We narrowed to 15,323 results for: pET
-
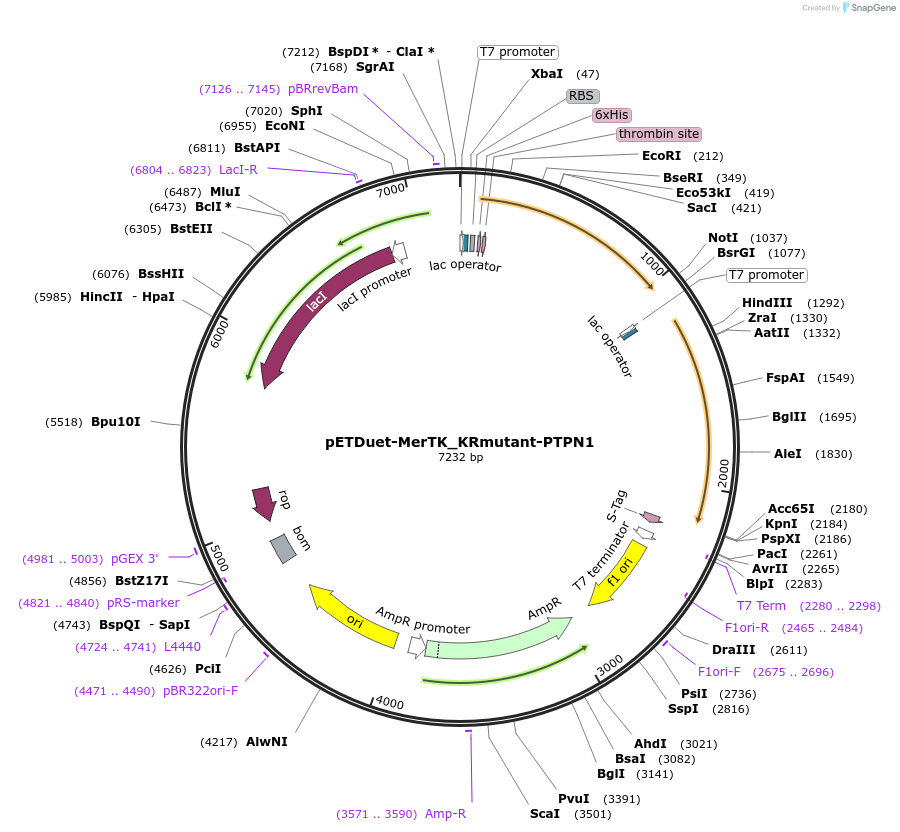
Plasmid#196447PurposeExpression of MerTK Kinase Domain in E. coli and surface entropy reduction mutant for crystallization, coexpress PTPN1DepositorExpressionBacterialMutationK591R, K639R, K702R, K865RPromoterT7 promoterAvailable SinceMarch 22, 2023AvailabilityAcademic Institutions and Nonprofits only
-
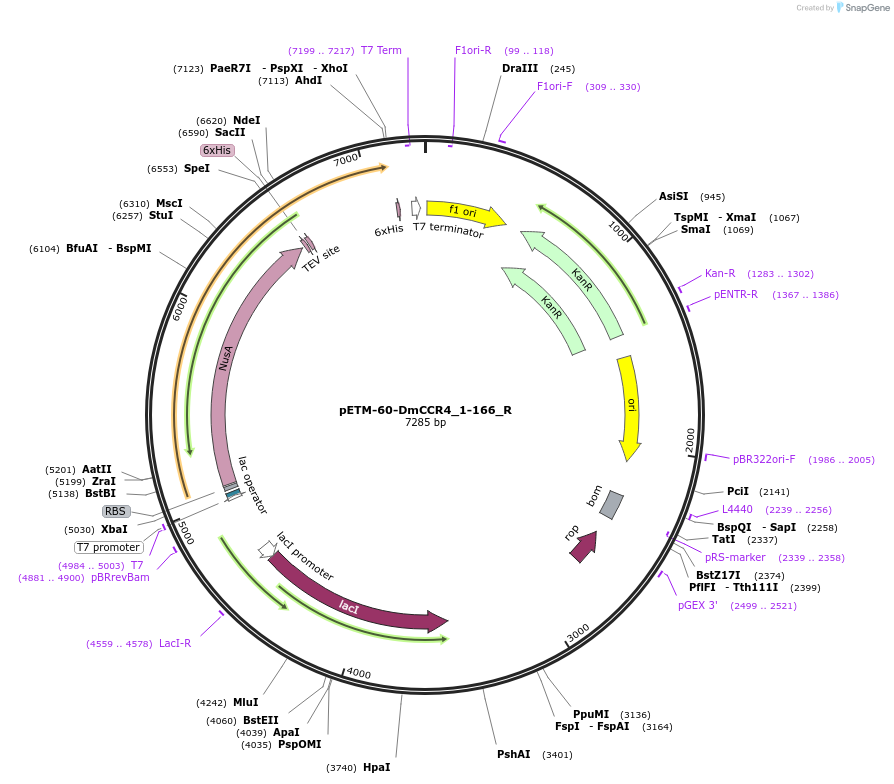
pETM-60-DmCCR4_1-166_R
Plasmid#147439PurposeBacterial Expression of DmCCR4_1-166DepositorInsertDmCCR4_1-166 (twin Fly)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
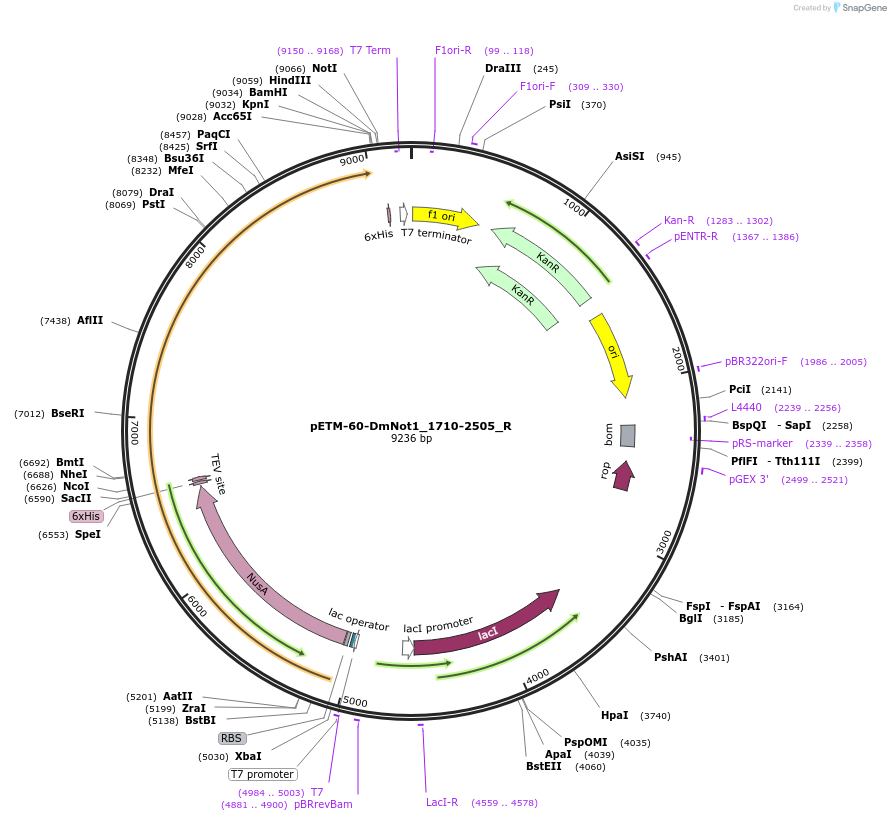
pETM-60-DmNot1_1710-2505_R
Plasmid#147440PurposeBacterial Expression of DmNot1_1710-2505DepositorInsertDmNot1_1710-2505 (Not1 Fly)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
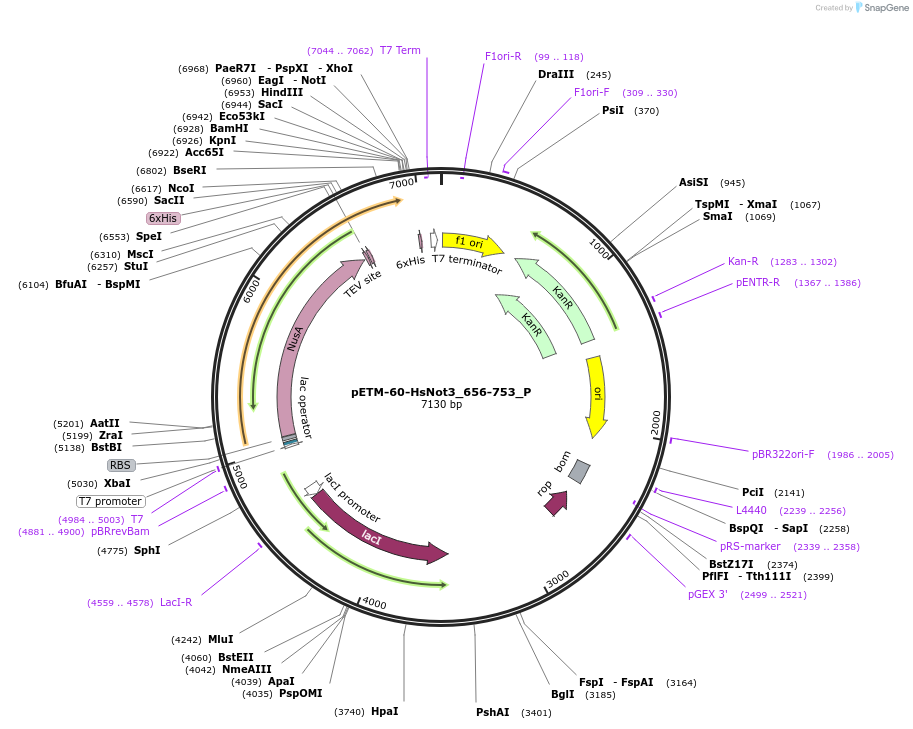
pETM-60-HsNot3_656-753_P
Plasmid#147223PurposeBacterial Expression of HsNot3_656-753DepositorInsertHsNot3_656-753 (CNOT3 Human)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
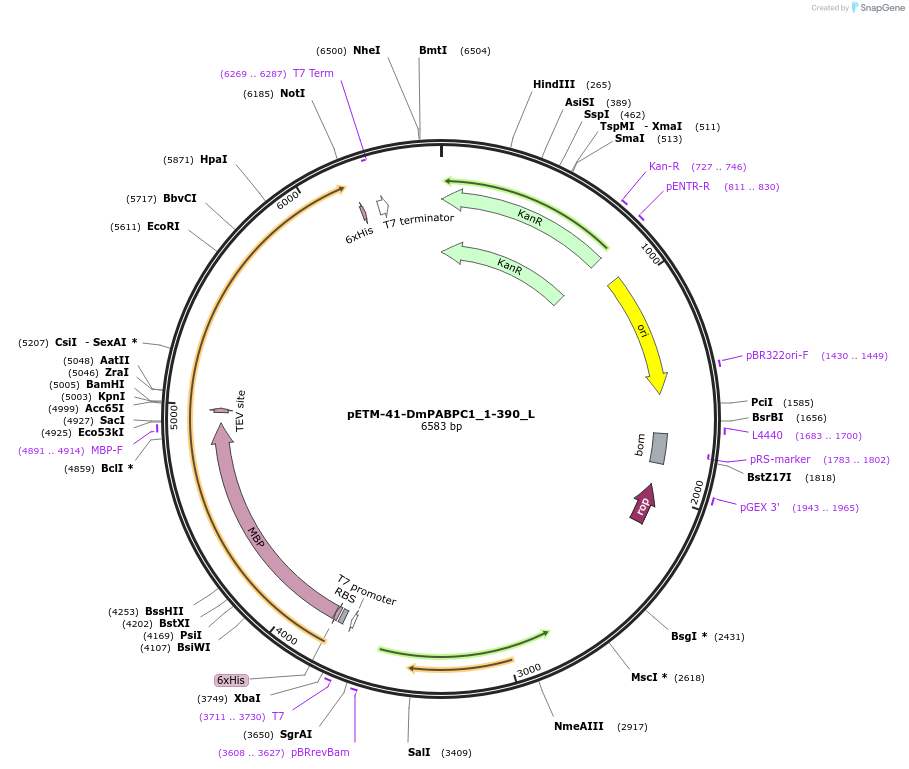
pETM-41-DmPABPC1_1-390_L
Plasmid#146866PurposeBacterial Expression of DmPABPC1_1-390DepositorInsertDmPABPC1_1-390 (pAbp Fly)
ExpressionBacterialMutation4 silent mutations compared to the sequence geven…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
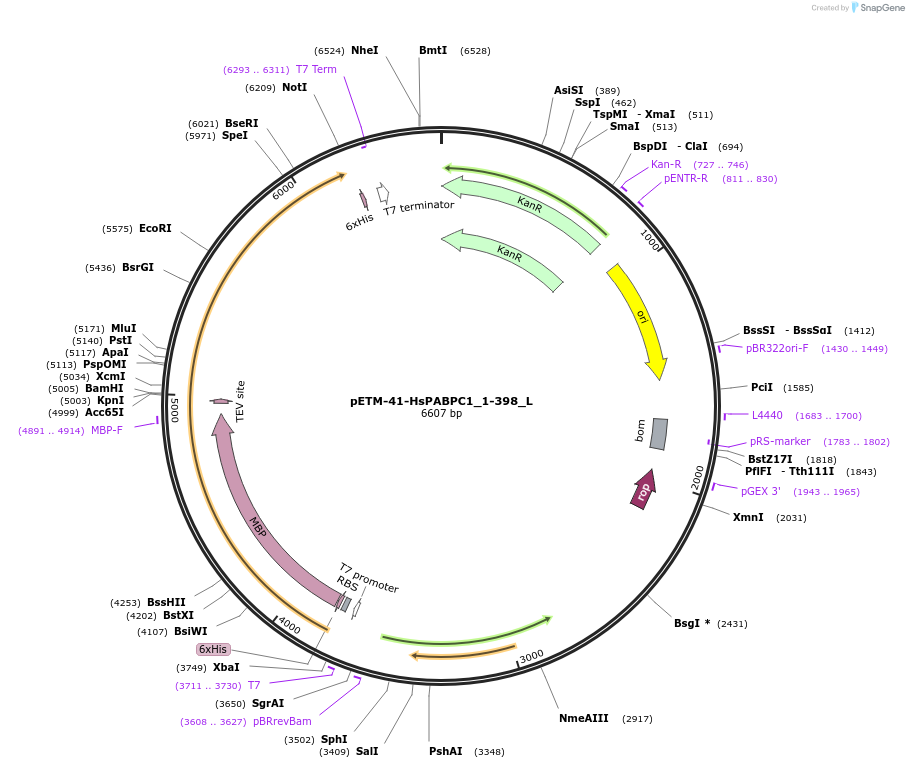
pETM-41-HsPABPC1_1-398_L
Plasmid#146867PurposeBacterial Expression of HsPABPC1_1-398DepositorInsertHsPABPC1_1-398 (PABPC1 Human)
ExpressionBacterialMutationone silent mutations compared to the sequence giv…Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
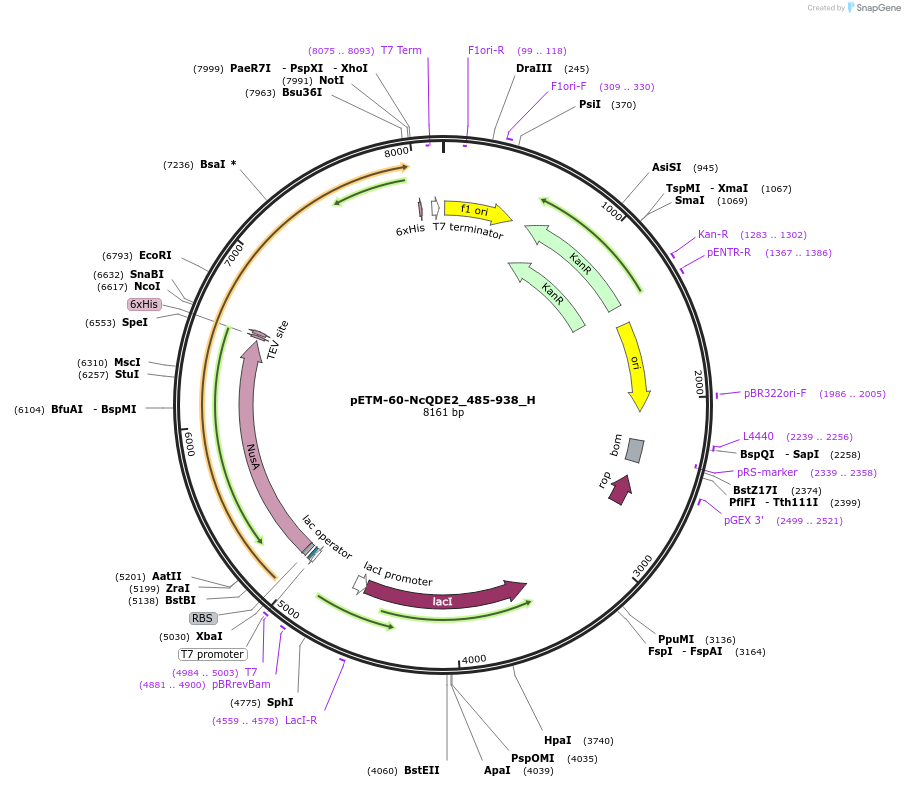
pETM-60-NcQDE2_485-938_H
Plasmid#146457PurposeBacterial Expression of NcQDE2_485-938DepositorInsertNcQDE2_485-938
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
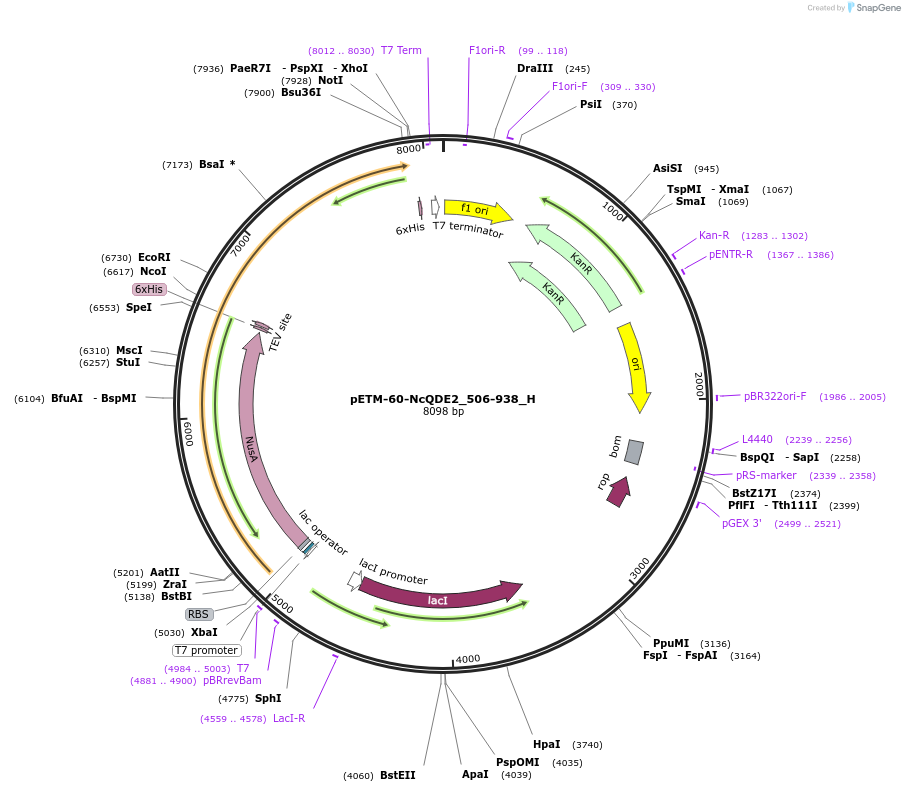
pETM-60-NcQDE2_506-938_H
Plasmid#146458PurposeBacterial Expression of NcQDE2_506-938DepositorInsertNcQDE2_506-938
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
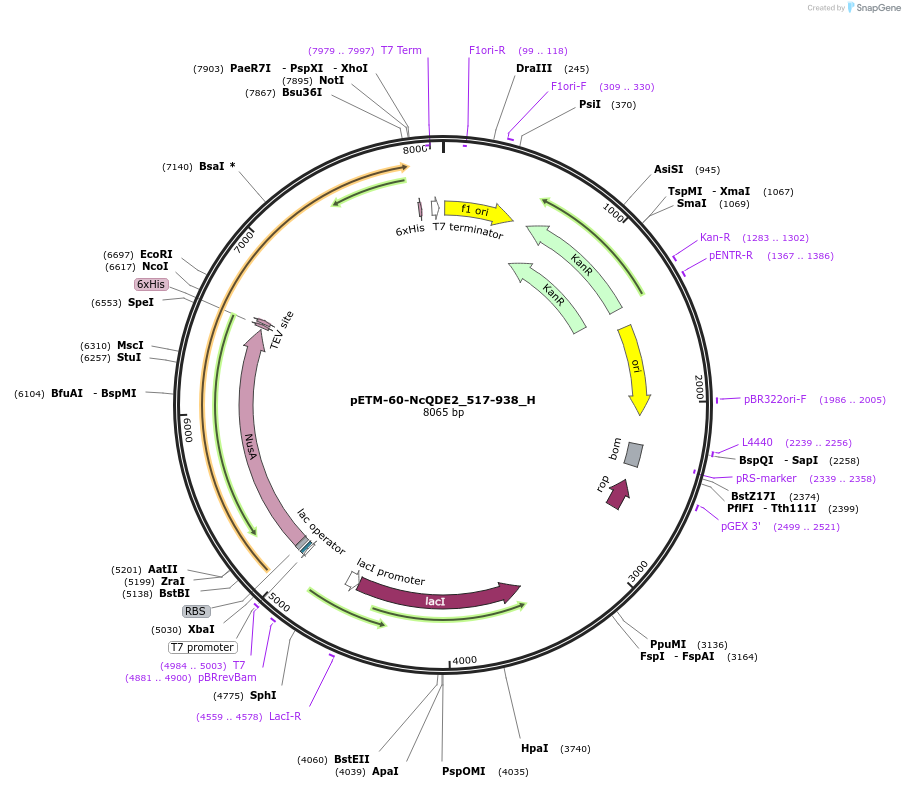
pETM-60-NcQDE2_517-938_H
Plasmid#146459PurposeBacterial Expression of NcQDE2_517-938DepositorInsertNcQDE2_517-938
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
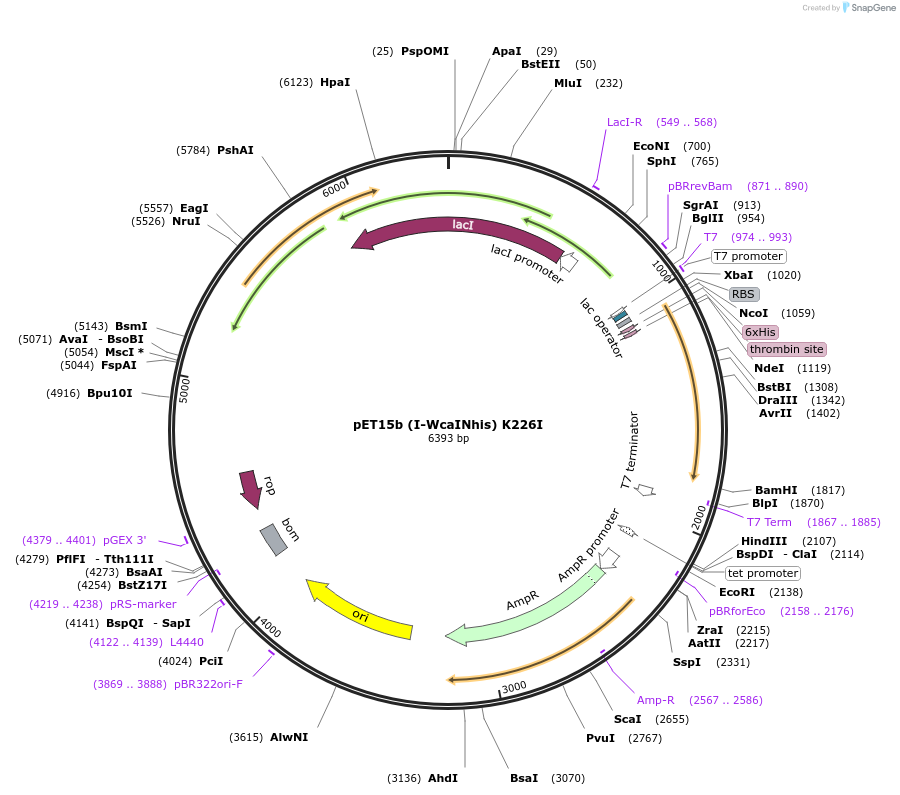
pET15b (I-WcaINhis) K226I
Plasmid#195620PurposeExpresses I-WcaI homing endonuclease from Wickerhamomyces canadensis containing the K226I mutationDepositorInsertI-WcaI
ExpressionBacterialMutationA1930T, A1931TPromoterT7Available SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
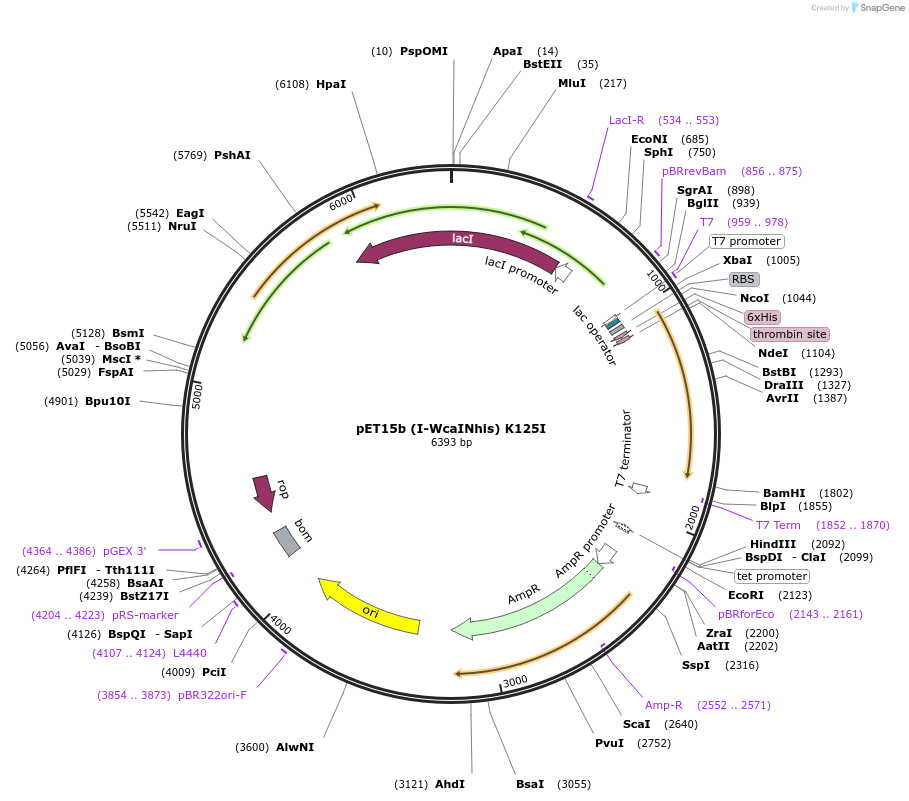
pET15b (I-WcaINhis) K125I
Plasmid#195619PurposeExpresses I-WcaI homing endonuclease from Wickerhamomyces canadensis containing the K125I mutationDepositorInsertI-WcaI
ExpressionBacterialMutationA2030T, A2031TPromoterT7Available SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
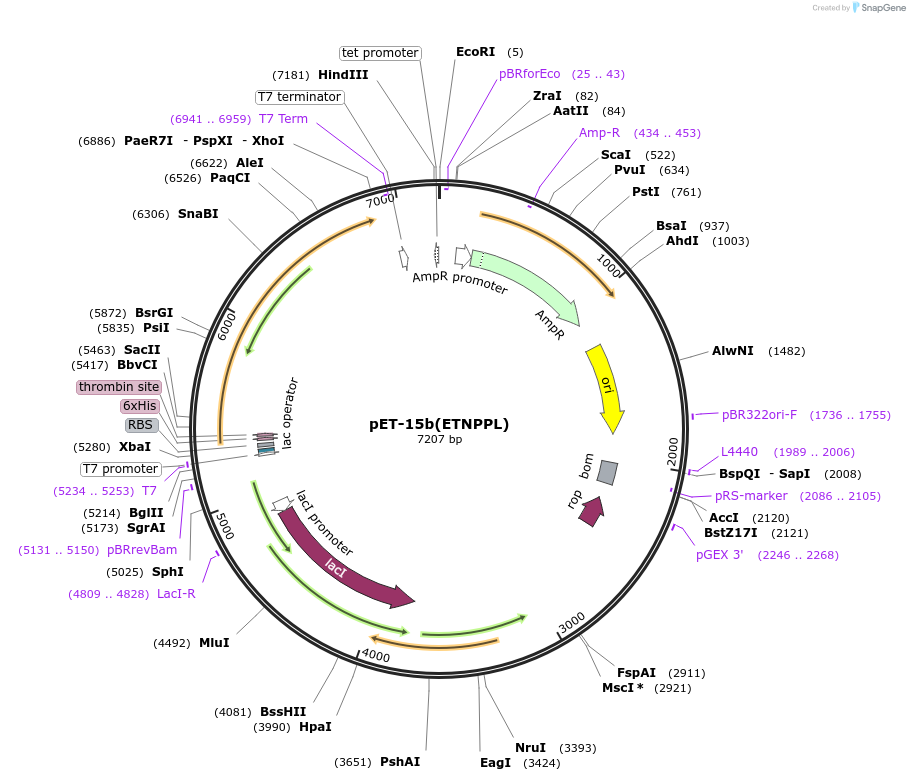
pET-15b(ETNPPL)
Plasmid#192264PurposeCoding sequence of mouse ETNPPL is inserted in pET-15b vectorDepositorAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
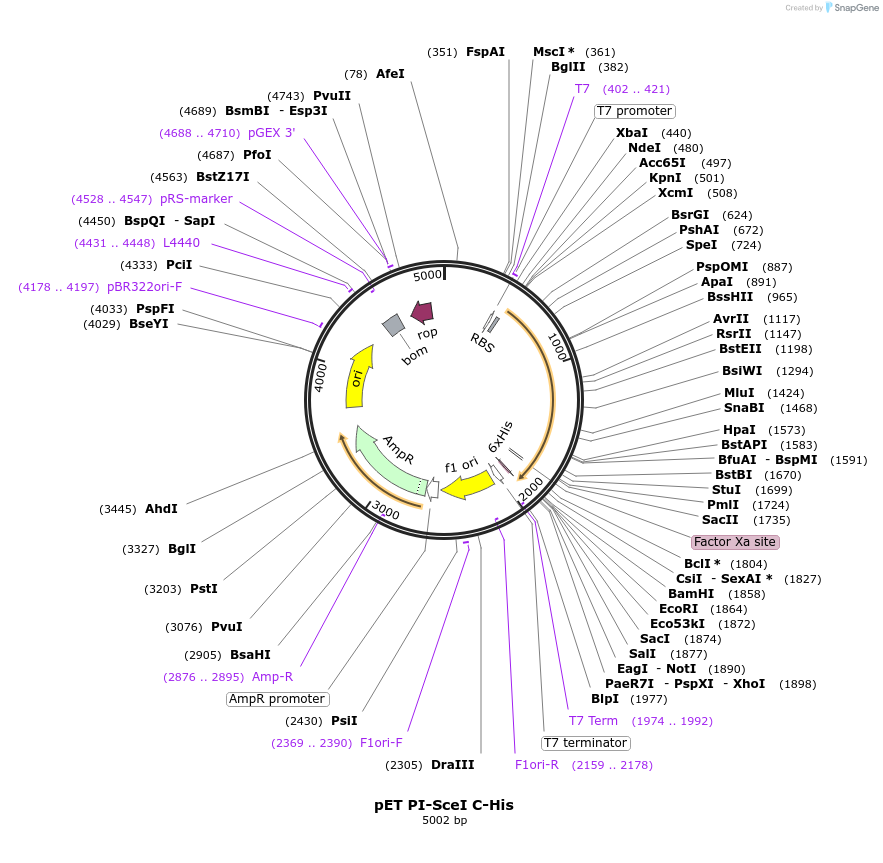
pET PI-SceI C-His
Plasmid#195610PurposeExpresses PI-SceI (VDE) homing endonuclease/intein from the T7 promoter with a carboxyl-terminal histidine tagDepositorInsertPI-SceI
ExpressionBacterialMutationT825C, C900G, A901C, A903C, T1131G, T1137C, T1227…PromoterT7Available SinceJan. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
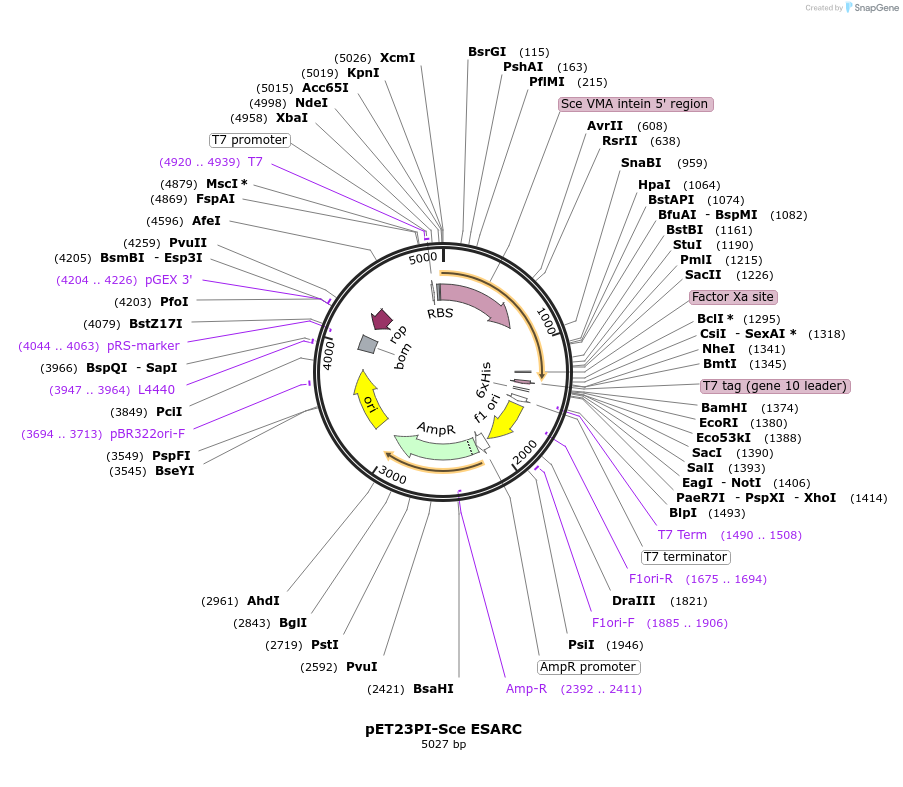
pET23PI-Sce ESARC
Plasmid#195607PurposeExpresses PI-SceI (VDE) homing endonuclease/intein from the T7 promoterDepositorInsertPI-SceI
ExpressionBacterialMutationC552T,C554A, A557T,G584T, C588A, G878T, C881T, C9…PromoterT7Available SinceJan. 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
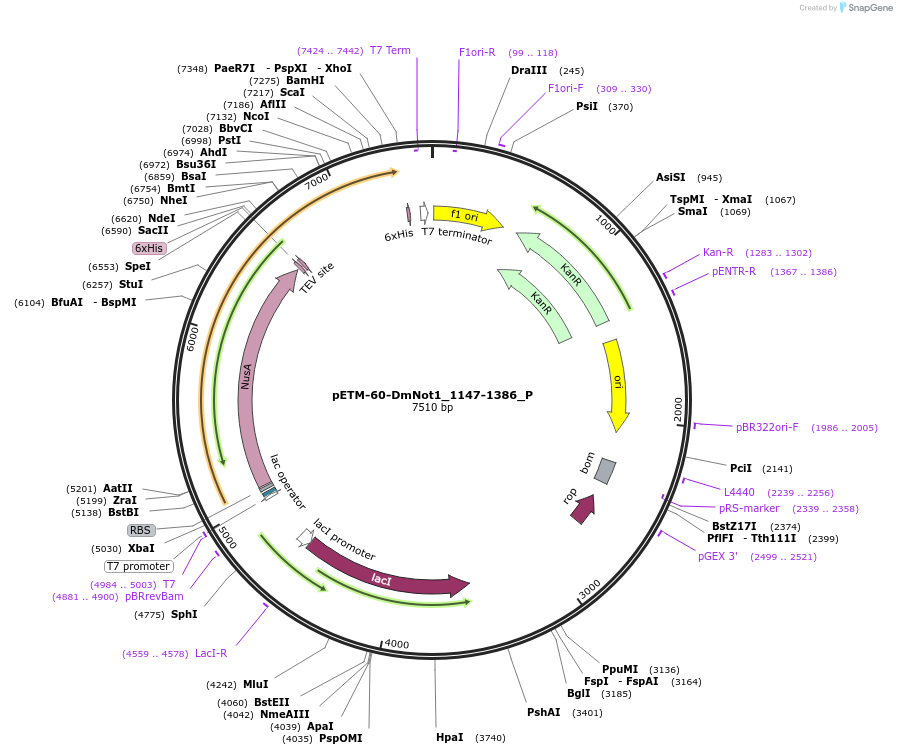
pETM-60-DmNot1_1147-1386_P
Plasmid#147175PurposeBacterial Expression of DmNot1_1147-1386DepositorInsertDmNot1_1147-1386 (Not1 Fly)
ExpressionBacterialMutation1 silent mutation compared to the sequence given …Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
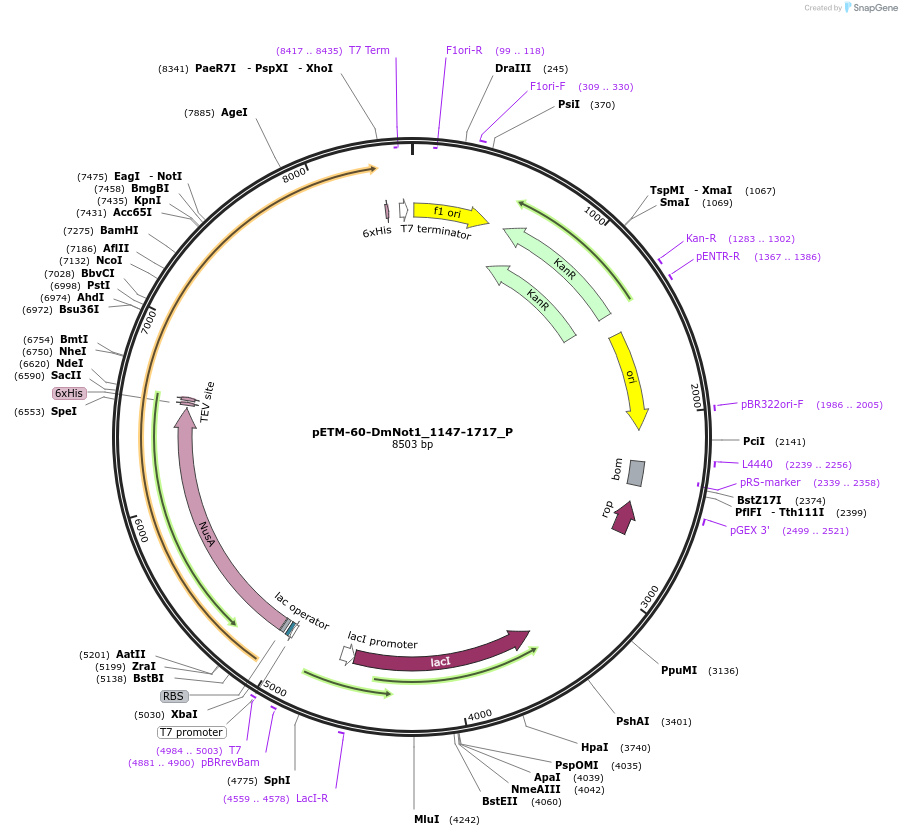
pETM-60-DmNot1_1147-1717_P
Plasmid#147176PurposeBacterial Expression of DmNot1_1147-1717DepositorInsertDmNot1_1147-1717 (Not1 Fly)
ExpressionBacterialMutation2 silent mutations compared to the sequence given…Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
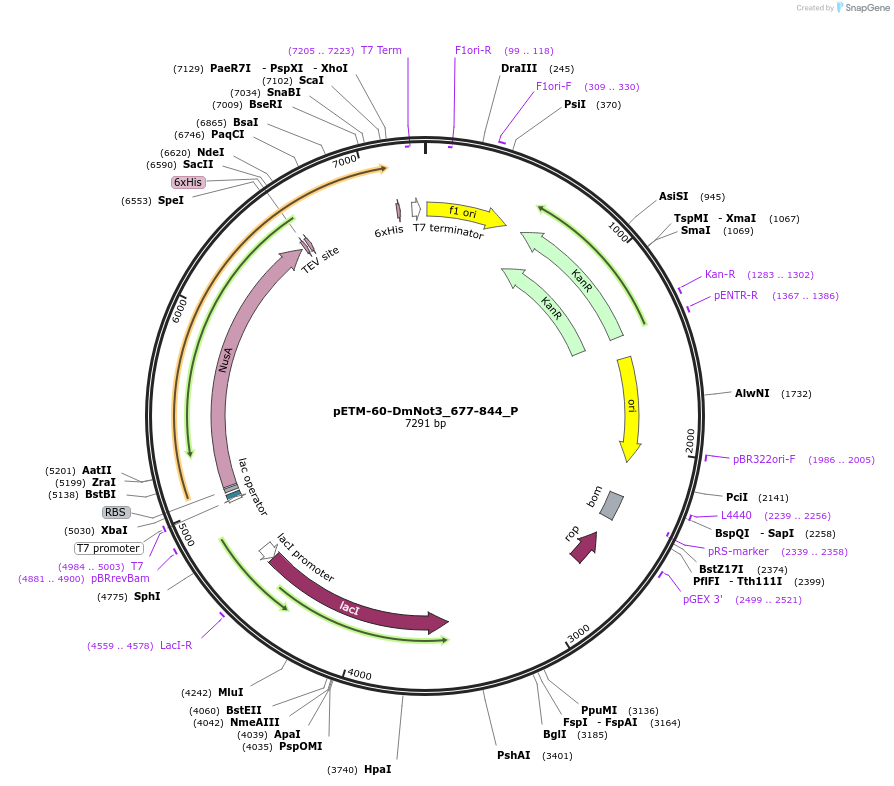
pETM-60-DmNot3_677-844_P
Plasmid#147177PurposeBacterial Expression of DmNot3_677-844DepositorInsertDmNot3_677-844 (Not3 Fly)
ExpressionBacterialAvailable SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
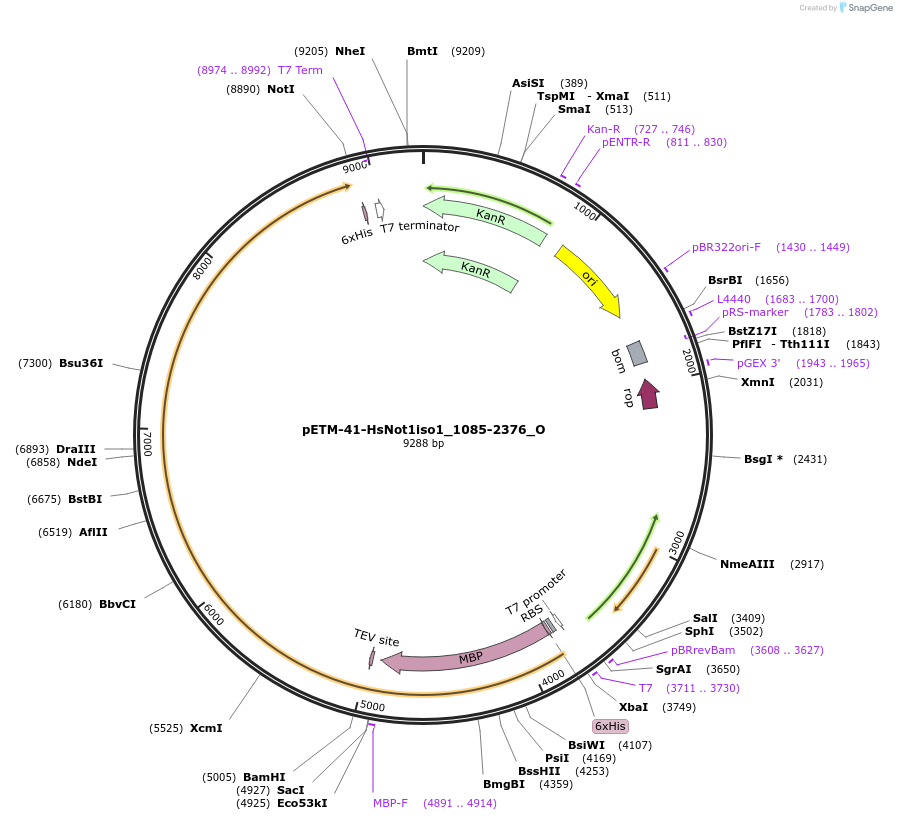
pETM-41-HsNot1iso1_1085-2376_O
Plasmid#147081PurposeBacterial Expression of HsNot1iso1_1085-2376DepositorInsertHsNot1iso1_1085-2376 (CNOT1 Human)
ExpressionBacterialMutation1 silent mutation compared to the sequence of iso…Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
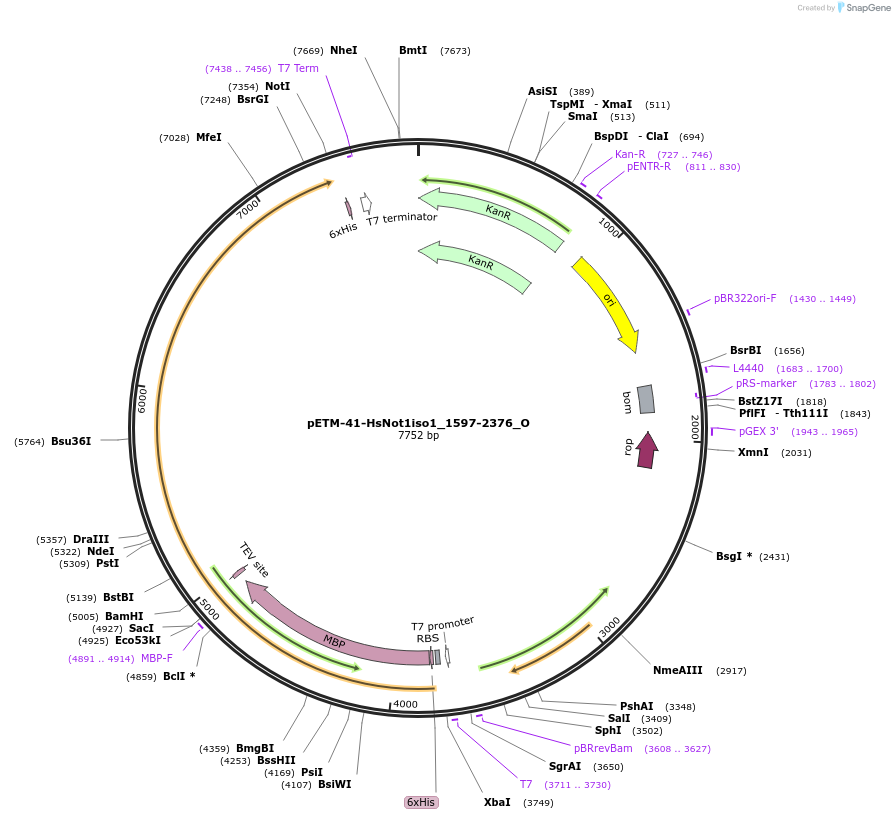
pETM-41-HsNot1iso1_1597-2376_O
Plasmid#147082PurposeBacterial Expression of HsNot1iso1_1597-2376DepositorInsertHsNot1iso1_1597-2376 (CNOT1 Human)
ExpressionBacterialAvailable SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
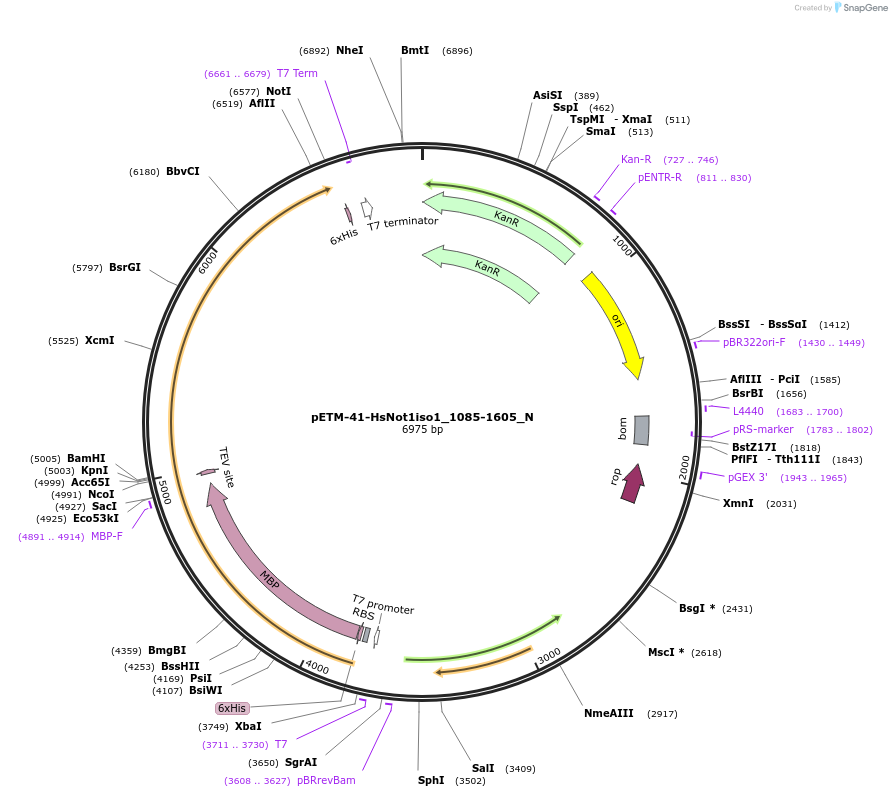
pETM-41-HsNot1iso1_1085-1605_N
Plasmid#147009PurposeBacterial Expression of HsNot1iso1_1085-1605DepositorInsertHsNot1iso1_1085-1605 (CNOT1 Human)
ExpressionBacterialMutation1 silent mutations compared to the sequence of is…Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
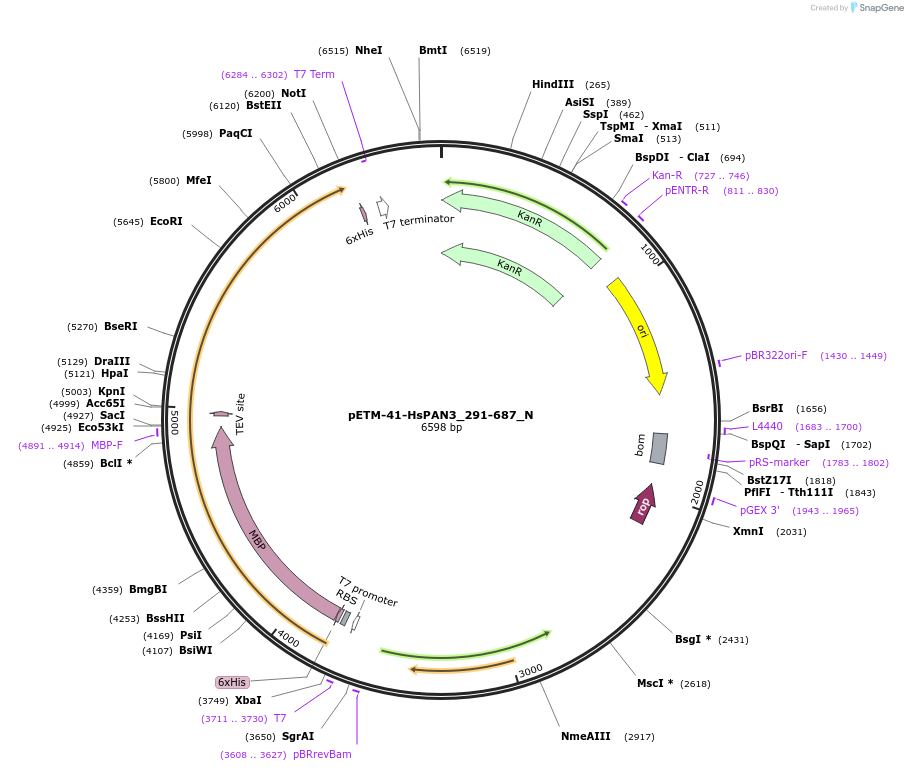
pETM-41-HsPAN3_291-687_N
Plasmid#147013PurposeBacterial Expression of HsPAN3_291-687DepositorInsertHsPAN3_291-687 (PAN3 Human)
ExpressionBacterialAvailable SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
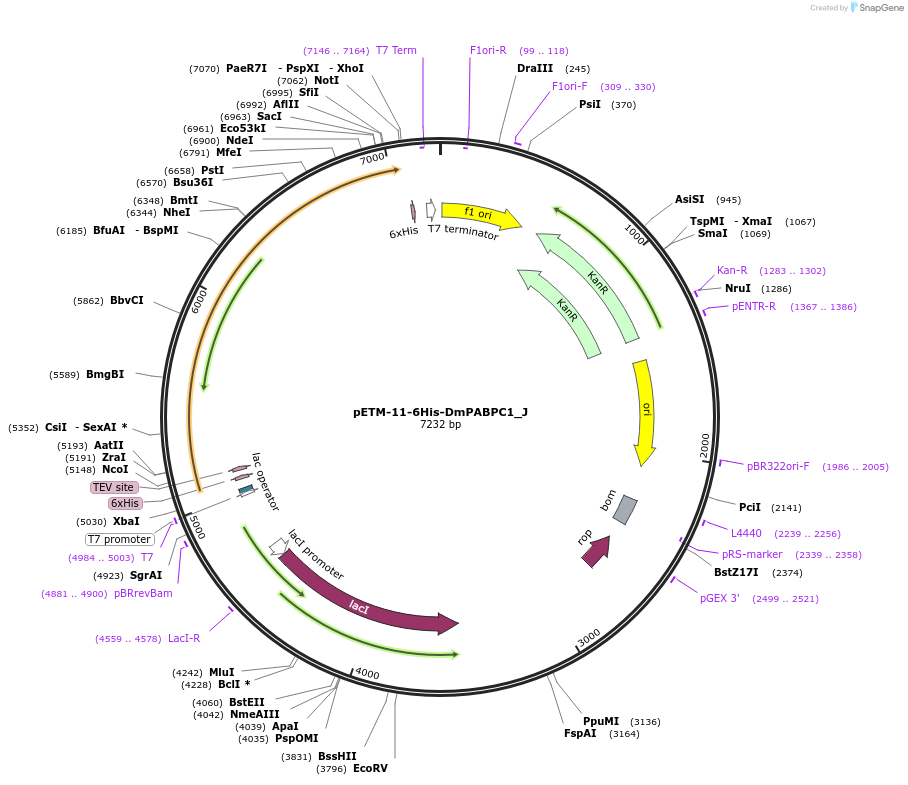
pETM-11-6His-DmPABPC1_J
Plasmid#146640PurposeBacterial Expression of DmPABPC1DepositorInsertDmPABPC1 (pAbp Fly)
ExpressionBacterialAvailable SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
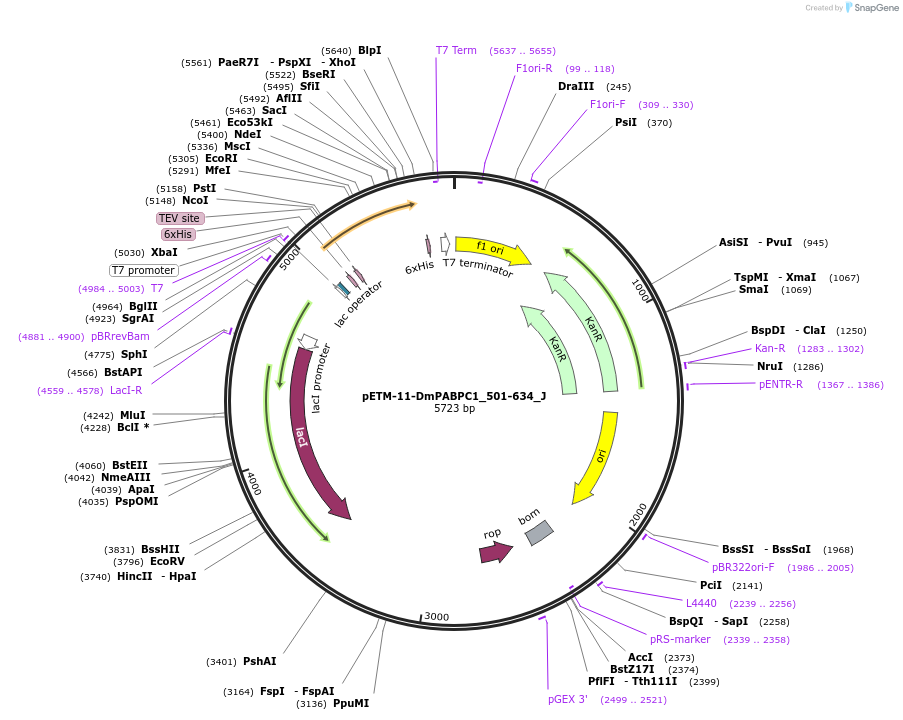
pETM-11-DmPABPC1_501-634_J
Plasmid#146641PurposeBacterial Expression of HsPABPC1_501-634DepositorInsertHsPABPC1_501-634
ExpressionBacterialMutation2 silent mutations compared to the sequence given…Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
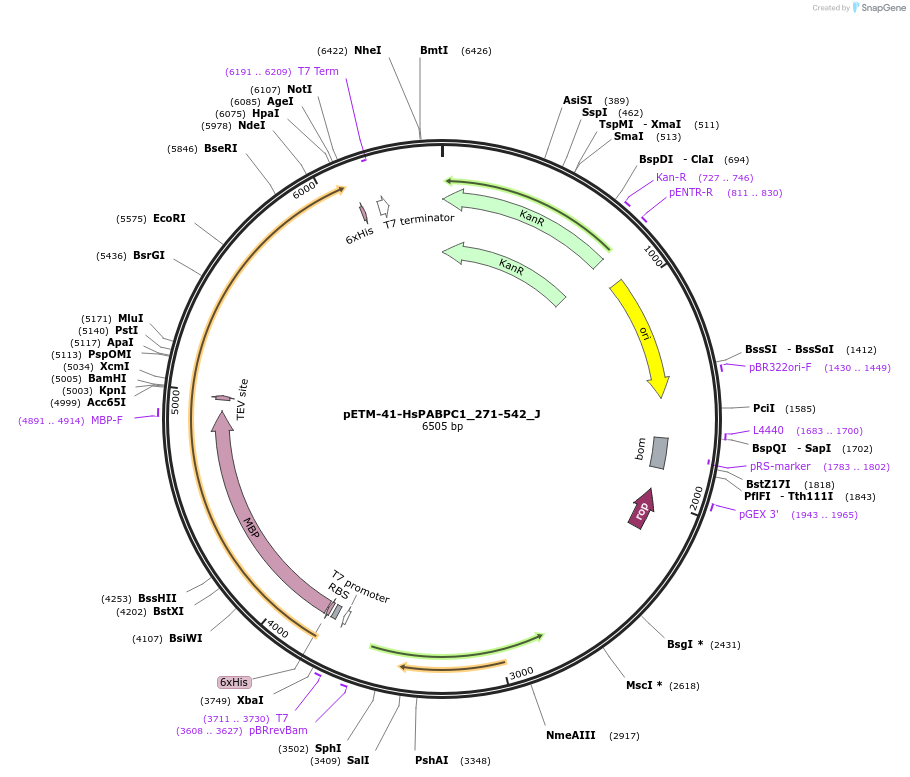
pETM-41-HsPABPC1_271-542_J
Plasmid#146645PurposeBacterial Expression of HsPABPC1_271-542DepositorInsertHsPABPC1_271-542 (PABPC1 Human)
ExpressionBacterialAvailable SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
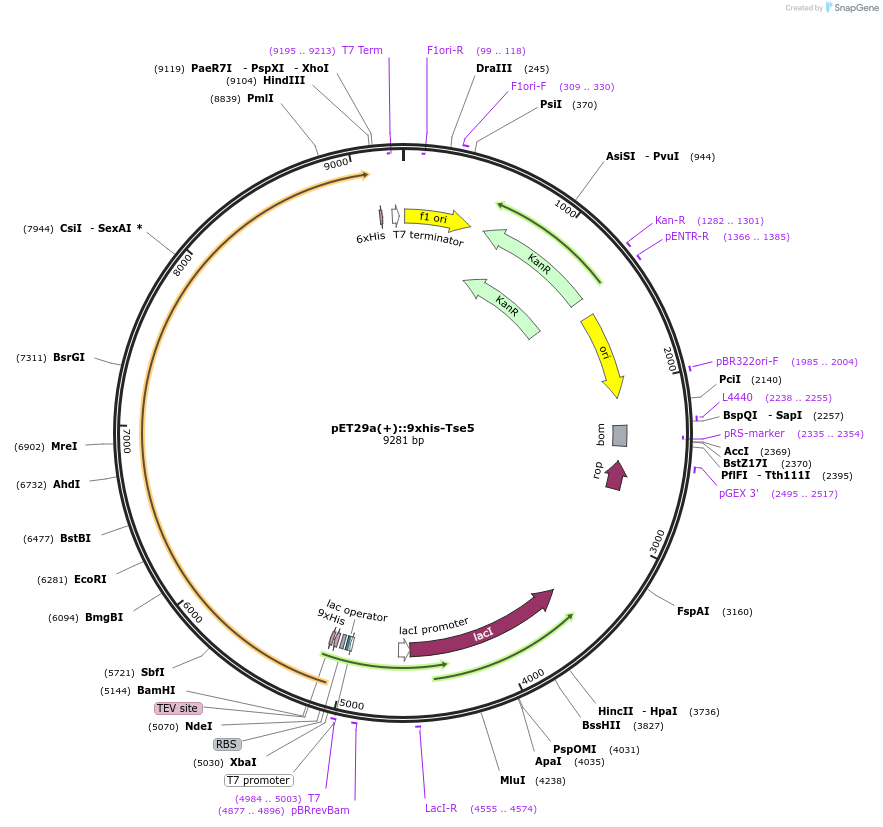
pET29a(+)::9xhis-Tse5
Plasmid#192962PurposeHeterologous expression of Tse5 in E. coli Lemo21 cells for purification of Tse5-CTDepositorInsertTse5
Tags9xhis + TEV tagExpressionBacterialAvailable SinceDec. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
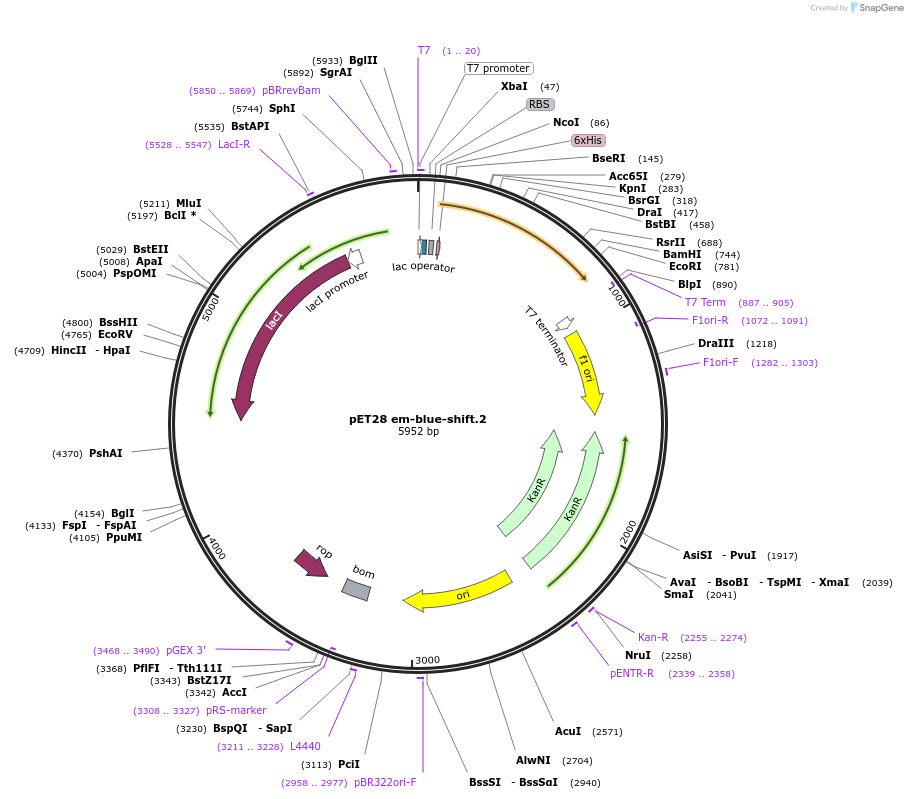
pET28 em-blue-shift.2
Plasmid#191901PurposeEncodes a GFP design, em-blue-shift.2DepositorInsertem-blue-shift.2
TagsHis TagExpressionBacterialMutationT65S S72A T108I Y145F V150IPromoterT7Available SinceDec. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
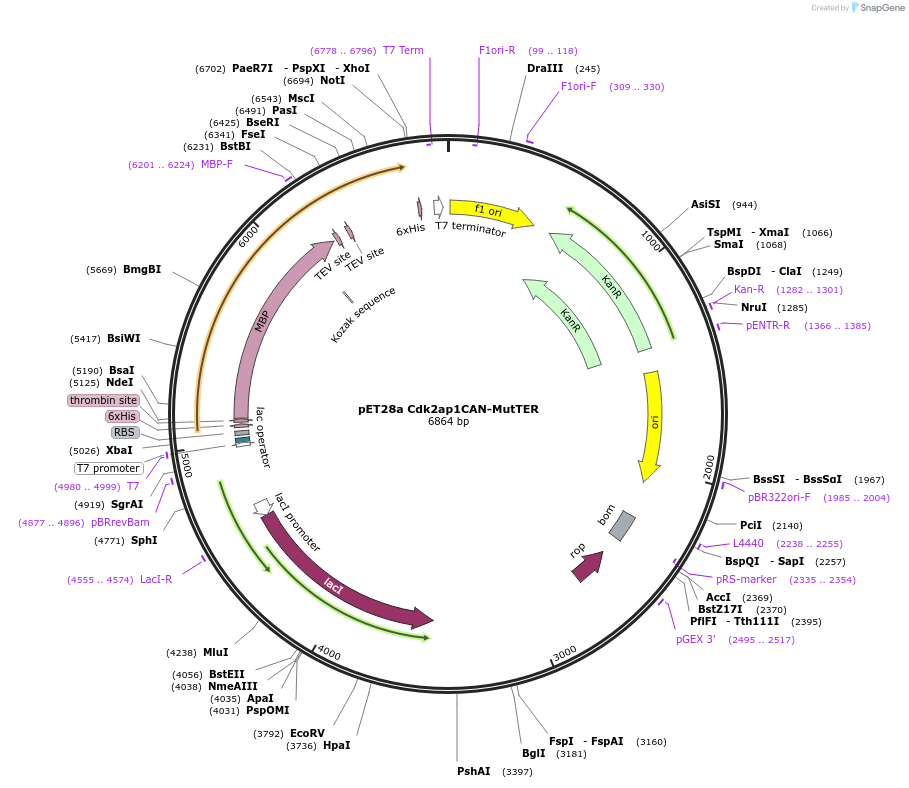
pET28a Cdk2ap1CAN-MutTER
Plasmid#178033PurposeExpression vector for the purpose of protein purification.DepositorInsertCdk2ap1 (Cdk2ap1 Mouse)
UseNonviralTags6xHIS - Thrombin - MBP- TEV-TRSExpressionBacterialMutationChanged T(108)A, E(109)A, R(110)APromoterT7LacIAvailable SinceNov. 16, 2022AvailabilityAcademic Institutions and Nonprofits only -
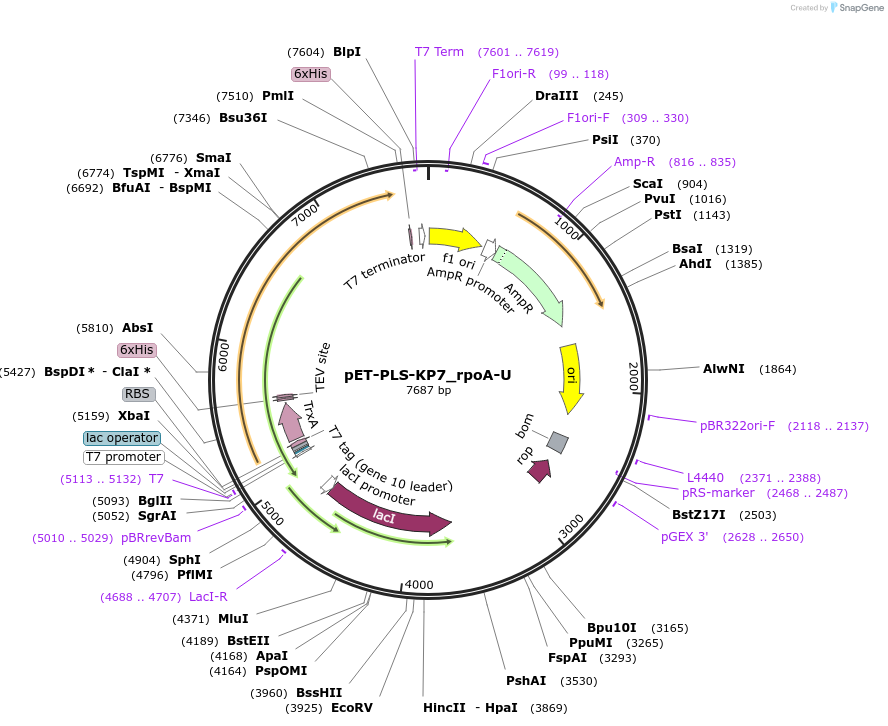
pET-PLS-KP7_rpoA-U
Plasmid#190974PurposeTest the U-to-C editing activity of KP7 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP7
TagsHisExpressionBacterialPromoterT7Available SinceOct. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
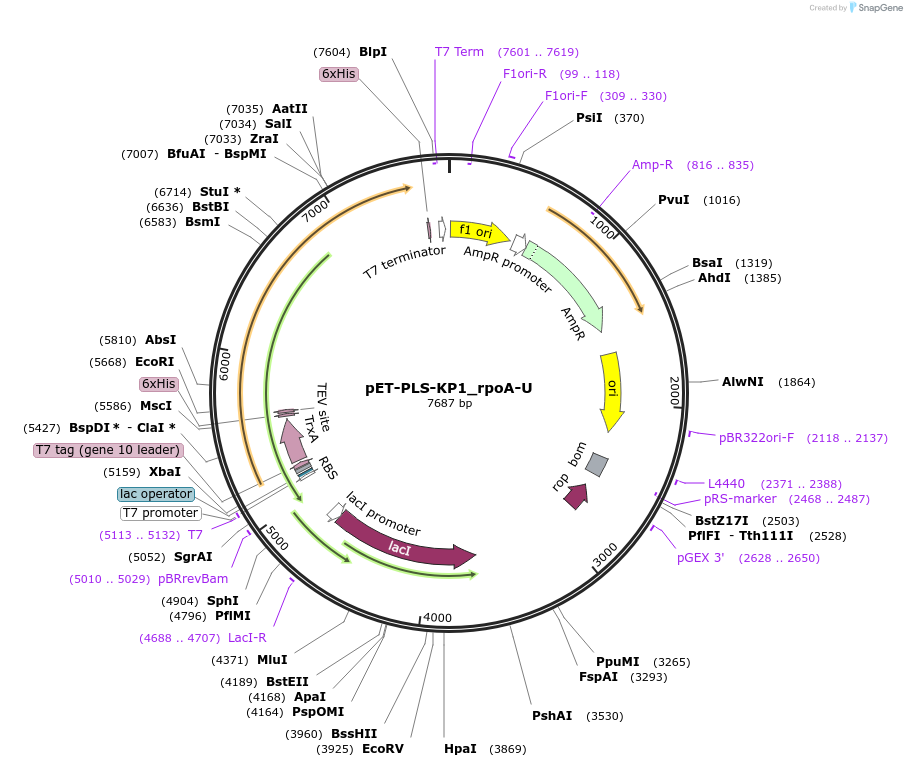
pET-PLS-KP1_rpoA-U
Plasmid#190955PurposeTest the U-to-C editing activity of KP1 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP1
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
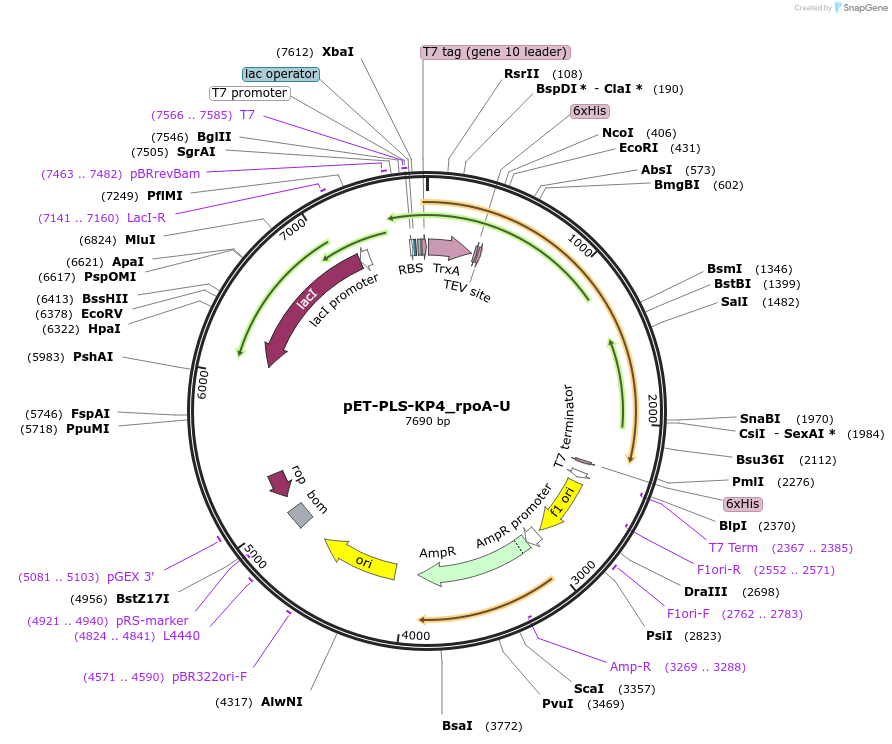
pET-PLS-KP4_rpoA-U
Plasmid#190964PurposeTest the U-to-C editing activity of KP4 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP4
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
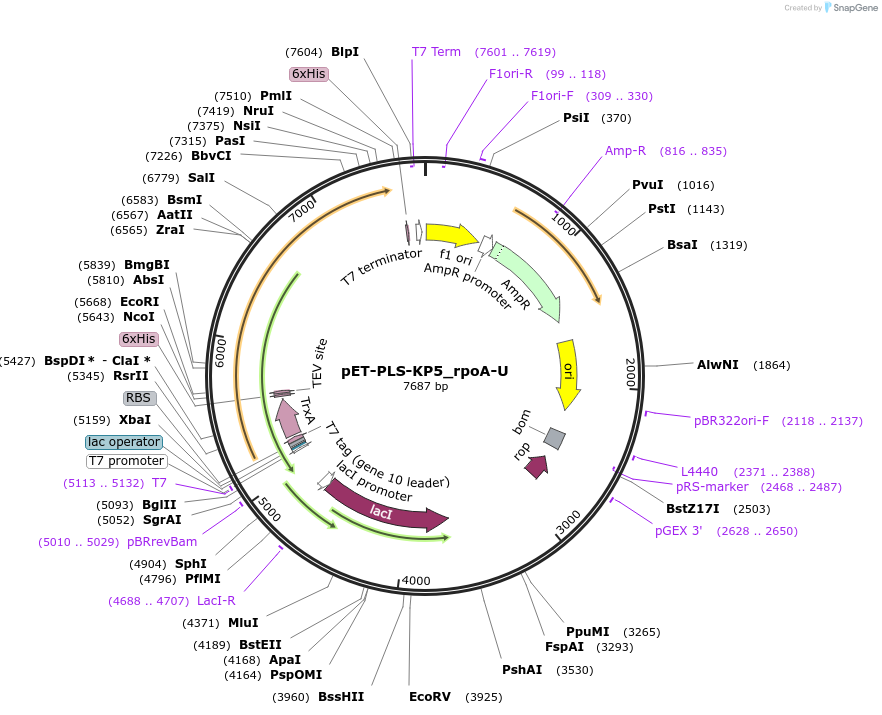
pET-PLS-KP5_rpoA-U
Plasmid#190965PurposeTest the U-to-C editing activity of KP5 on AtrpoA editing site, in E. coliDepositorInsertPLS-KP5
TagsHisExpressionBacterialPromoterT7Available SinceOct. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
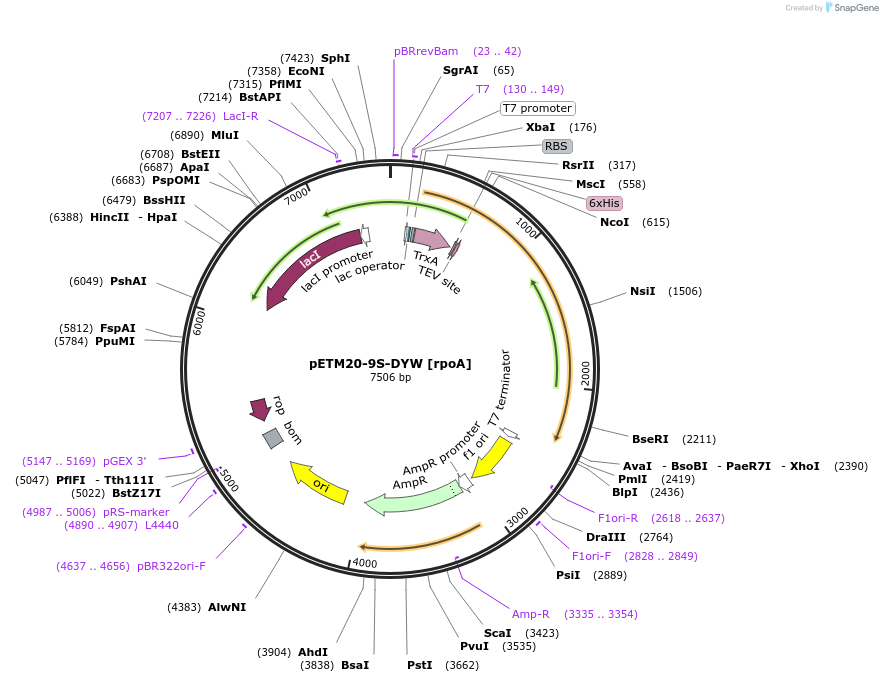
pETM20-9S-DYW [rpoA]
Plasmid#176220PurposeExpression of TRX-9S-DYW [rpoA]DepositorInsertTRX-9S-DYW [rpoA]
UseSynthetic BiologyTagsTRX-HisPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
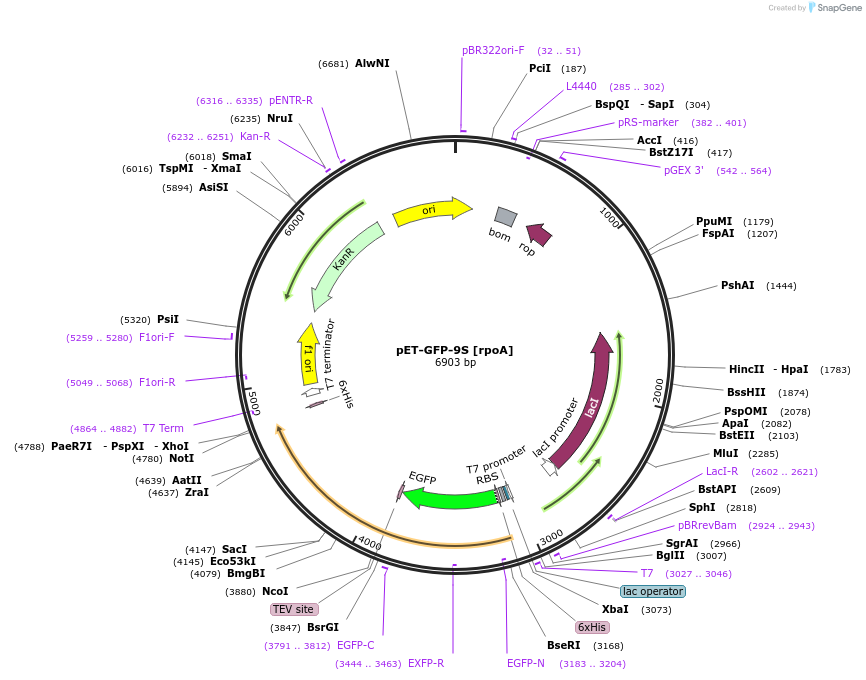
pET-GFP-9S [rpoA]
Plasmid#176221PurposeExpression of GFP-9S [rpoA]DepositorInsertGFP-9S [rpoA]
UseSynthetic BiologyTagsHis-GFPPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
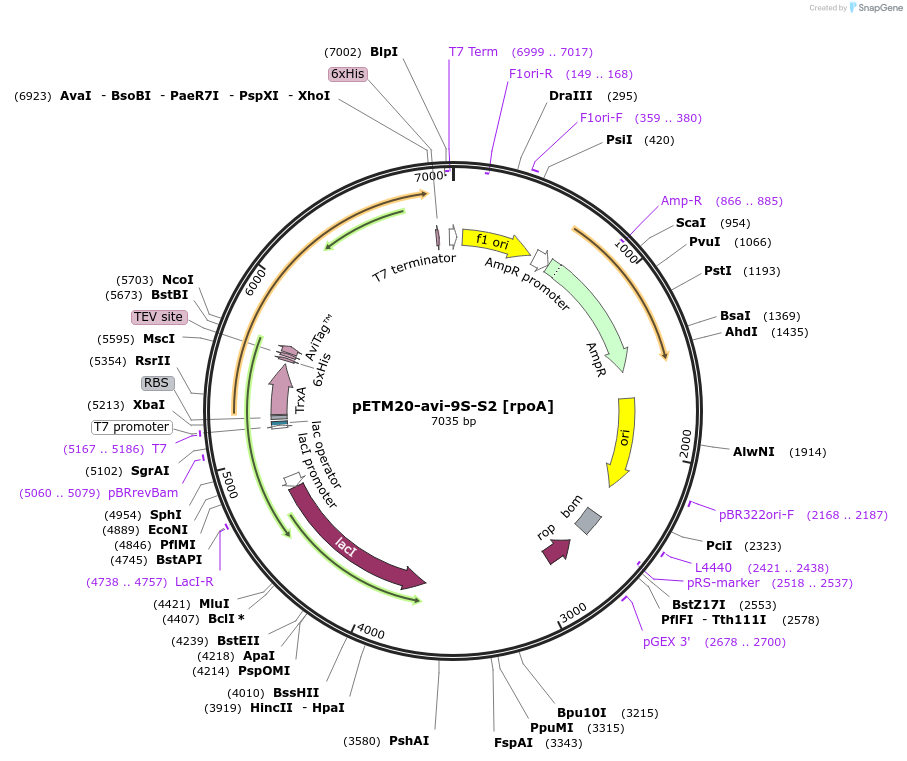
pETM20-avi-9S-S2 [rpoA]
Plasmid#176223PurposeExpression of TRX-AviTag-9S-S2 [rpoA]DepositorInsertTRX-Avi-9S-S2 [rpoA]
UseSynthetic BiologyTagsTRX-His-AviTagPromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
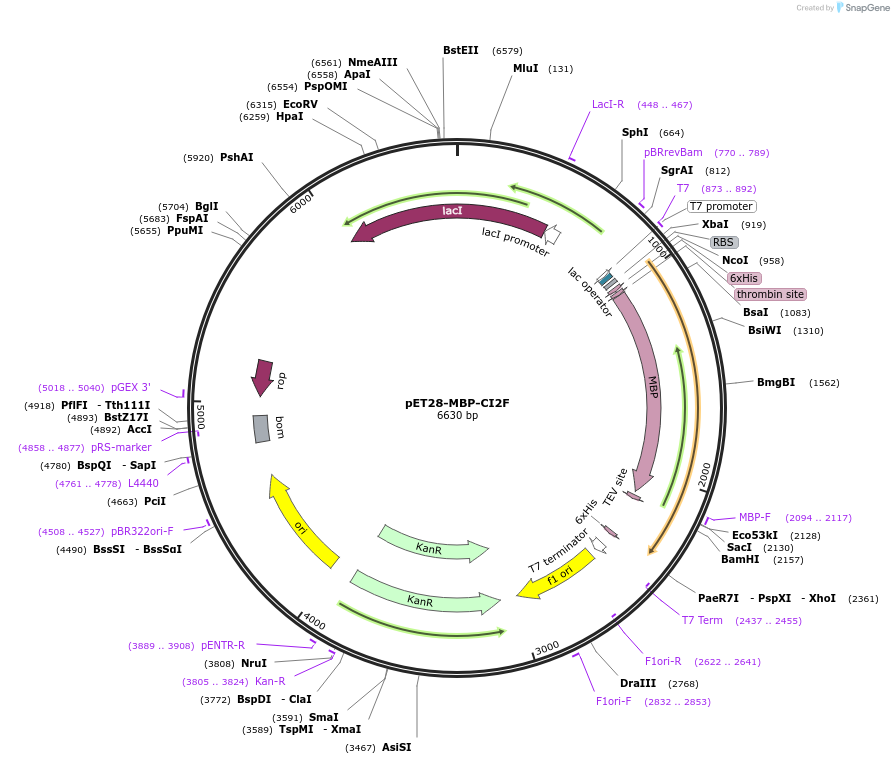
pET28-MBP-CI2F
Plasmid#191254PurposeExpresses chymotrypsin inhibitor with an N-terminal MBP fusion proteinDepositorInsertChymotrypsin Inhibitor 2
TagsMaltose Binding ProteinExpressionBacterialAvailable SinceOct. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
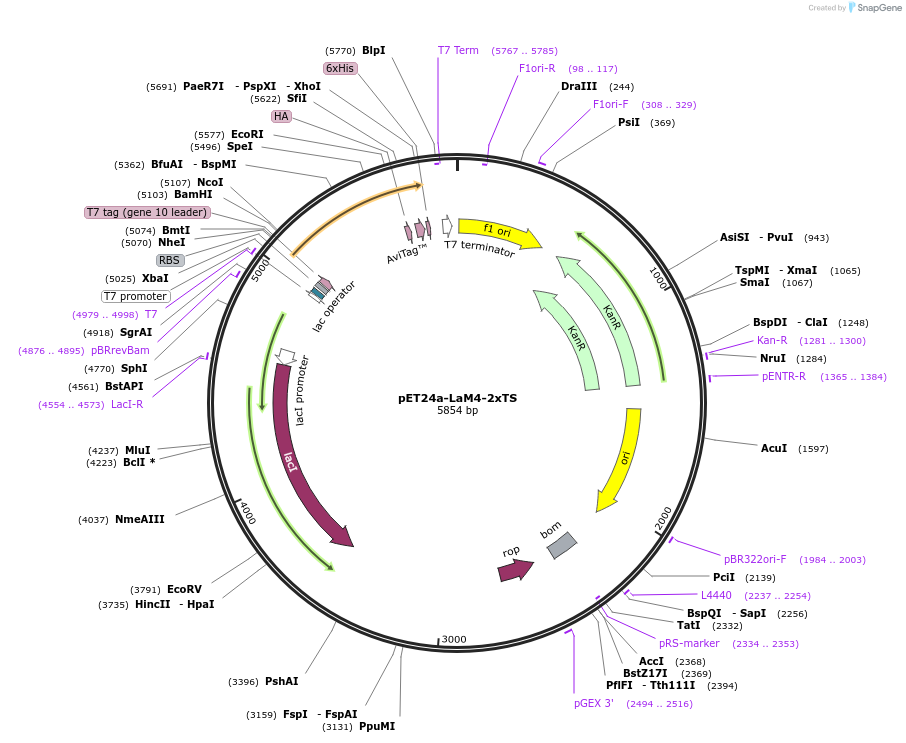
pET24a-LaM4-2xTS
Plasmid#162779PurposeBacterial expression of a functionalized anti-mCherry (LaM4) nanobody fused to two tyrosine sulfation (TS) motifs. LaM4-2xTS also contains a T7, HA, BAP and His6 epitopeDepositorInsertanti-mCherry nanobody fused to two TS sites, T7, HA, BAP and His6 epitope
TagsBAP, HA, His6, T7, and TS site from proCCKExpressionBacterialPromoterT7Available SinceOct. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
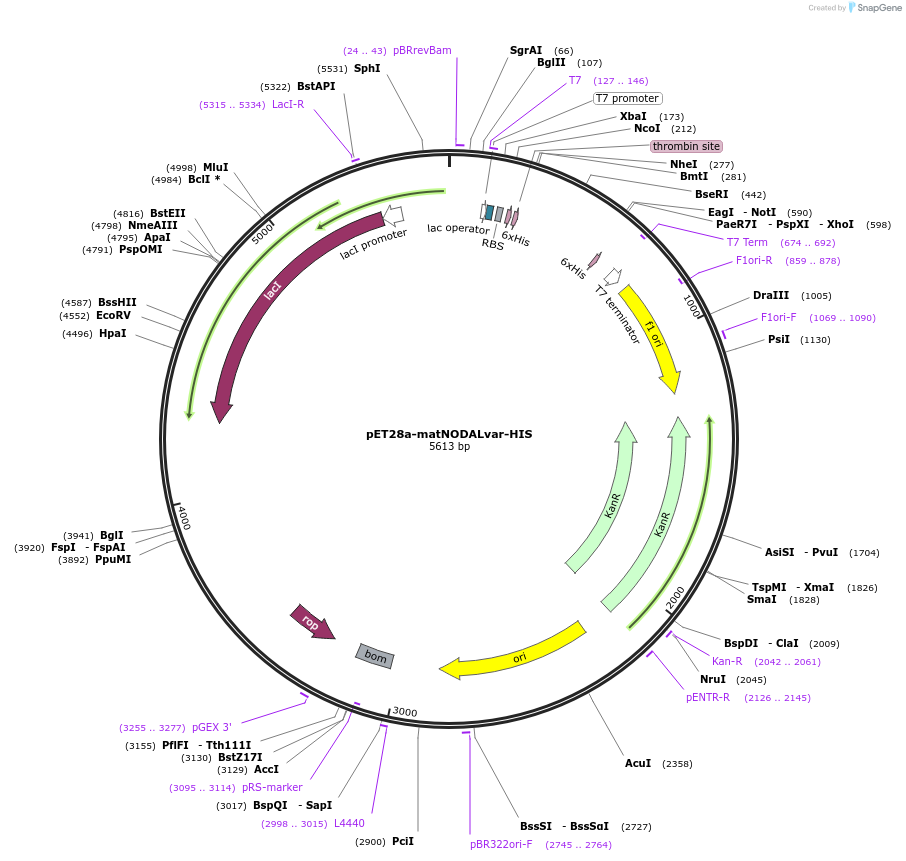
pET28a-matNODALvar-HIS
Plasmid#115265PurposeFor bacterial recombinant protein production of the mature peptide of a human NODAL splice variantDepositorAvailable SinceSept. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
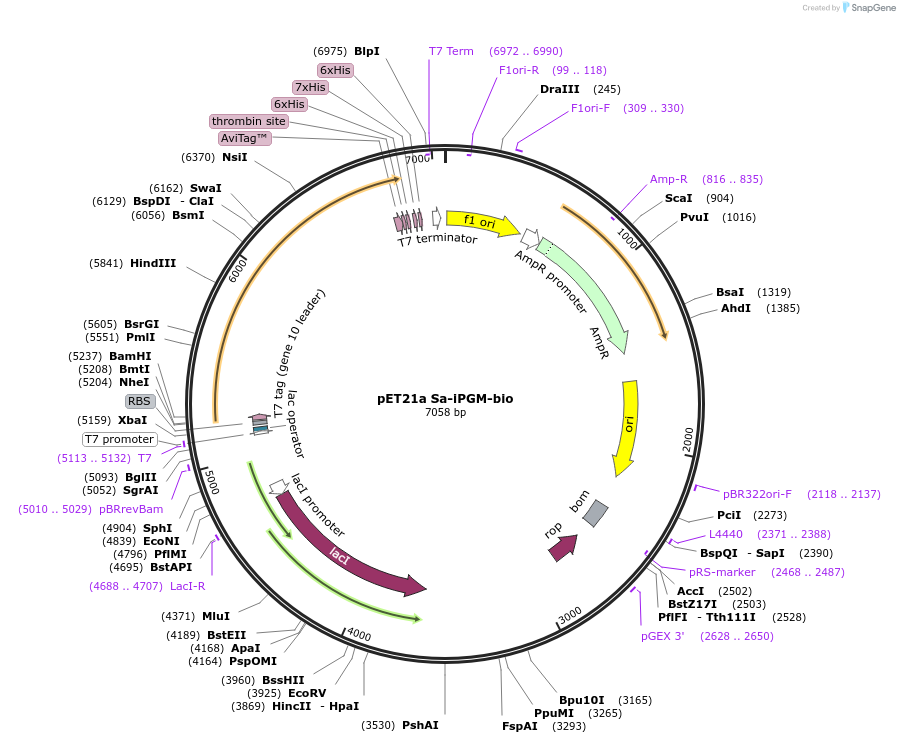
pET21a Sa-iPGM-bio
Plasmid#179523PurposeExpresses S. aureus iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertS. aureus iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
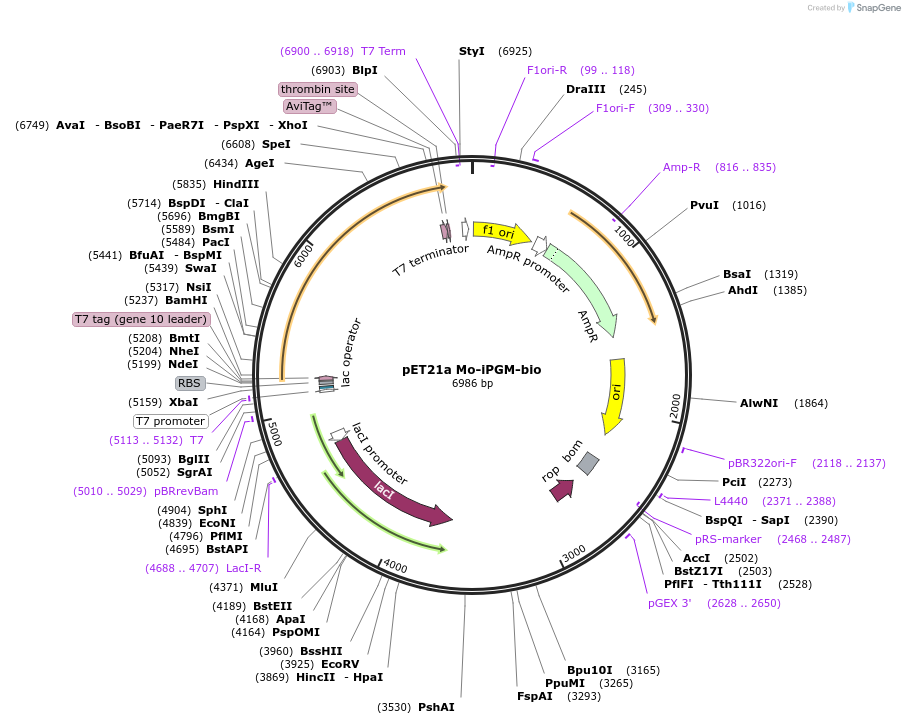
pET21a Mo-iPGM-bio
Plasmid#179614PurposeExpresses M. orale iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertM. orale iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
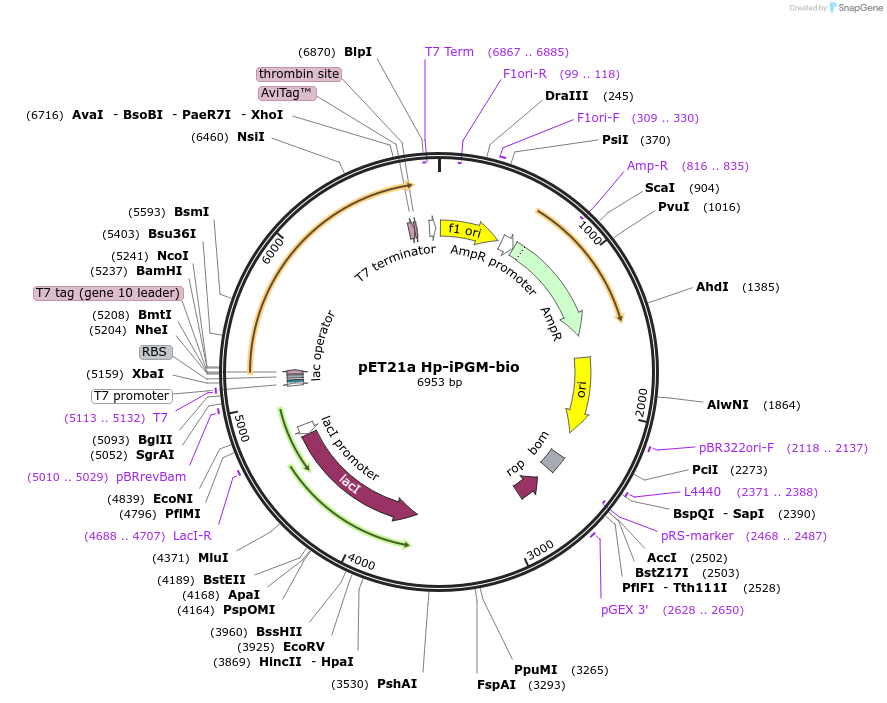
pET21a Hp-iPGM-bio
Plasmid#179615PurposeExpresses H. pylori iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertH. pylori iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only