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter.DepositorInsertmurine Nr1d1 promoter+mScarlet-I3-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-Enhancer+OriAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
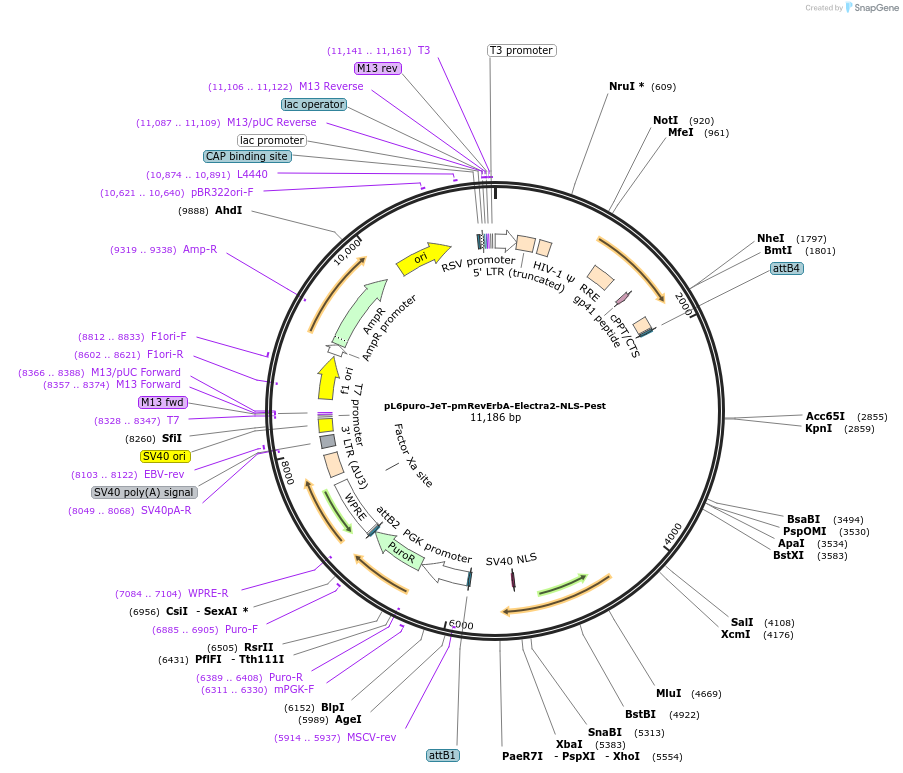
pL6puro-JeT-pmRevErbA-Venus-NLS-Pest
Plasmid#240106PurposeA lentiviral (yellow) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by synthtic JeT promoter.DepositorInsertmurine Nr1d1 promoter+Venus-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded JeT PromoterAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
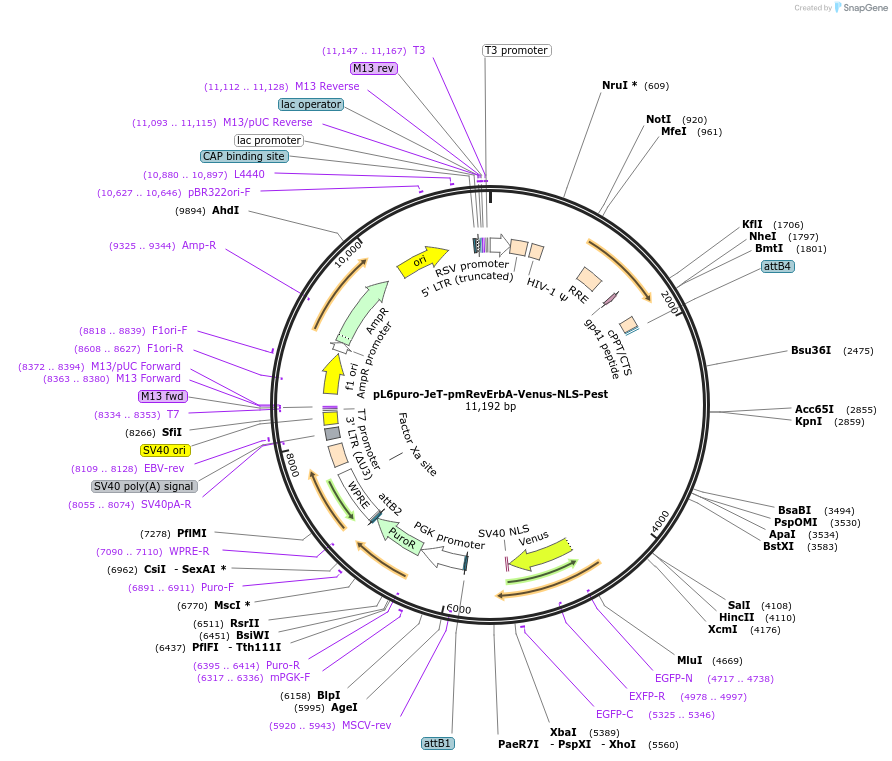
pL6puro-pSV40(1)-pmRevErbA-Venus-NLS-Pest
Plasmid#240112PurposeA lentiviral (yellow) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter fragment.DepositorInsertmurine Nr1d1 promoter+Venus-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-EnhancerAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
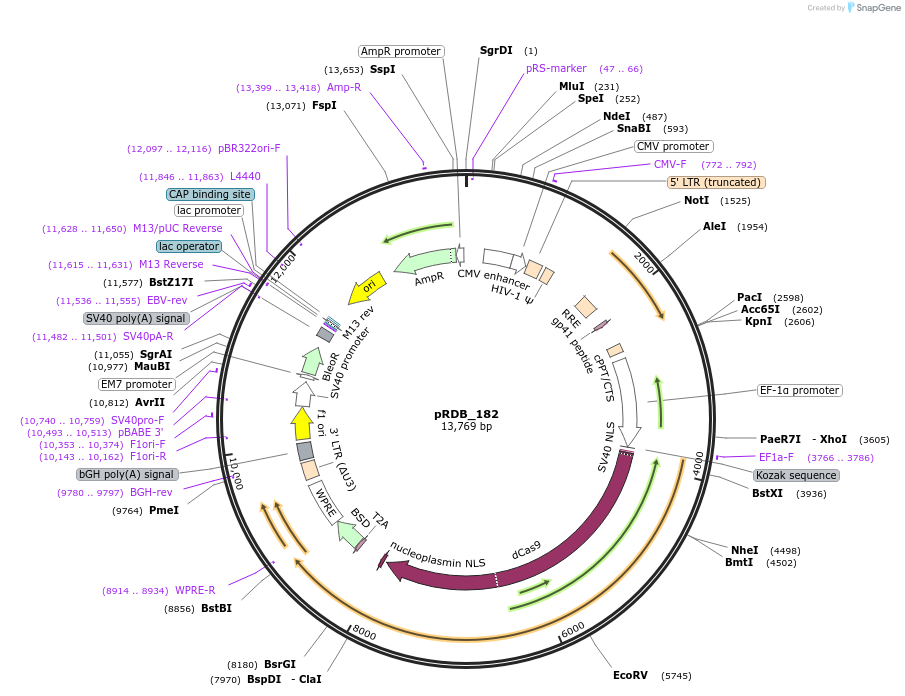
pRDB_182
Plasmid#216103PurposeCRISPRi, EF1a-driven dCas9-KRAB (ZIM3) (Cas only)DepositorInsertCas9 [Sp]
UseCRISPR and Lentiviral; Assembled vectorAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
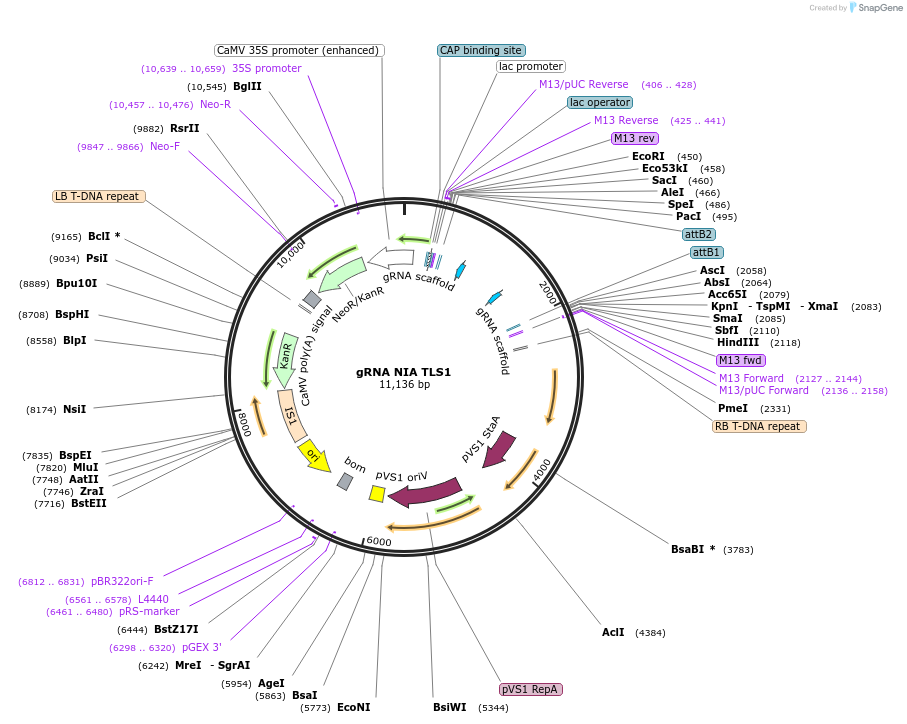
gRNA NIA TLS1
Plasmid#196980PurposeExpresses two gRNAs targeting Arabidopsis NIA1 gene for knockout. Each gRNA is fused to TLS1 mobility signal that confers graft mobility.DepositorInsertAtNIA1 gRNAs fused to TLS1 tags
ExpressionPlantPromoterpU6-26, pU6-29Available SinceMay 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
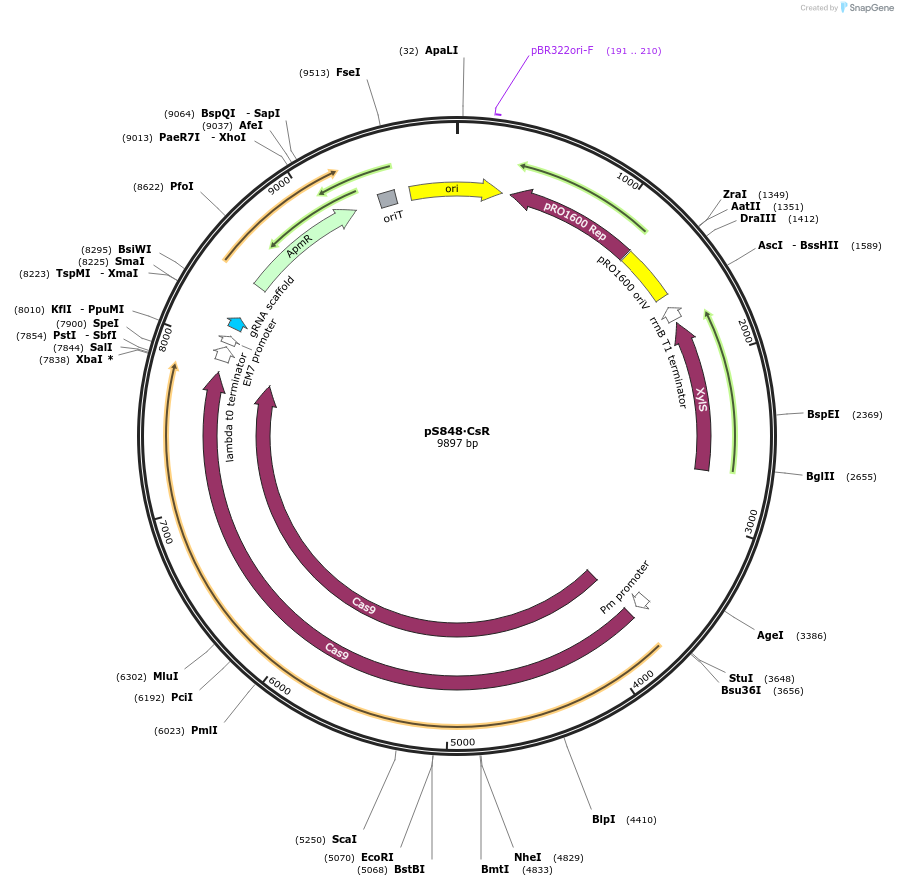
pS848∙CsR
Plasmid#199240PurposeExpresses Streptococcus pyogenes' cas9 and a tailored sgRNA for counterselection in gram-negative bacteriaDepositorInsertgRNA scaffold - Gm resistance cassette - incP oriT
UseCRISPR and Synthetic BiologyPromoterPEM7Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
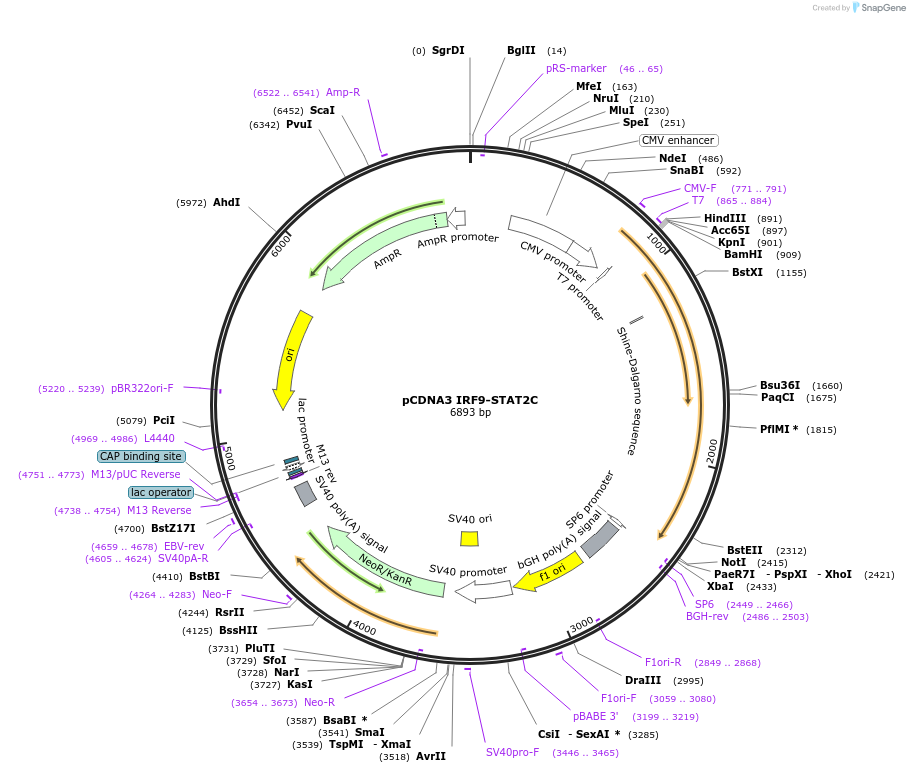
pCDNA3 IRF9-STAT2C
Plasmid#37544DepositorAvailable SinceMarch 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
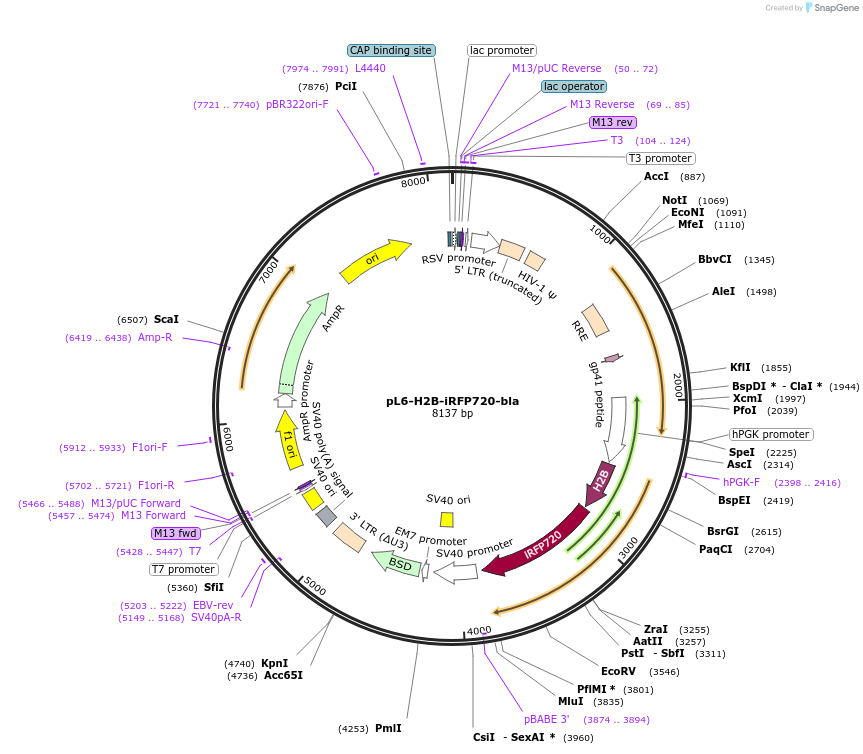
pL6-H2B-iRFP720-bla
Plasmid#189991PurposeLentivirial plasmid for nuclear iRFP-720 expression with blasticidin resistanceDepositorInsertHuman histone H2B
UseLentiviralTagsiRFP720ExpressionMammalianAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
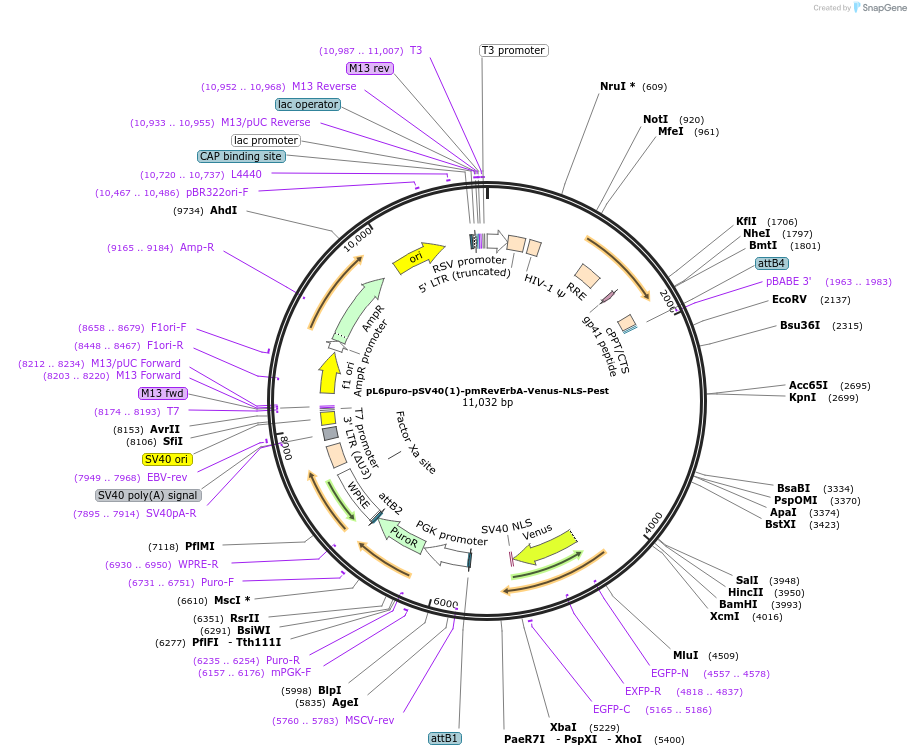
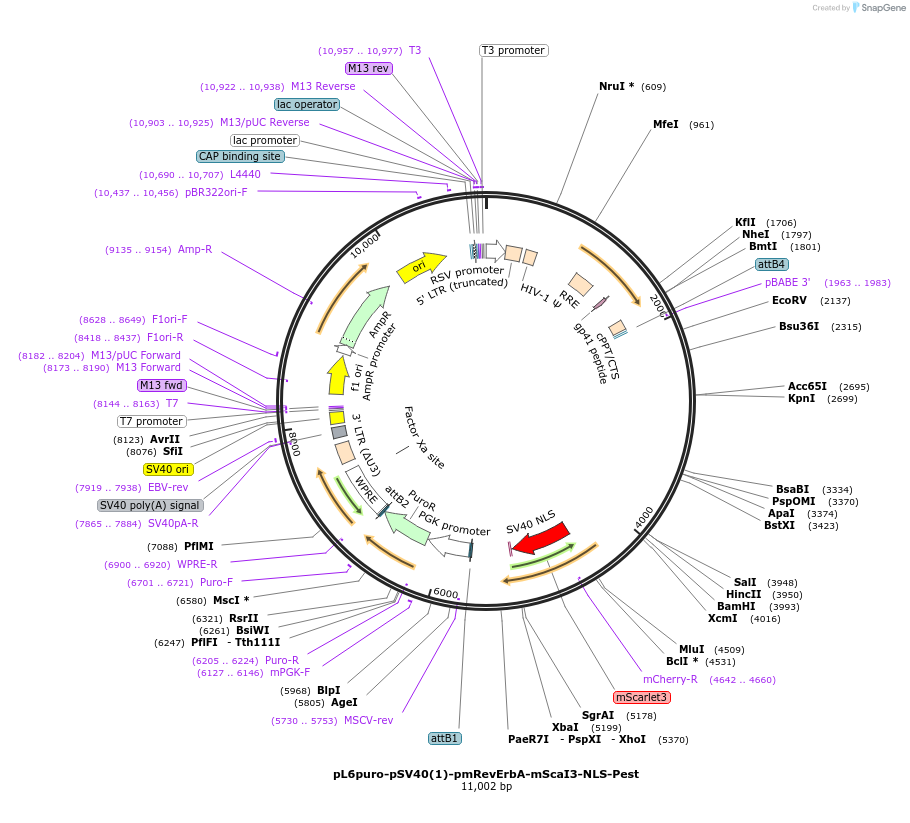
pL6puro-pSV40(1)-pmRevErbA-mScaI3-NLS-Pest
Plasmid#240116PurposeA lentiviral (red) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter fragment.DepositorInsertmurine Nr1d1 promoter+mScarlet-I3-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-EnhancerAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
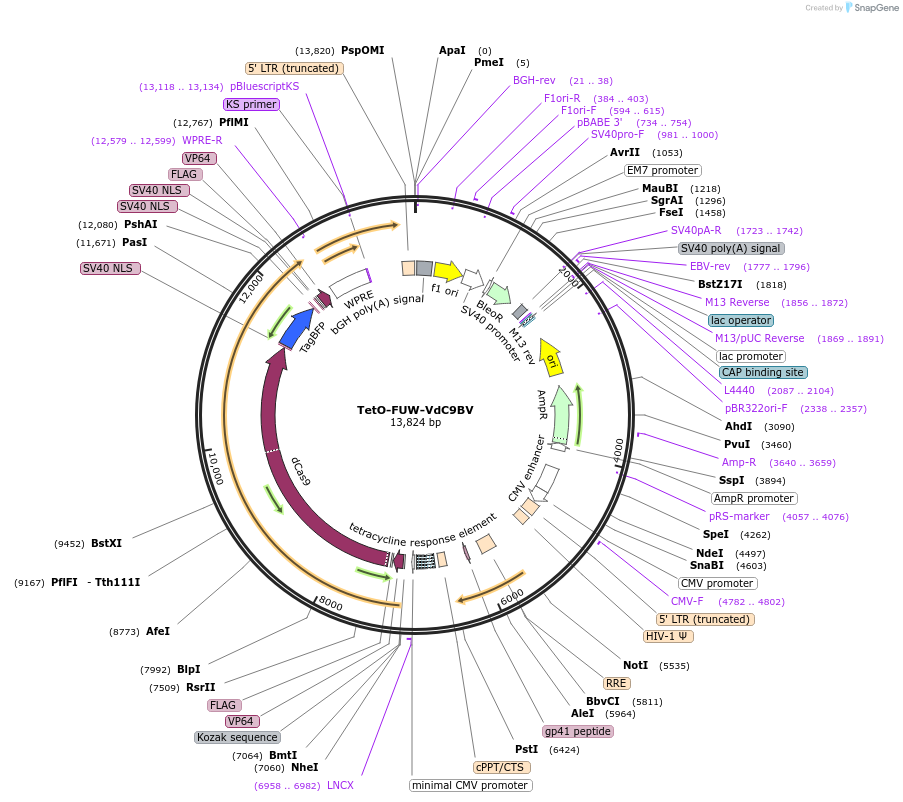
TetO-FUW-VdC9BV
Plasmid#62195PurposeExpresses RNA-Guided, Nuclease-Inactive VP64:dCas9-BFP:VP64—VdC9BV—Fusion Protein to Enable Transactivation of Endogenous GenesDepositorInsertVP64dCas9BFPVP64
UseLentiviralTagsTwo VP64s tagged to dCas9 fused to BFPExpressionMammalianMutationD10A H840A (catalytically inactive) Cas9 (dCas9)Available SinceJune 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
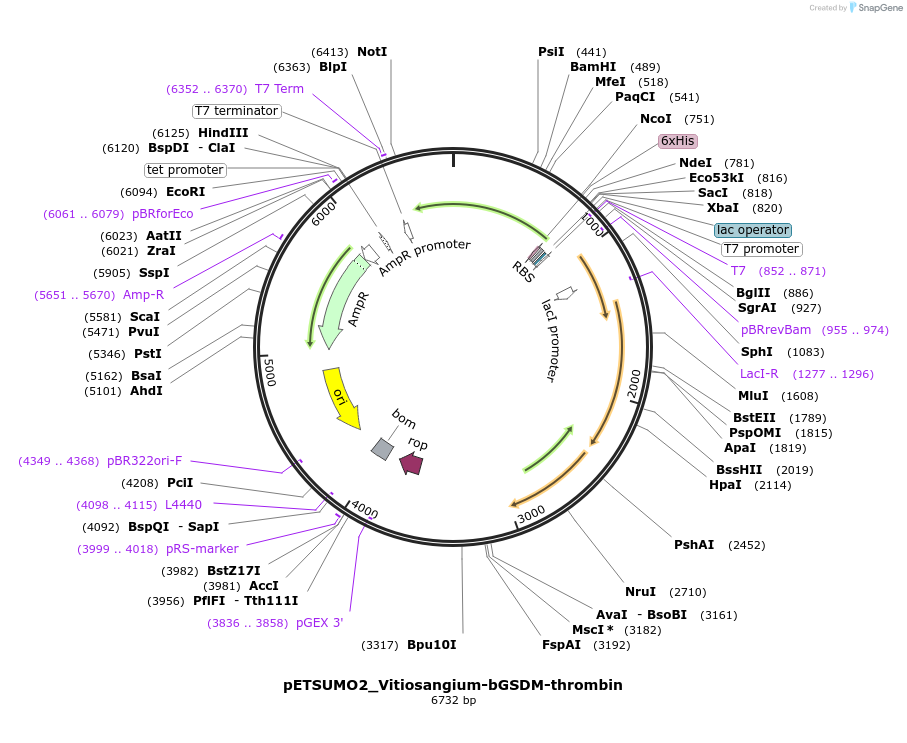
pETSUMO2_Vitiosangium-bGSDM-thrombin
Plasmid#222099PurposeFor expression of a mutant bacterial gasdermin (bGSDM) encoded by a Vitiosangium species with a thrombin protease cleavage site for controlled activation.DepositorInsertVitiosangium-bGSDM-thrombin ORF
Tags6xHis-SUMO2ExpressionBacterialMutationN-terminal methionine deleted and replaced by sin…PromoterT7Available SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
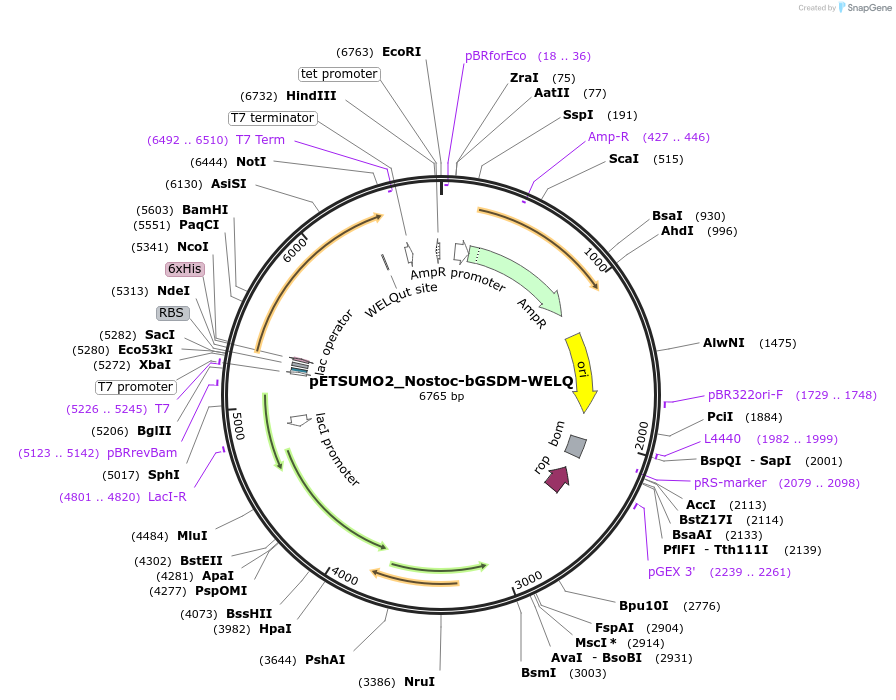
pETSUMO2_Nostoc-bGSDM-WELQ
Plasmid#222100PurposeFor expression of a mutant bacterial gasdermin (bGSDM) encoded by a Nostoc species with a WELQut protease cleavage site for controlled activation.DepositorInsertNostoc-bGSDM-WELQ ORF
Tags6xHis-SUMO2ExpressionBacterialMutationN-terminal methionine deleted and replaced by sin…PromoterT7Available SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
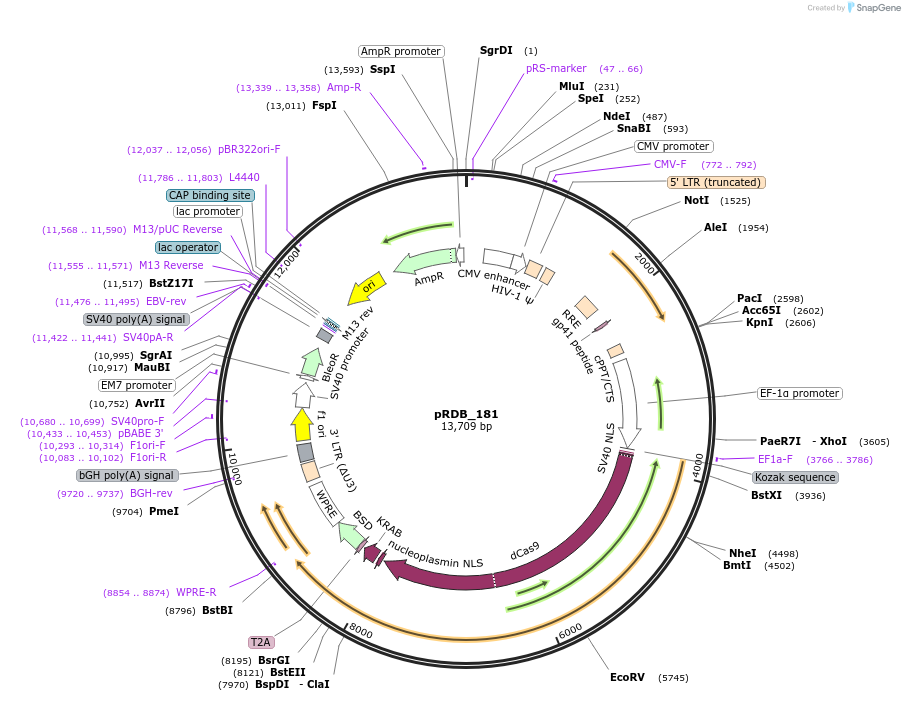
pRDB_181
Plasmid#216102PurposeCRISPRi, EF1a-driven dCas9-KRAB (ZNF10) (Cas only)DepositorInsertCas9 [Sp]
UseCRISPR and Lentiviral; Assembled vectorAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
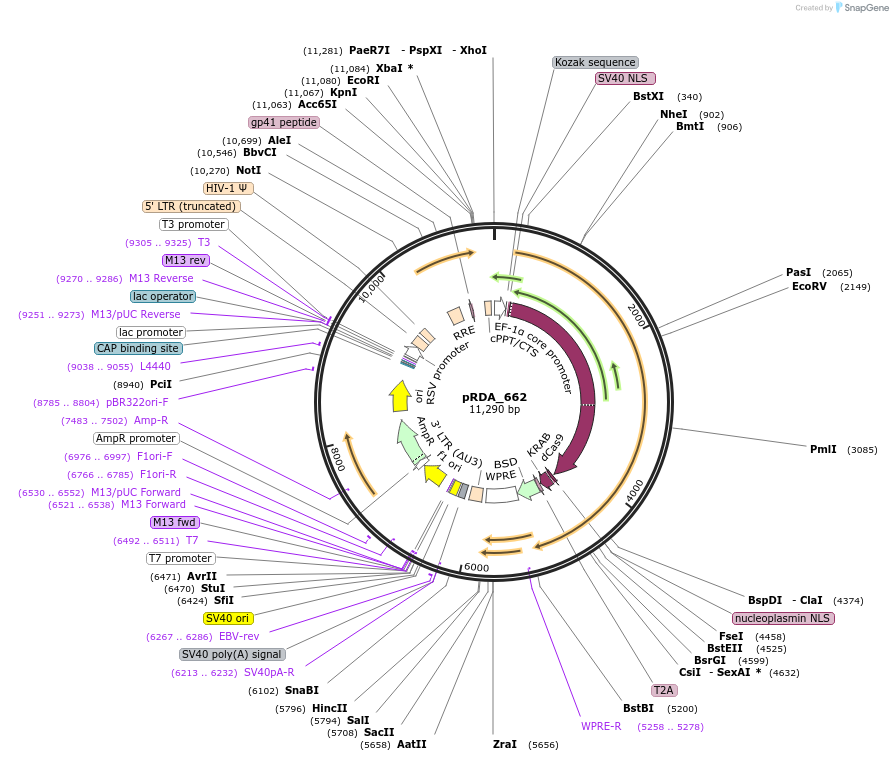
pRDA_662
Plasmid#216101PurposeCRISPRi, EFS-driven dCas9-KRAB (ZNF10) (Cas only)DepositorInsertCas9 [Sp]
UseCRISPR and Lentiviral; Assembled vectorAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
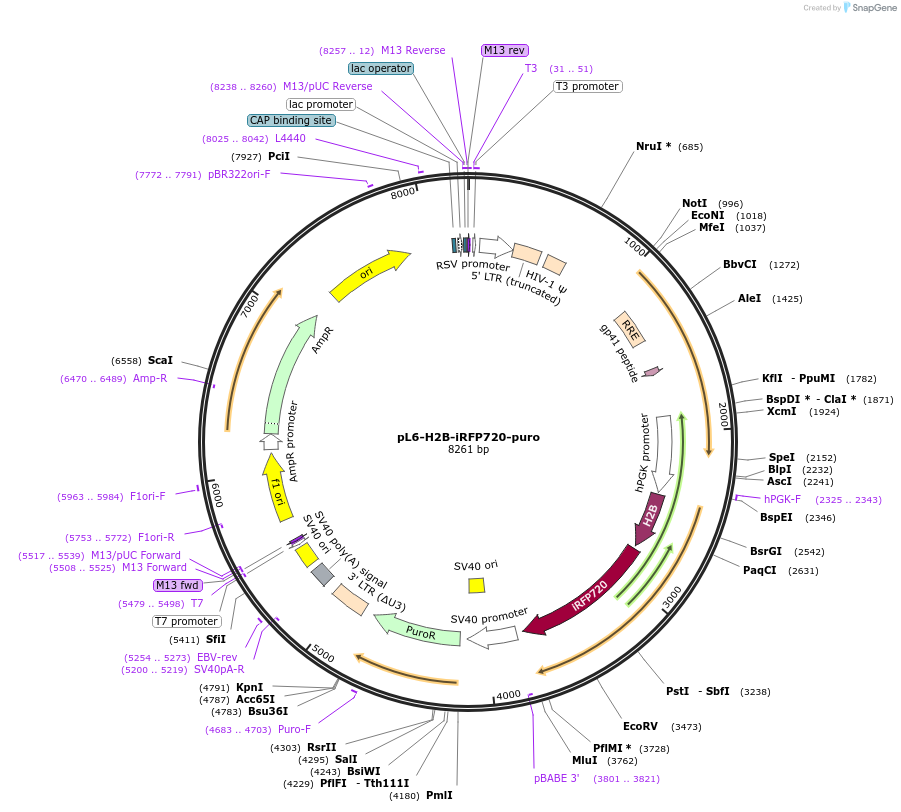
pL6-H2B-iRFP720-puro
Plasmid#189992PurposeLentivirial plasmid for nuclear iRFP-720 expression with puromycin resistanceDepositorInsertHuman histone H2B
UseLentiviralTagsiRFP720ExpressionMammalianAvailable SinceFeb. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET23-PBD(65-150)-N-Cys
Plasmid#13722DepositorInsertPBD(65-150)-N-Cys (PAK1 Human)
Tags6xHisExpressionBacterialMutationMet-Cys-Gly-PBD(65-150)-Available SinceMarch 28, 2007AvailabilityAcademic Institutions and Nonprofits only -
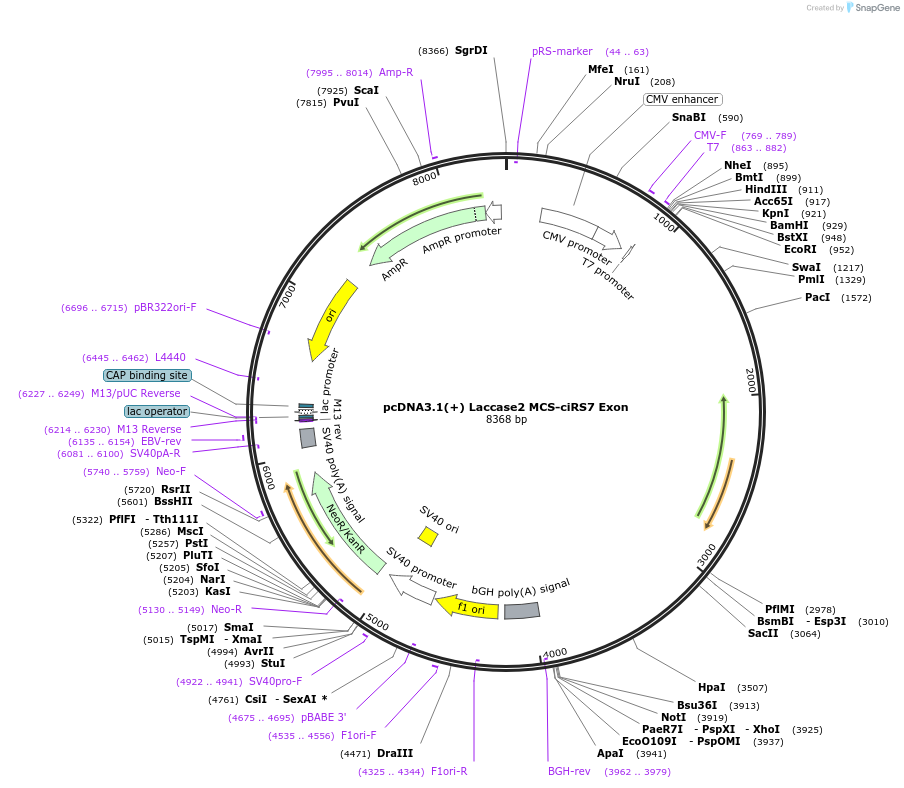
pcDNA3.1(+) Laccase2 MCS-ciRS7 Exon
Plasmid#69900PurposeExpresses the human ciRS7/CDR1as circular RNA in mammalian cellsDepositorAvailable SinceOct. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
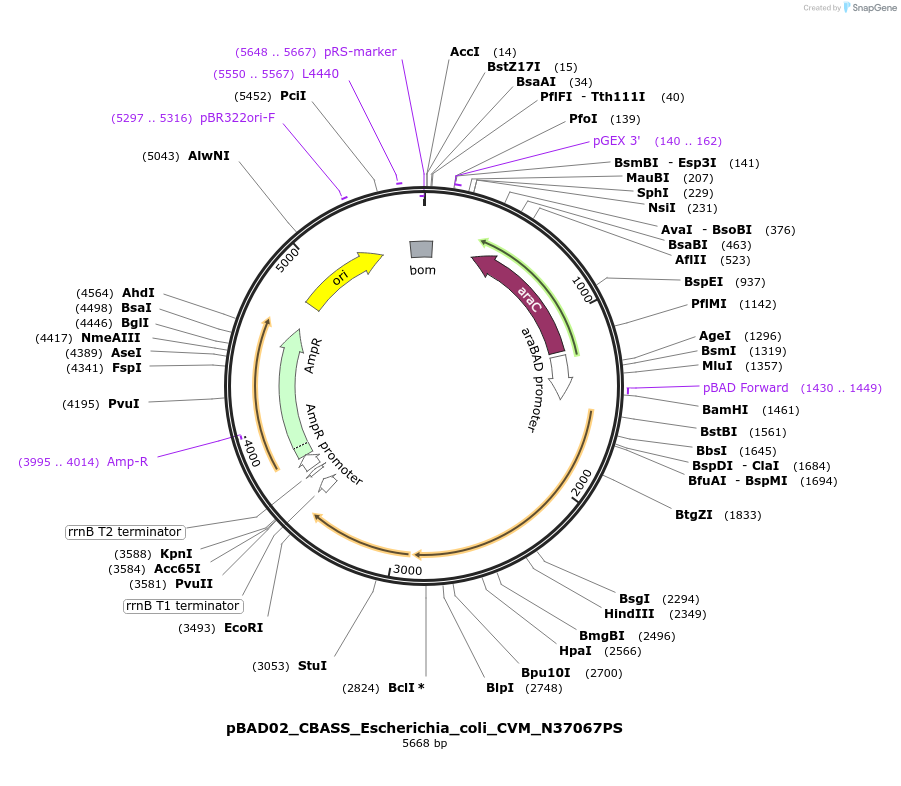
pBAD02_CBASS_Escherichia_coli_CVM_N37067PS
Plasmid#224403PurposeFor expression of the type I CBASS system encoded by Escherichia coli CVM N37067PSDepositorInsertEcCdnF_AGS-C_SLATT6
ExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
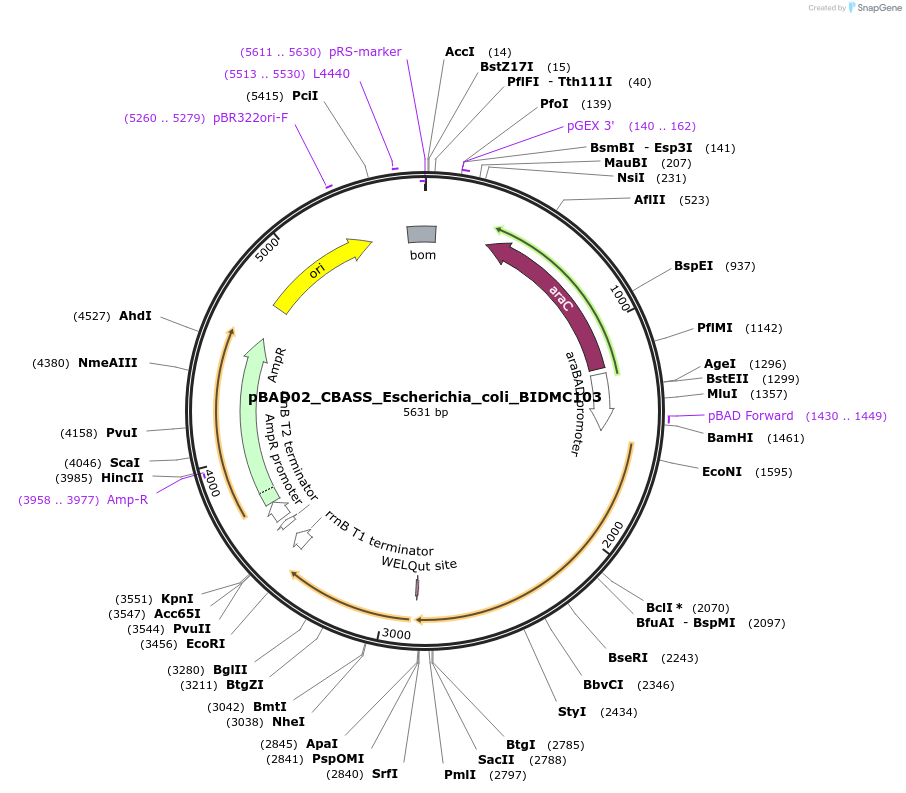
pBAD02_CBASS_Escherichia_coli_BIDMC103
Plasmid#224402PurposeFor expression of the type I CBASS system encoded by Escherichia coli BIDMC103DepositorInsertEcCdnF_AGS-C_SLATT6
ExpressionBacterialAvailable SinceSept. 5, 2024AvailabilityAcademic Institutions and Nonprofits only