We narrowed to 10,180 results for: yeast
-
Plasmid#200712PurposePromoter region from ATPase PMA1 as Pro5U promoter module for heterologous expression of plasma membrane localized transporters in yeastDepositorInsertPMA1 promoter (PMA1 Budding Yeast)
UseSynthetic Biology; Modular cloningMutationdomesticated for MoClo cloning (removal of intern…Available SinceAug. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAG306-pTet07-synYFP[TTG/AGA]-4xArg[CGA]-nLuc
Plasmid#199759PurposeElongation reporter construct to quantify elongation duration of 4_CGA stallingDepositorInsertsynYFP[TTG/AGA]-4xArg[CGA]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
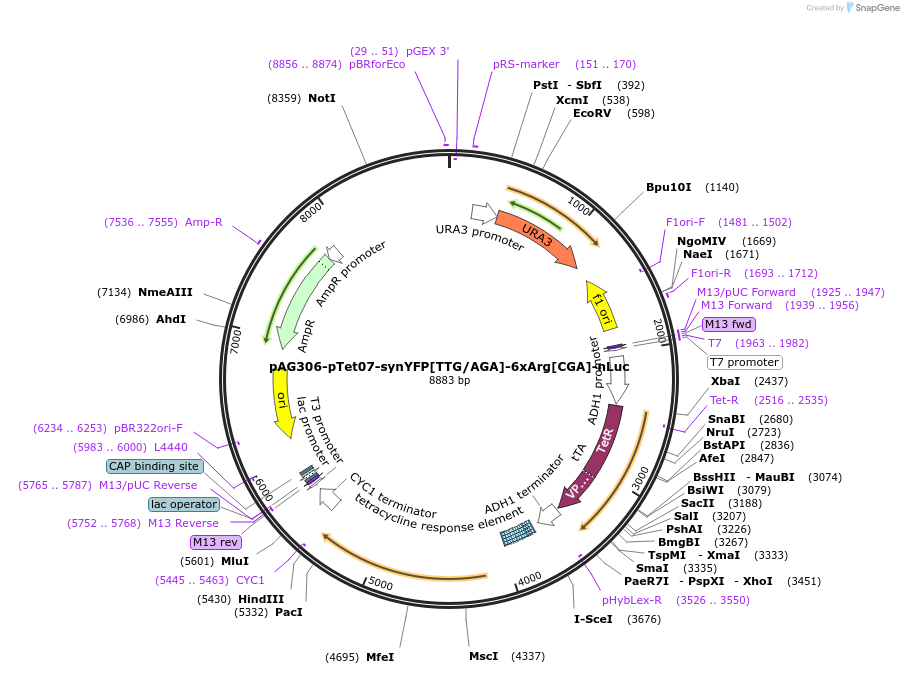
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[CGA]-nLuc
Plasmid#199760PurposeElongation reporter construct to quantify elongation duration of 6_CGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[CGA]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
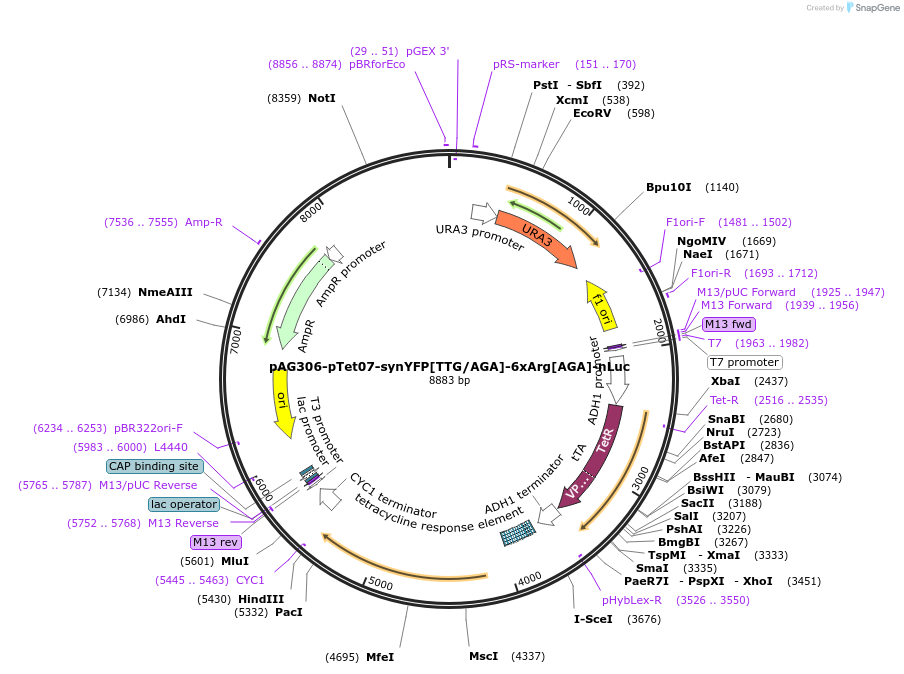
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[AGA]-nLuc
Plasmid#199761PurposeElongation reporter construct to quantify elongation duration of 6_AGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[AGA]-cLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
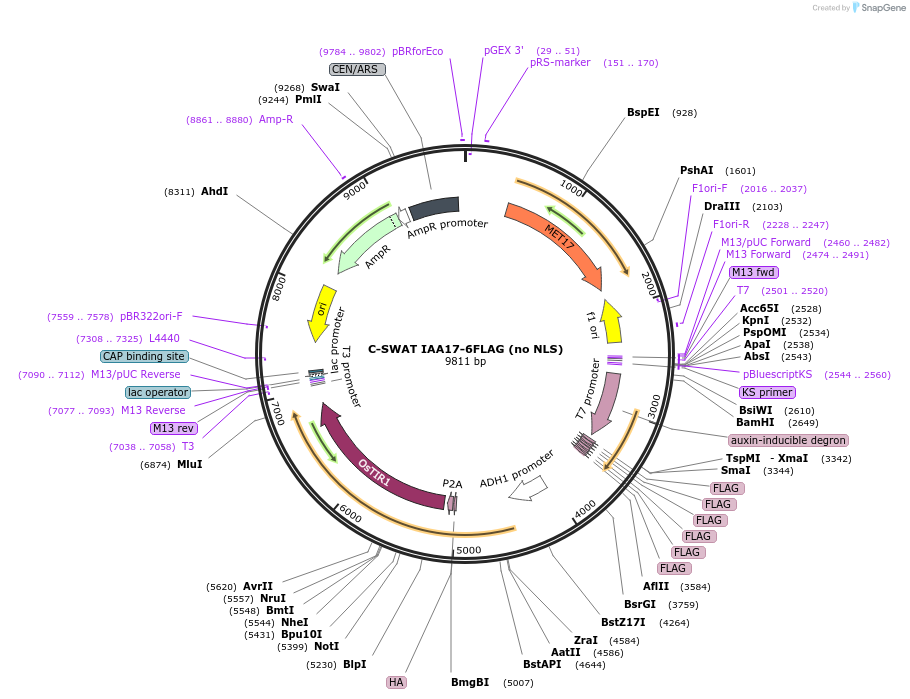
C-SWAT IAA17-6FLAG (no NLS)
Plasmid#193322PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin-Inducible Degron) and 6FLAG. ARF16(PB1)-P2A-osTIR1 has SIR4 homology arms for genome integration and does not have NLS tags..DepositorInsertIAA17
Tags6FLAGExpressionYeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
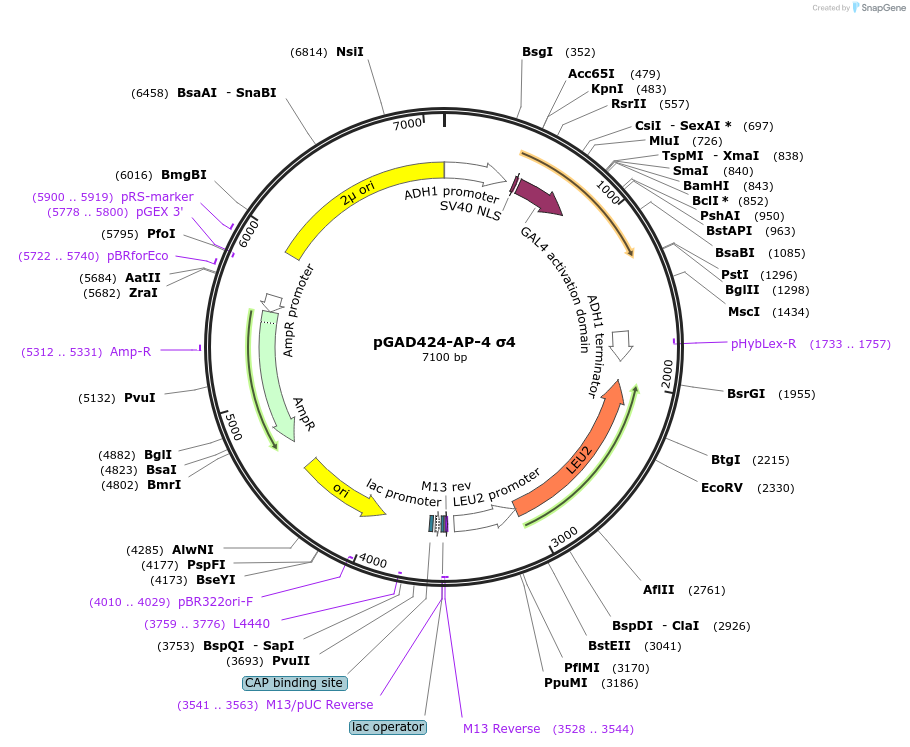
pGAD424-AP-4 σ4
Plasmid#198328PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-4 σ4 fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-4 σ4
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastPromoterADH1Available SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
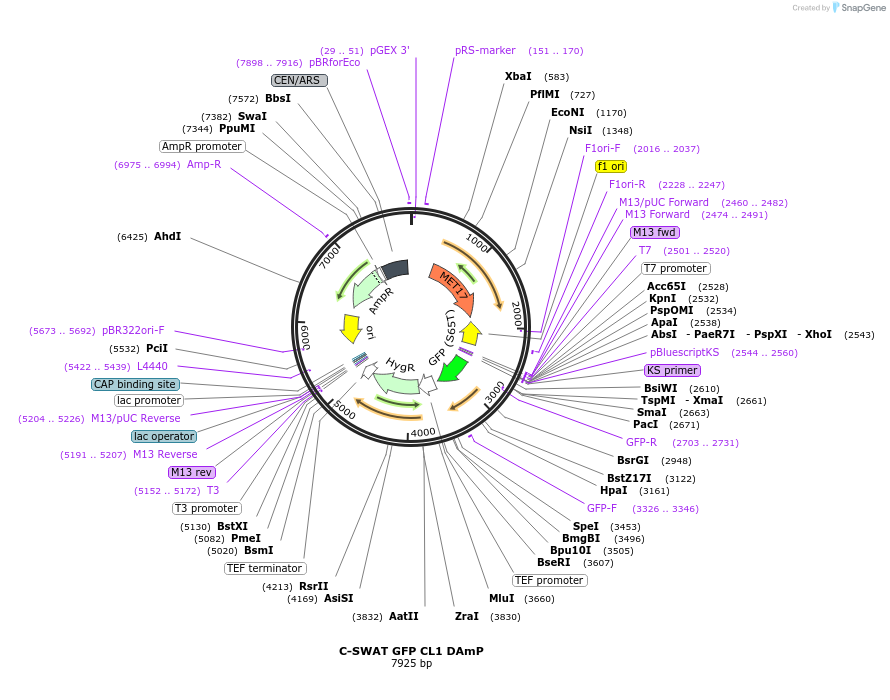
C-SWAT GFP CL1 DAmP
Plasmid#182513PurposeC-SWAT compatible vector for C-terminal tagging with GFP CL1 followed by the DAmP cassette. The stop codon after the CL1 degron is immediately followed by the Hygromycin B (Hph) resistance cassette.DepositorInsertCL1 and DAmP
TagsGFPExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
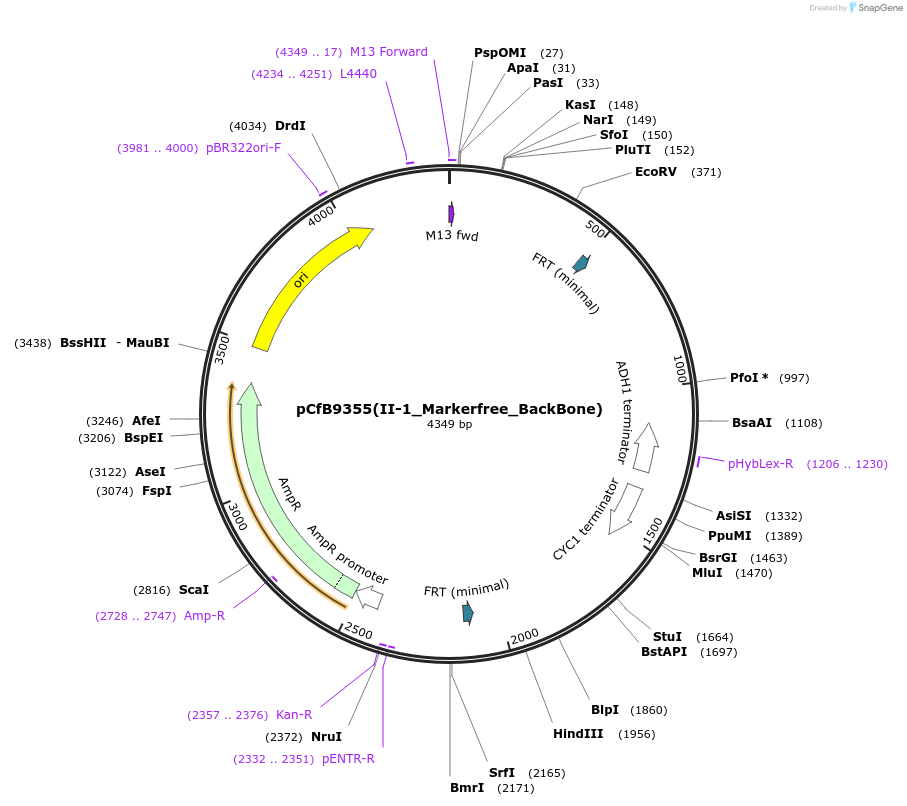
pCfB9355(II-1_Markerfree_BackBone)
Plasmid#161597PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site II-1, (Chr II: 341622..341641)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
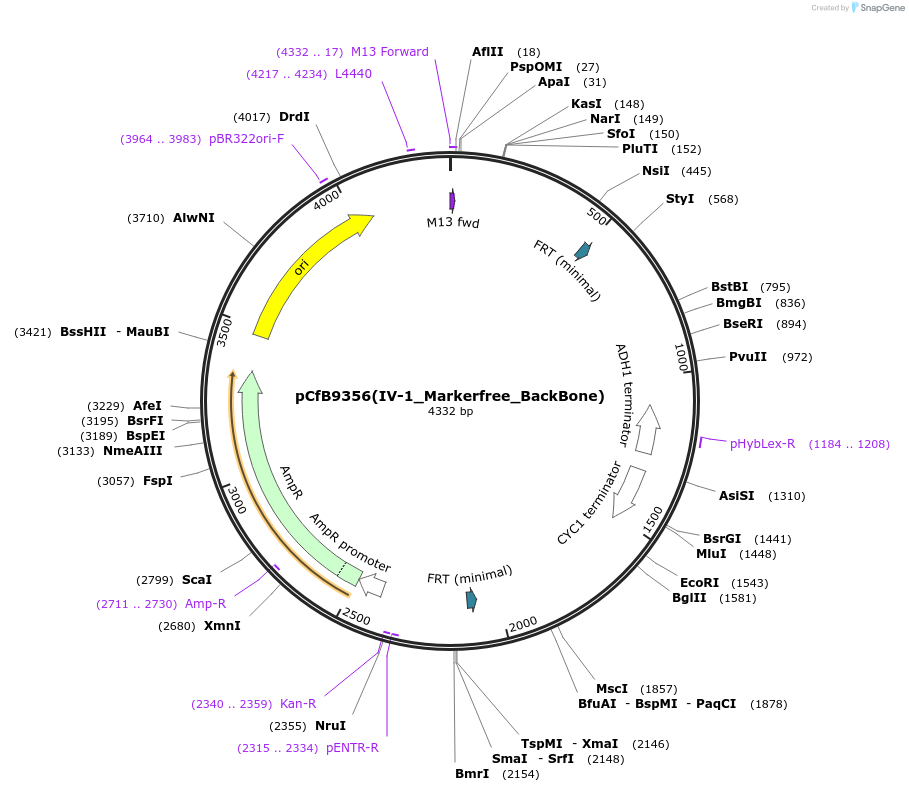
pCfB9356(IV-1_Markerfree_BackBone)
Plasmid#161598PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site IV-1, (Chr IV: 132921..132940)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
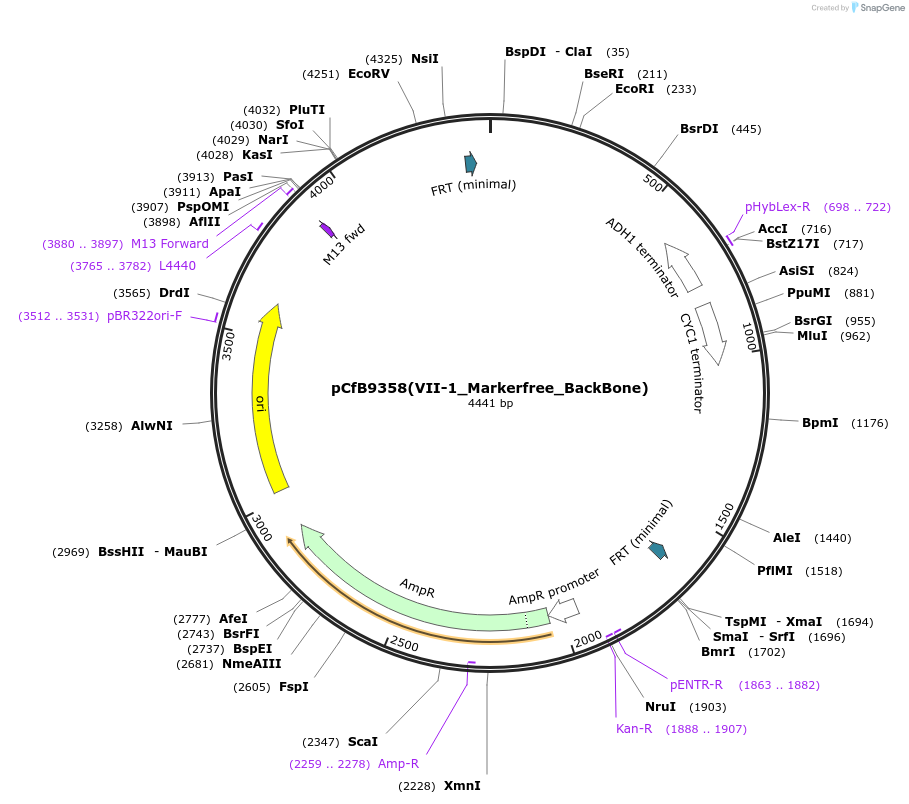
pCfB9358(VII-1_Markerfree_BackBone)
Plasmid#161599PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site VII-1, (Chr VII: 438509..438490)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
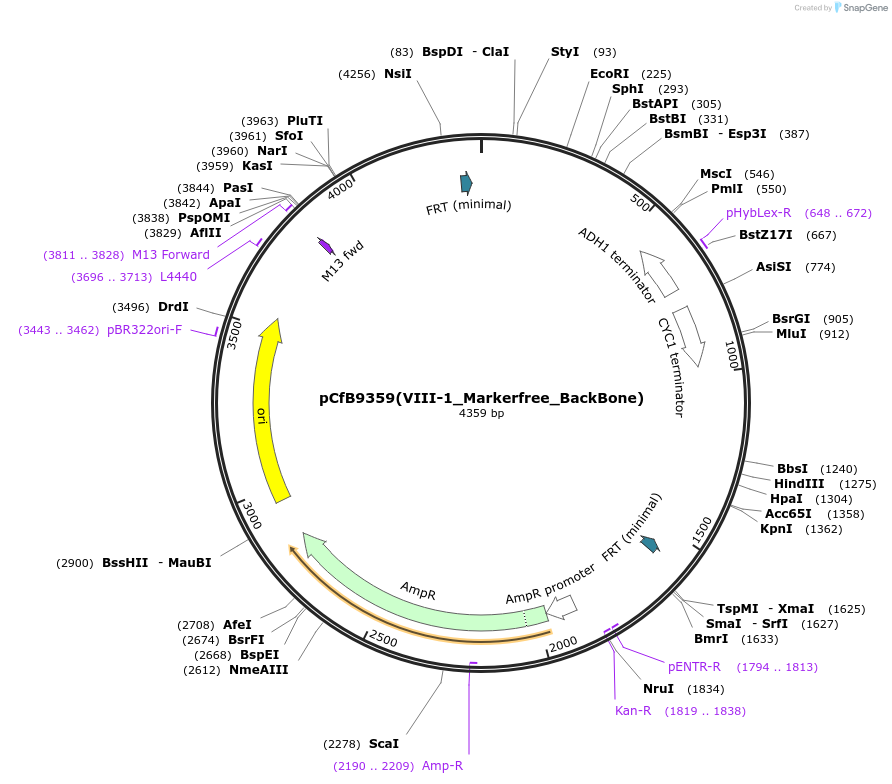
pCfB9359(VIII-1_Markerfree_BackBone)
Plasmid#161600PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site VIII-1, (Chr VIII: 293615..293634)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
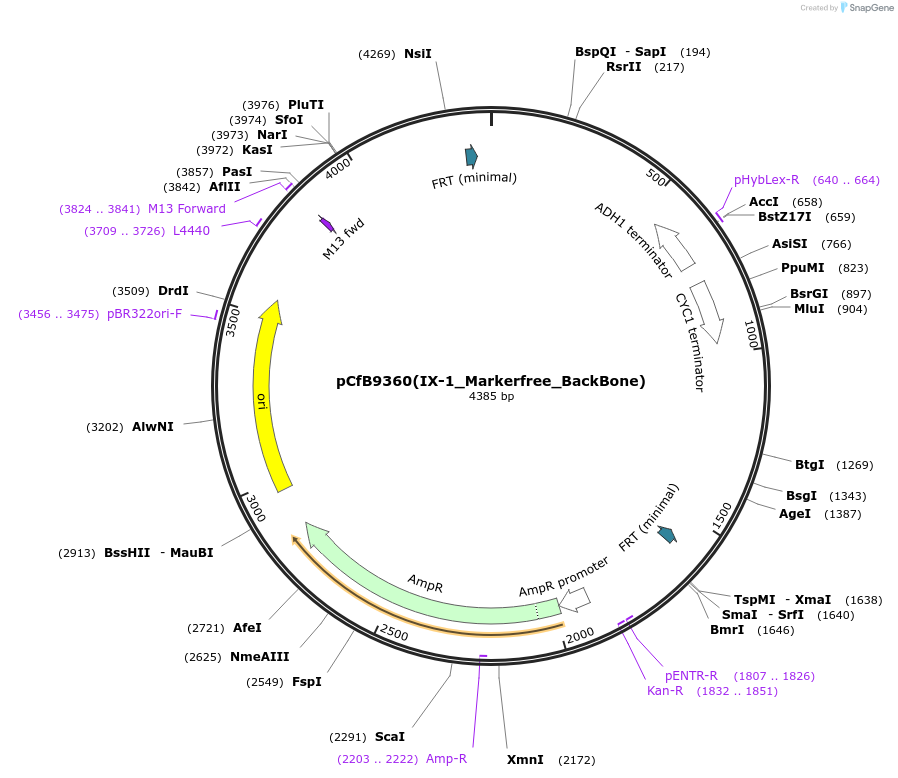
pCfB9360(IX-1_Markerfree_BackBone)
Plasmid#161601PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site IX-1, (Chr IX: 388864..388883)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
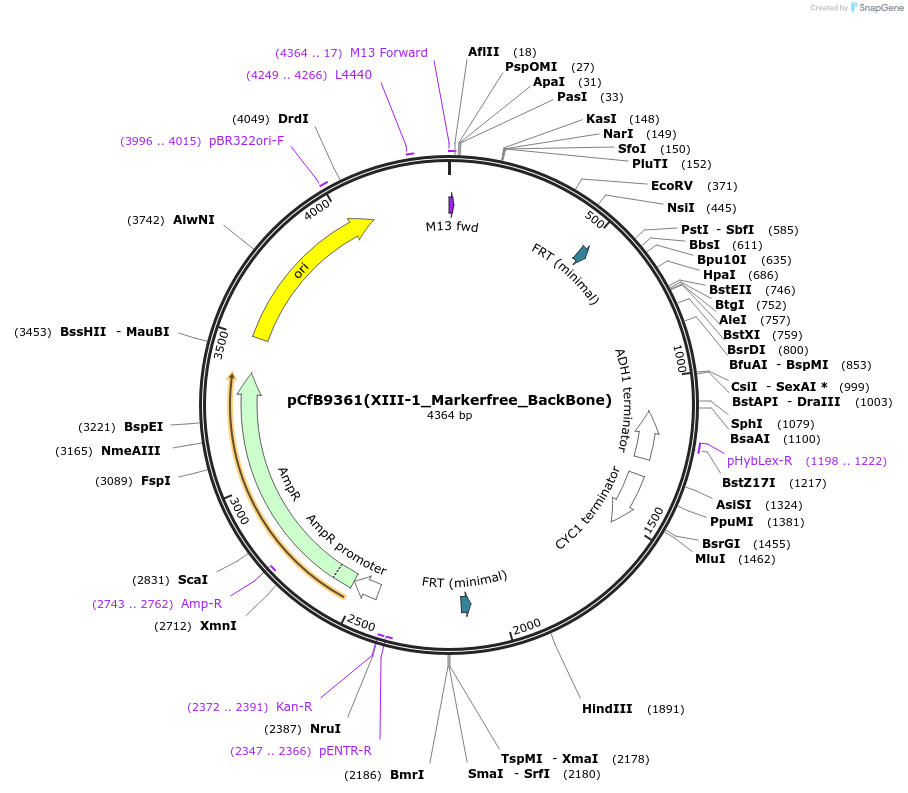
pCfB9361(XIII-1_Markerfree_BackBone)
Plasmid#161602PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site XIII-1, (Chr XIII: 306804..306823)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
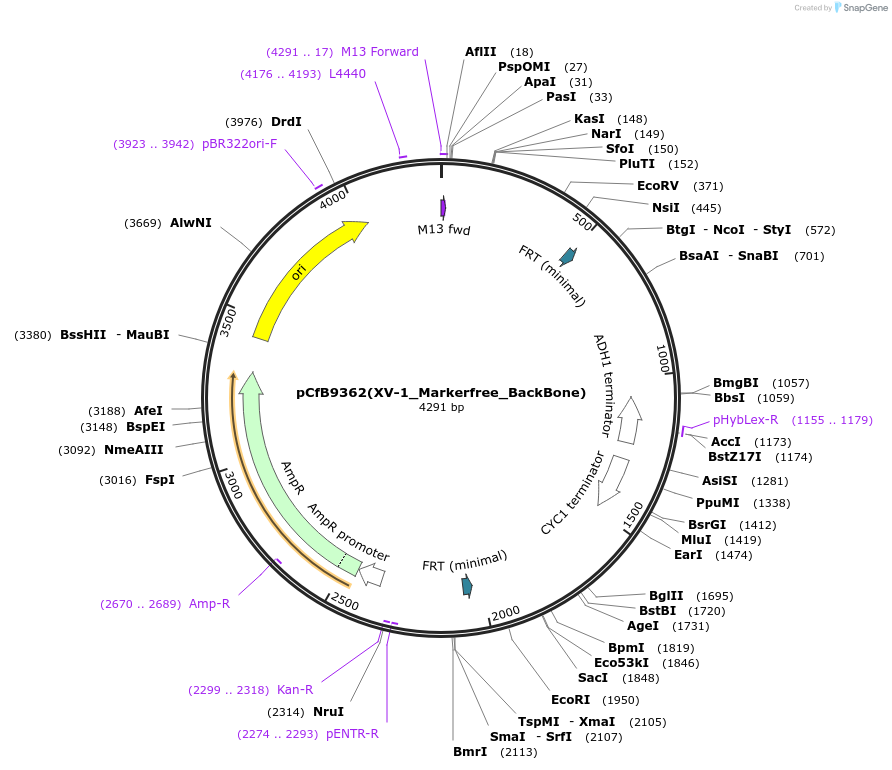
pCfB9362(XV-1_Markerfree_BackBone)
Plasmid#161603PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site XV-1, (Chr XV: 93186..93205)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
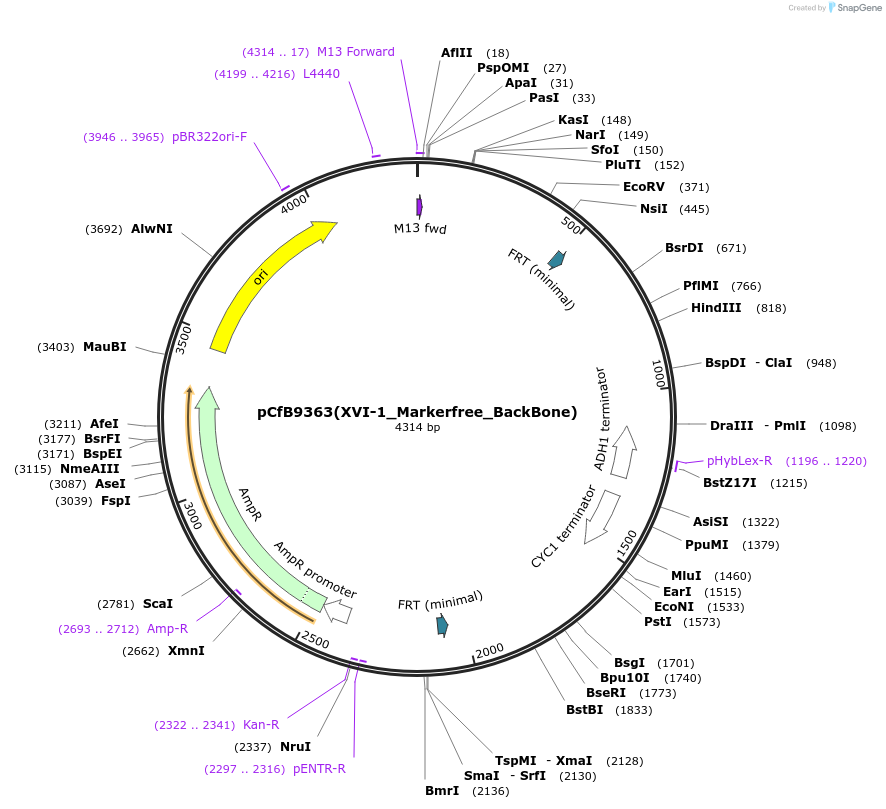
pCfB9363(XVI-1_Markerfree_BackBone)
Plasmid#161604PurposeEasyClone-MarkerFree backbone vector for the addition of 1 or 2 genes and promoters for integration into site XVI-1, (Chr XVI: 406267..406286)DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceAug. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
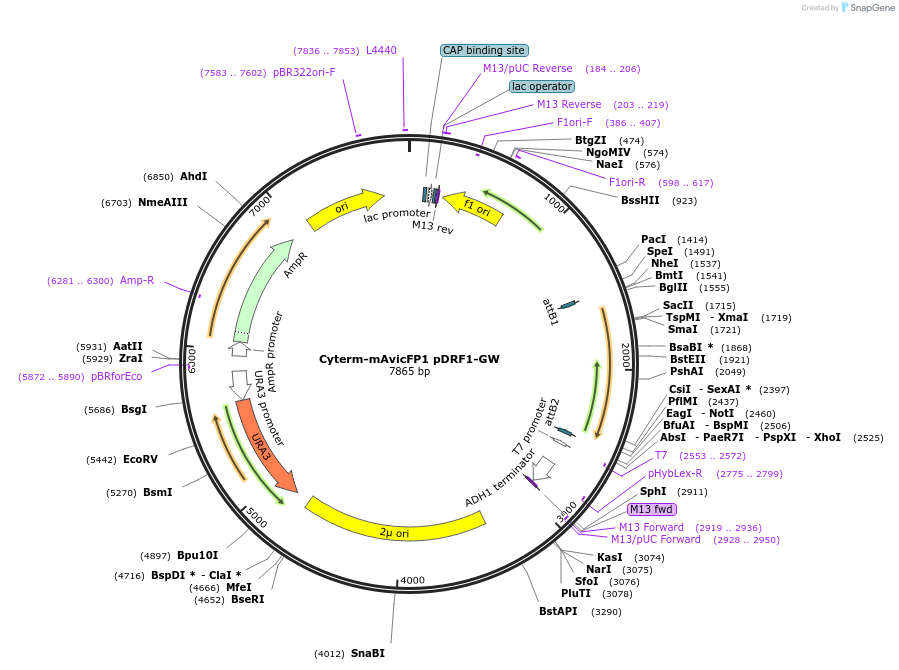
Cyterm-mAvicFP1 pDRF1-GW
Plasmid#168092PurposeExpresses Cyterm-mAvicFP1 to check for monomericity.DepositorInsertmAvicFP1
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
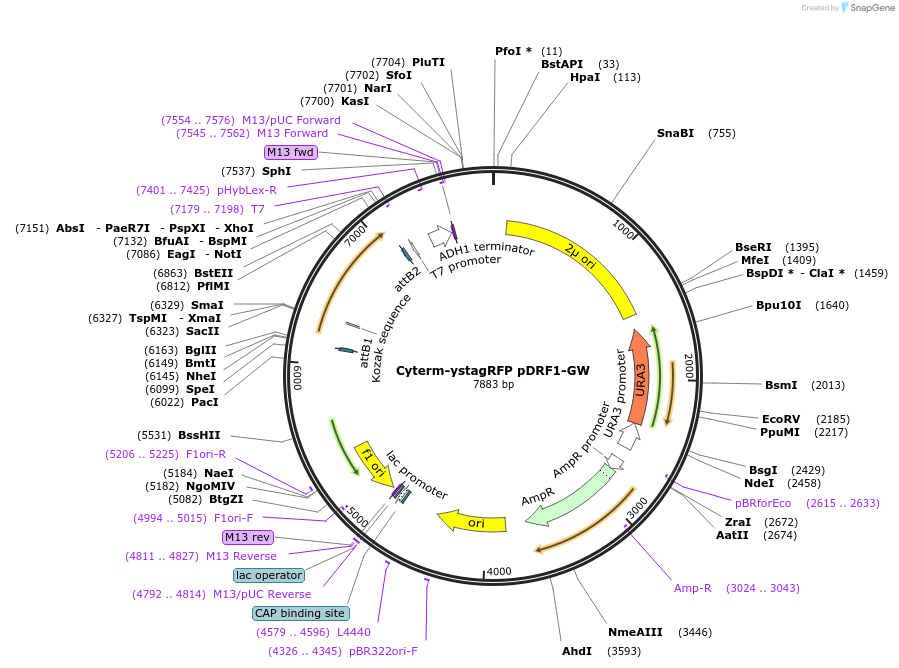
Cyterm-ystagRFP pDRF1-GW
Plasmid#168061PurposeExpresses Cyterm-ystagRFP to check for monomericity.DepositorInsertCyterm-ystagRFP
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
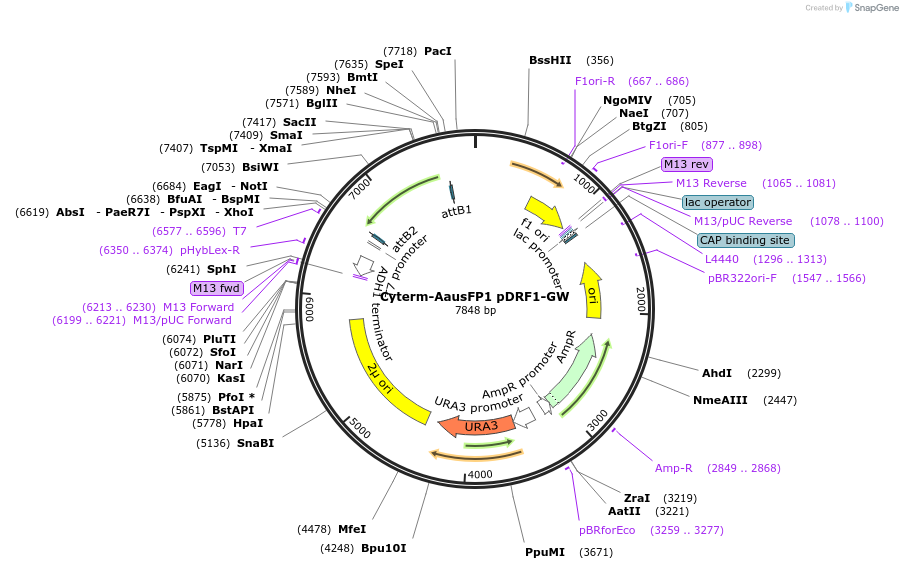
Cyterm-AausFP1 pDRF1-GW
Plasmid#168062PurposeExpresses Cyterm-AausFP1 to check for monomericity.DepositorInsertCyterm-AausFP1
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
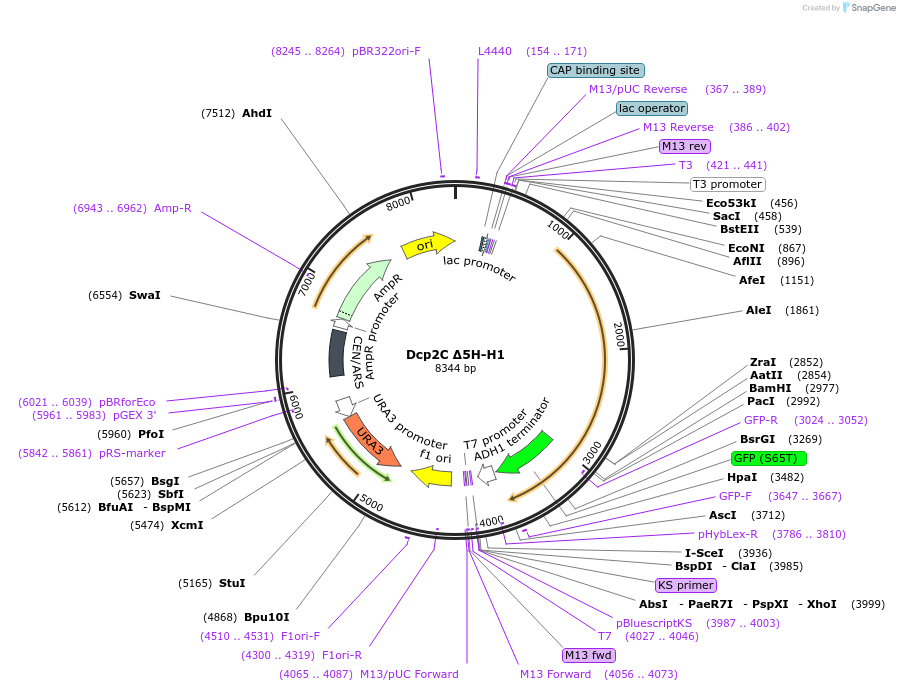
Dcp2C Δ5H-H1
Plasmid#163584Purposeexpress Dcp2 FL-H1 at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationTruncation of aa1-326; the first HLM (EDQLKSYAEEQ…PromoterDCP2 promoterAvailable SinceApril 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
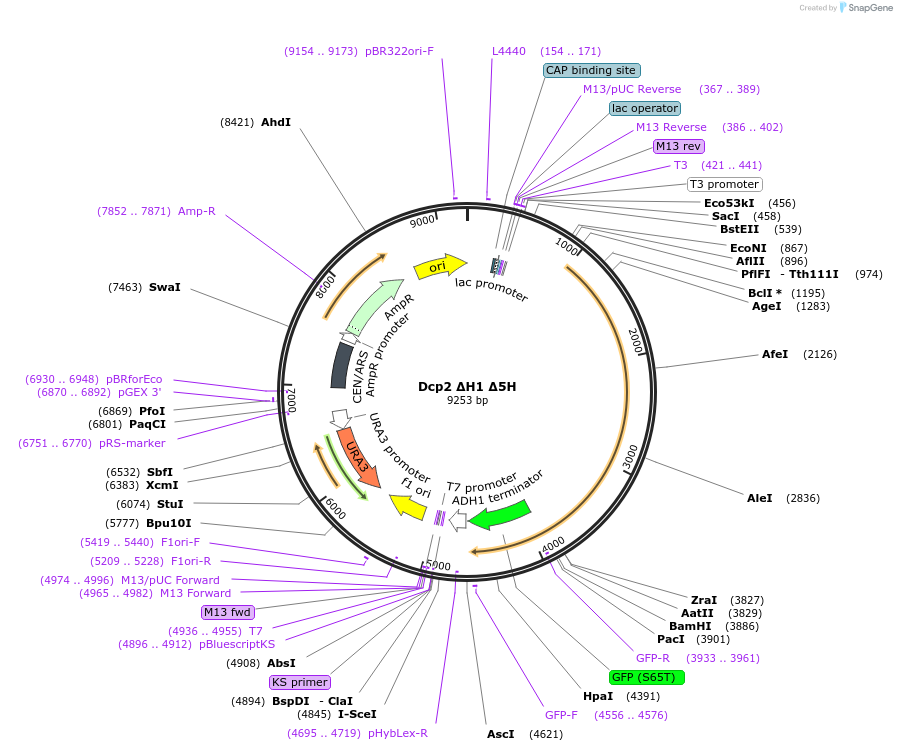
Dcp2 ΔH1 Δ5H
Plasmid#163582Purposeexpress Dcp2 ΔH1 Δ5H at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationL255A; mutate 440-450, 489-500, 701-708, 827-831,…PromoterDCP2 promoterAvailable SinceApril 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
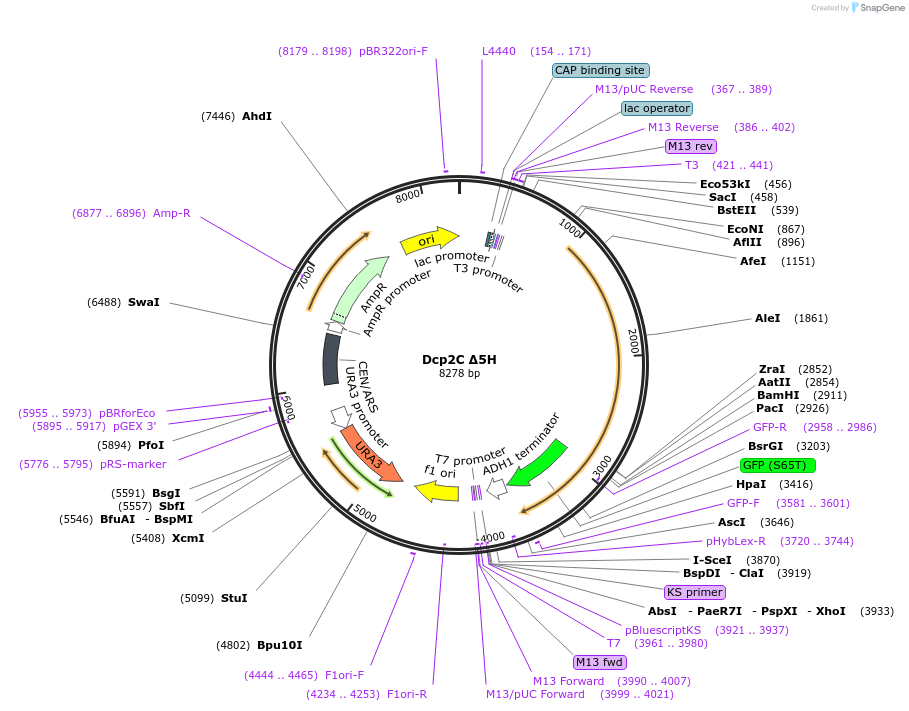
Dcp2C Δ5H
Plasmid#163581Purposeexpress Dcp2C Δ5H at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationTruncation of aa1-300; mutate aa440-450, aa489-50…PromoterDCP2 promoterAvailable SinceApril 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
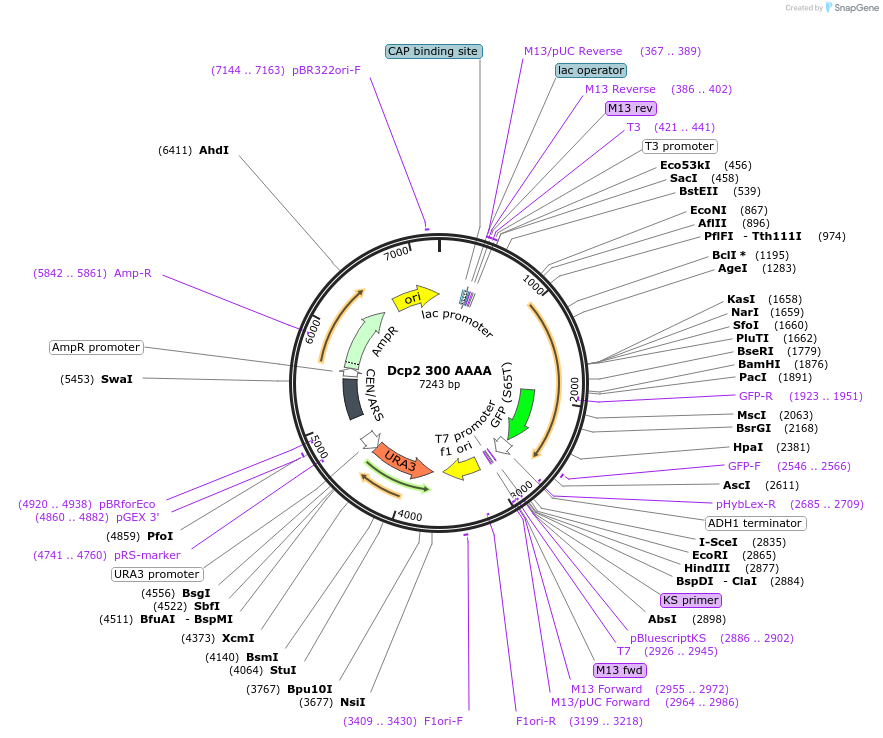
Dcp2 300 AAAA
Plasmid#163579Purposeexpress Dcp2 300 AAAA at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationTruncation of C-terminal domain (301-970); mutati…PromoterDCP2 promoterAvailable SinceApril 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
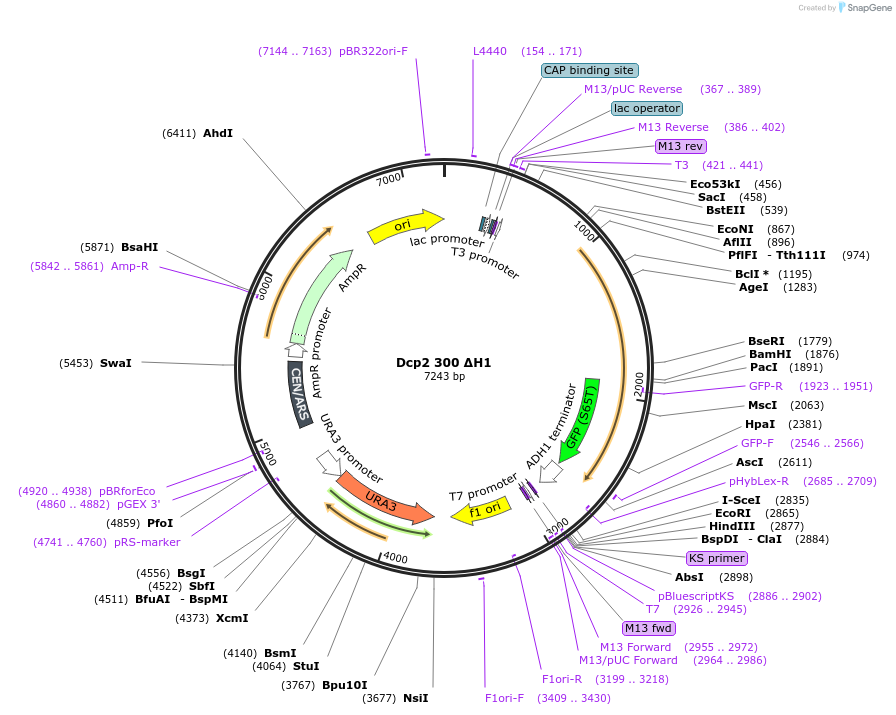
Dcp2 300 ΔH1
Plasmid#163578Purposeexpress Dcp2 300 ΔH1 at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationTruncation of C-terminal domain (301-970); Lysine…PromoterDCP2 promoterAvailable SinceApril 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
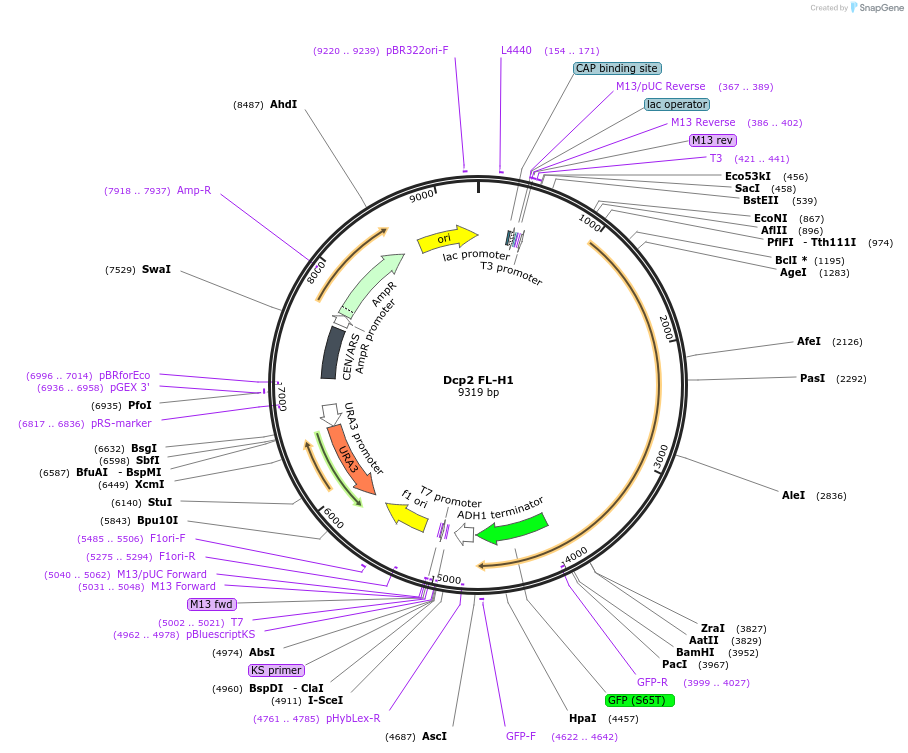
Dcp2 FL-H1
Plasmid#163583Purposeexpress Dcp2 FL-H1 at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationThe first HLM (EDQLKSYAEEQLKLLLGITKEE) is added t…PromoterDCP2 promoterAvailable SinceApril 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
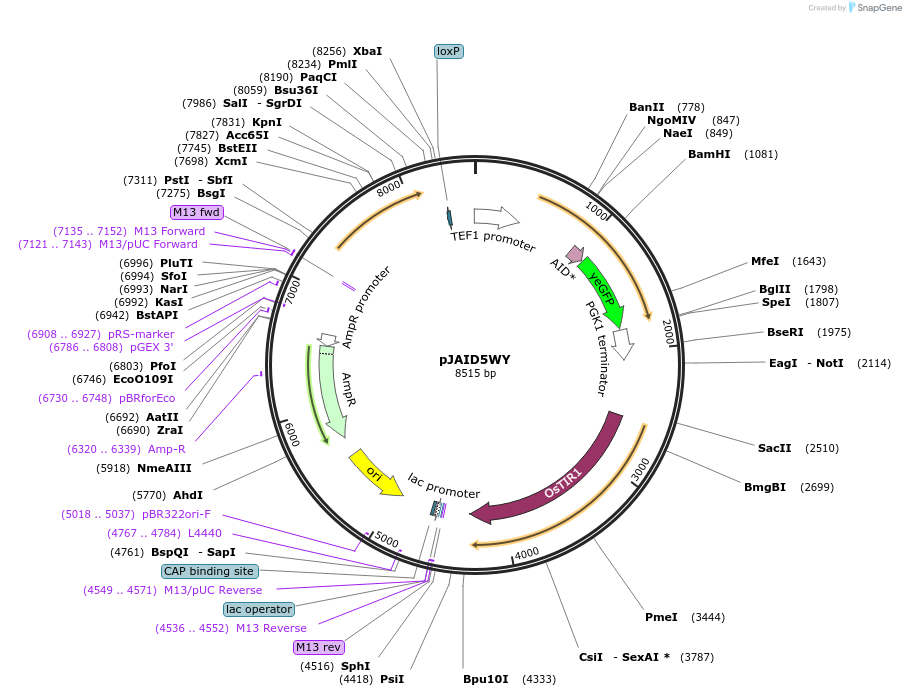
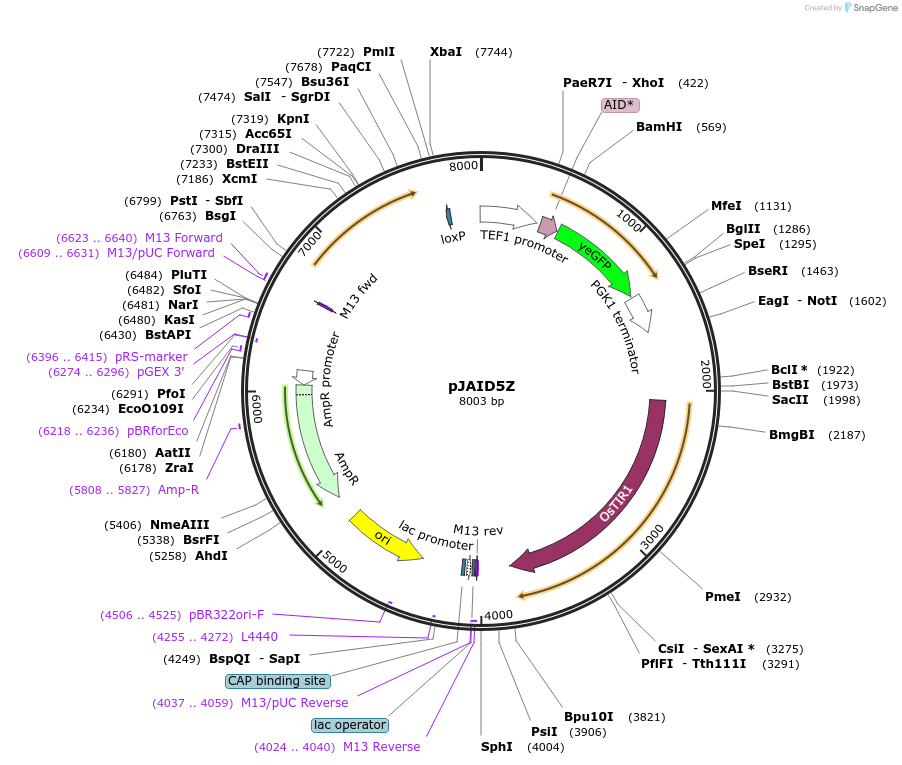
pJAID5WY
Plasmid#165050PurposeIntegrative expression of SOD1-AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>SOD1-AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationOsTIR1 is codon-optimised for expression in S. ce…Available SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
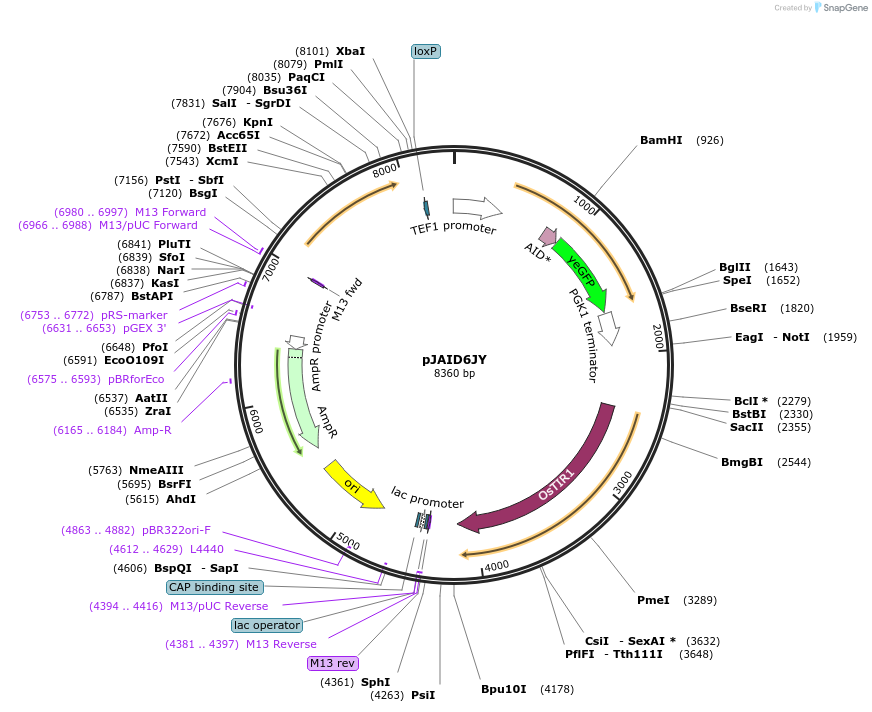
pJAID6JY
Plasmid#165049PurposeIntegrative expression of TRX1-AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>TRX1-AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
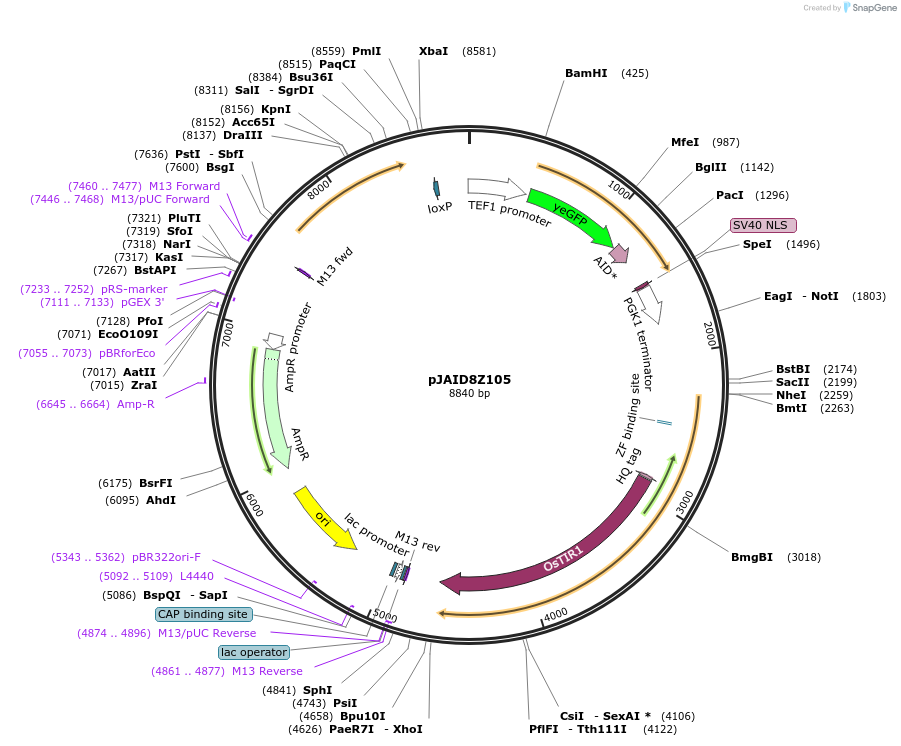
pJAID8Z105
Plasmid#165057PurposeIntegrative expression of nucleus-targeting yEGFP-AID*-CUP1 and SKP1-fusing rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>AID*>CUP1>SV40NSL>T-PGK1-P-ACS2>SKP1>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
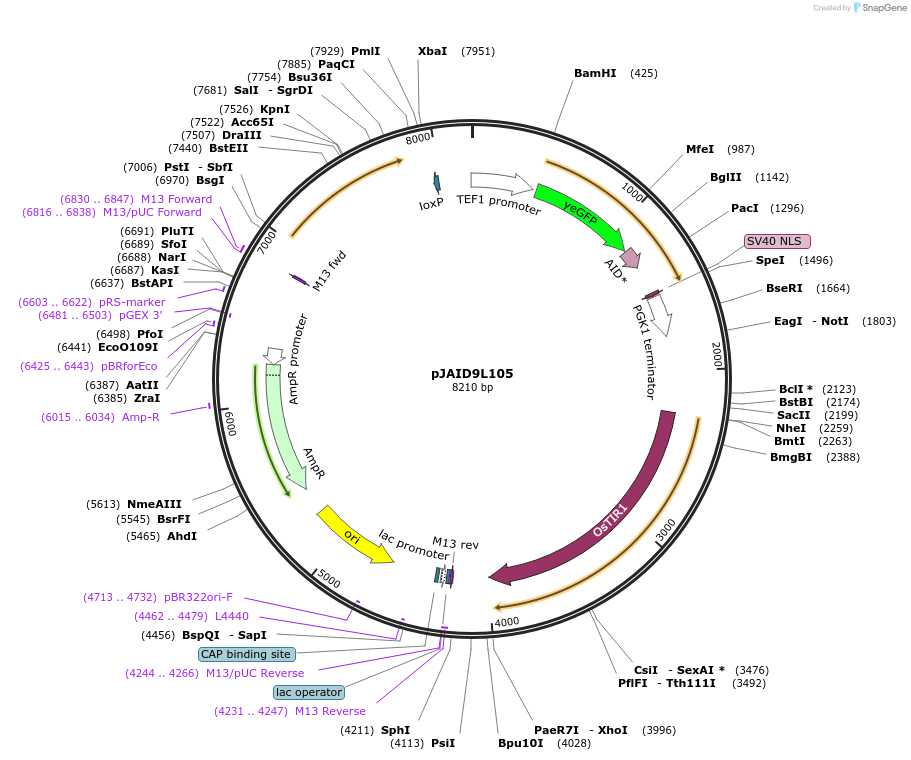
pJAID9L105
Plasmid#165055PurposeIntegrative expression of nucleus-targeting yEGFP-AID*-CUP1 and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>AID*>CUP1>SV40NSL>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
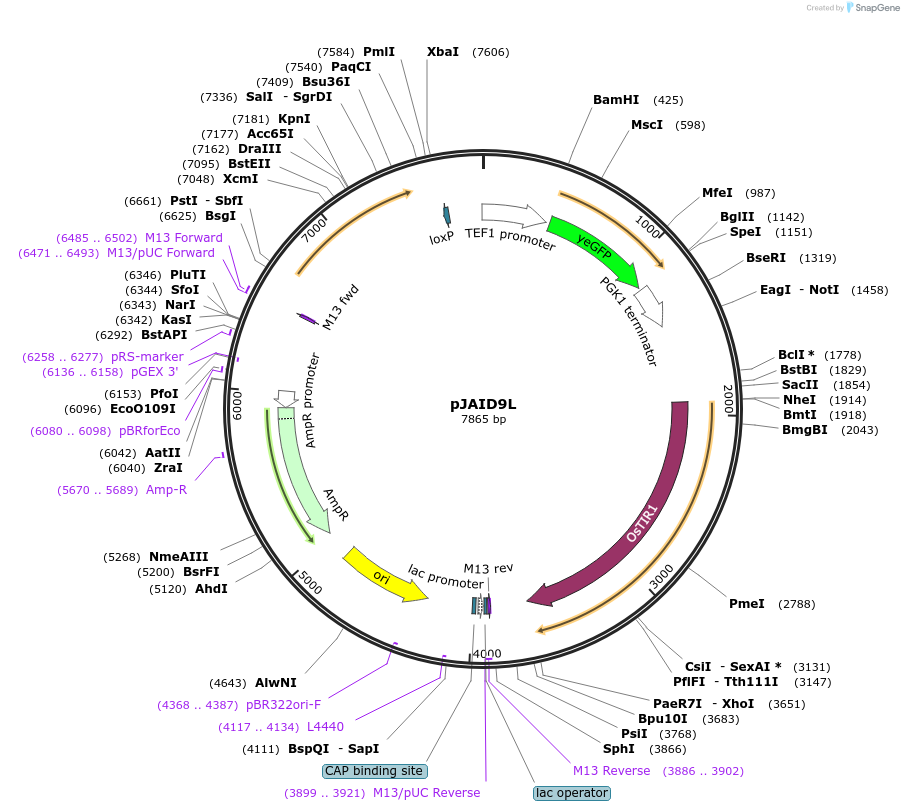
pJAID9L
Plasmid#165052PurposeIntegrative expression of yEGFP and rice auxin receptor gene OsTIR1. OsTIR1 is codon-optimised for expression in S. cerevisiaeDepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJAID5Z
Plasmid#165047PurposeIntegrative expression of AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationOsTIR1 is codon-optimised for expression in S. ce…Available SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
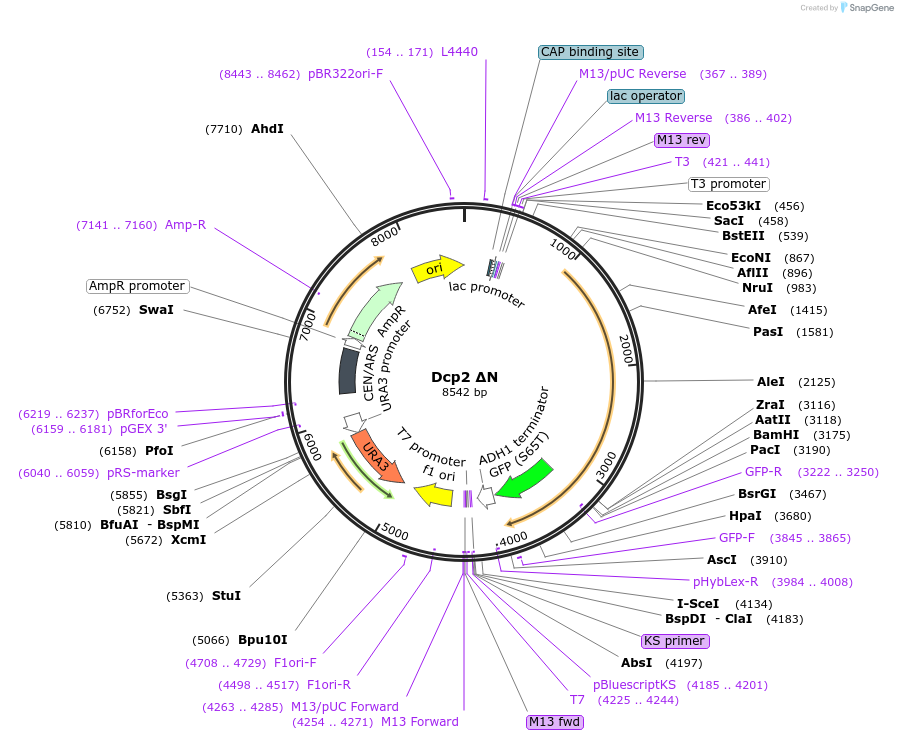
Dcp2 ΔN
Plasmid#163575Purposeexpress wild type Dcp2 ΔN(243-970) at endogenous level in S.cerevisiaeDepositorInsertDcp2 (DCP2 Budding Yeast)
TagsGFPExpressionYeastMutationTruncation of N-terminal domain (1-242) of Dcp2PromoterDCP2 promoterAvailable SinceMarch 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
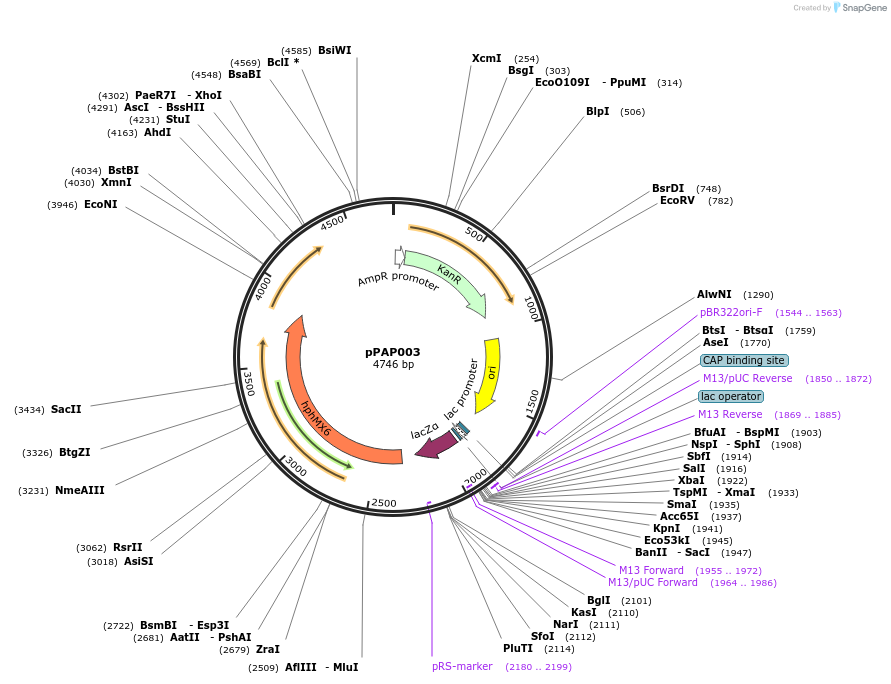
pPAP003
Plasmid#153490PurposeLevel 2 integrative plasmid for stable expression in P. pastorisDepositorTypeEmpty backboneExpressionYeastAvailable SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
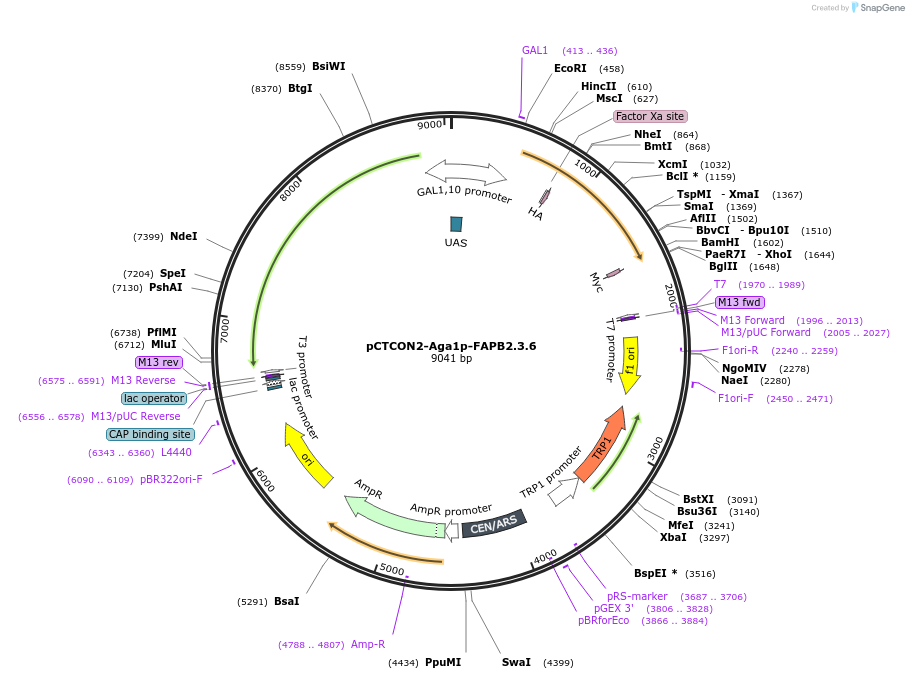
pCTCON2-Aga1p-FAPB2.3.6
Plasmid#158142PurposeYeast display reporter for non-display yeast strains (wild-type construct)DepositorInsertFAPB2.3.6
ExpressionYeastPromoterGAL1-10Available SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
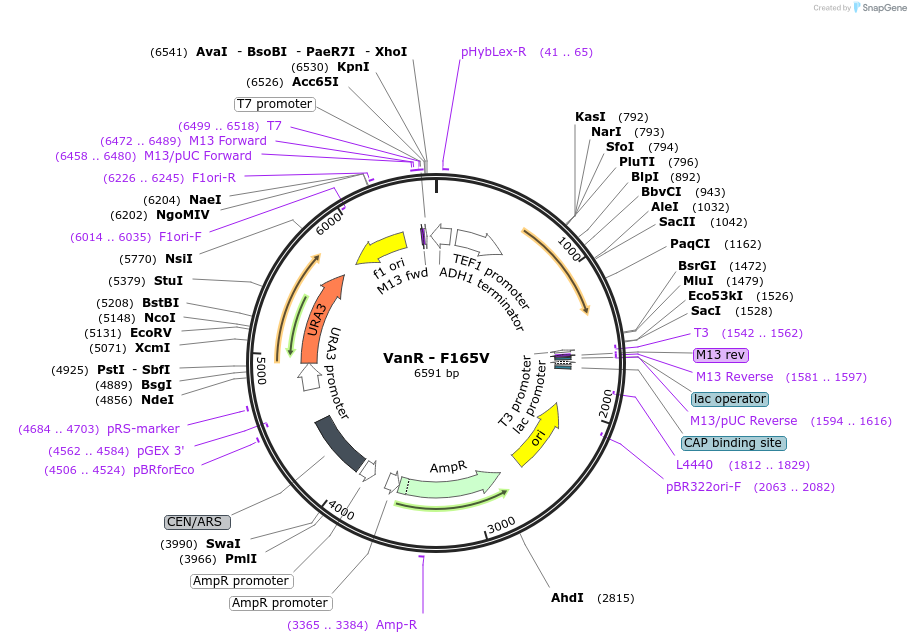
VanR - F165V
Plasmid#158965PurposeWild type VanR where the residue in position 165 was mutated from Phenylalanine to Valine. This mutation abolished the response to vanillic acid. The gene is under the control of the TEF1 promoter.DepositorInsertVanR - F165V
ExpressionYeastMutationWild type VanR from Caulobacter crescentus with m…PromoterTEF1Available SinceJan. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
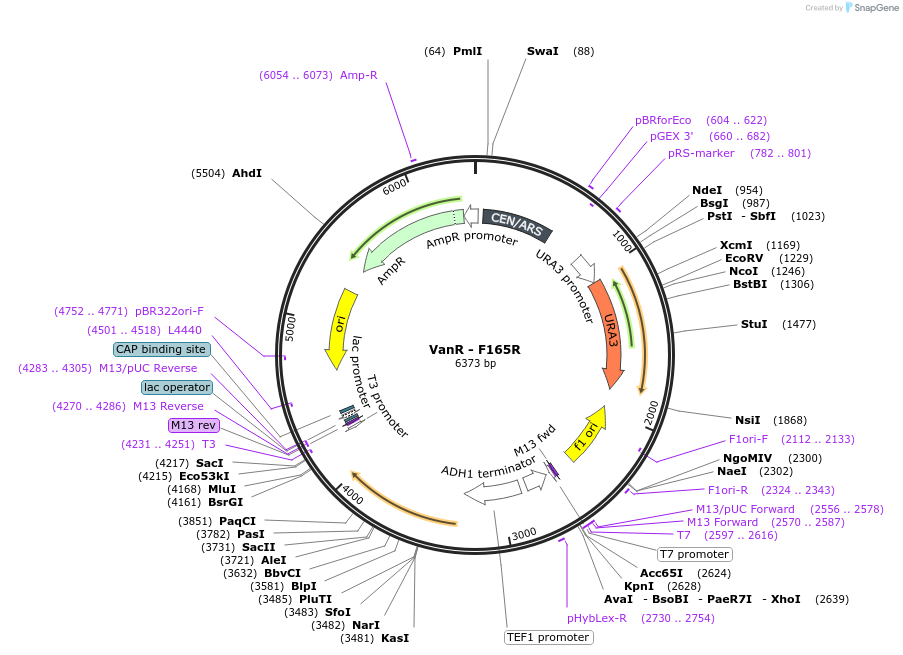
VanR - F165R
Plasmid#158966PurposeWild type VanR where the residue in position 165 was mutated from Phenylalanine to Arginine. This mutation abolished the response to vanillic acid. The gene is under the control of the TEF1 promoter.DepositorInsertVanR - F165R
ExpressionYeastMutationWild type VanR from Caulobacter crescentus with m…PromoterTEF1Available SinceJan. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
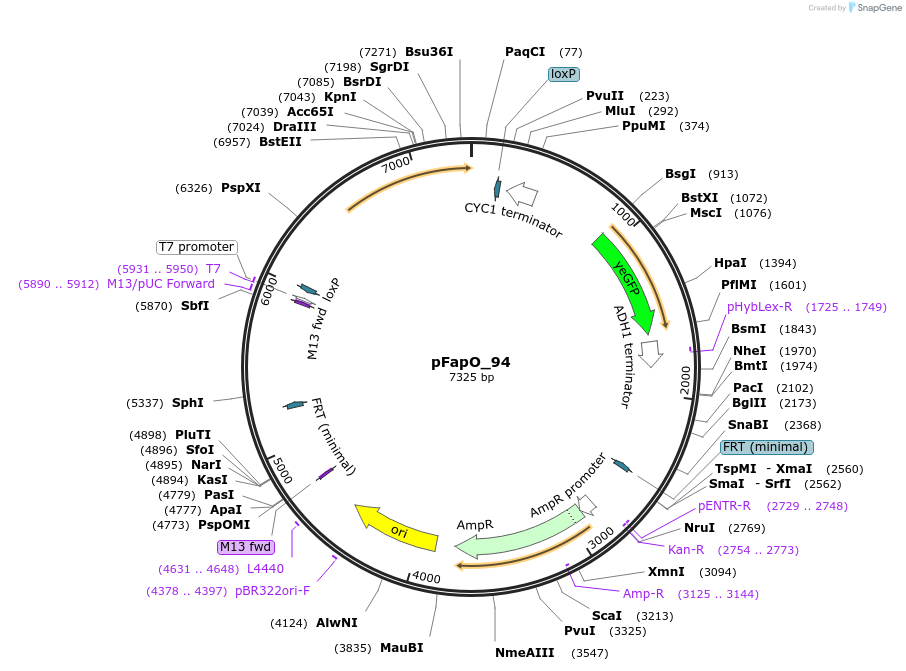
pFapO_94
Plasmid#149597PurposepXI-3-LoxP-KlURA3-pTEF1- FapO_94-yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastPromoterTEF1-FapO_94Available SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
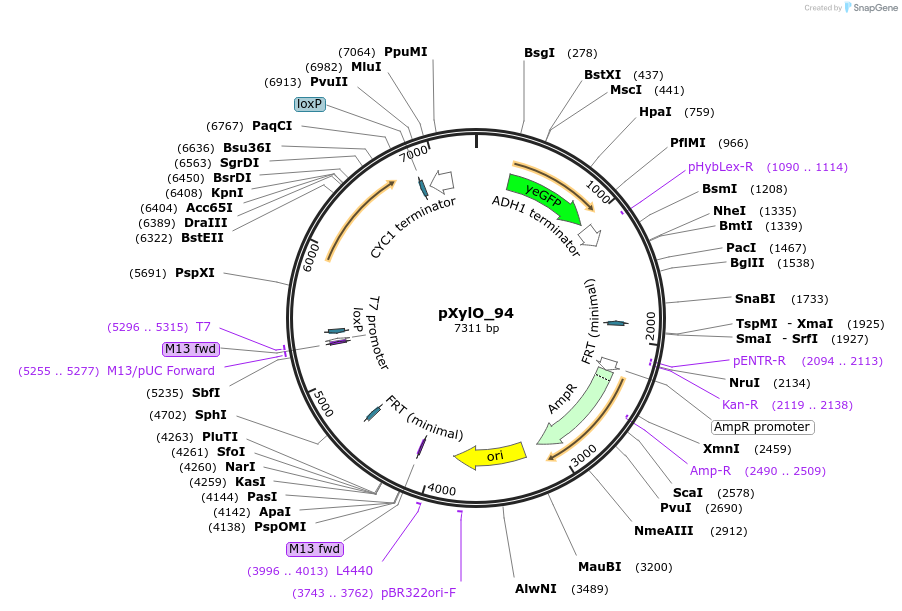
pXylO_94
Plasmid#149598PurposepXI - 3 -LoxP -KlURA3 -pTEF1 - XylO_94 -yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastPromoterTEF1-XylO_94Available SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
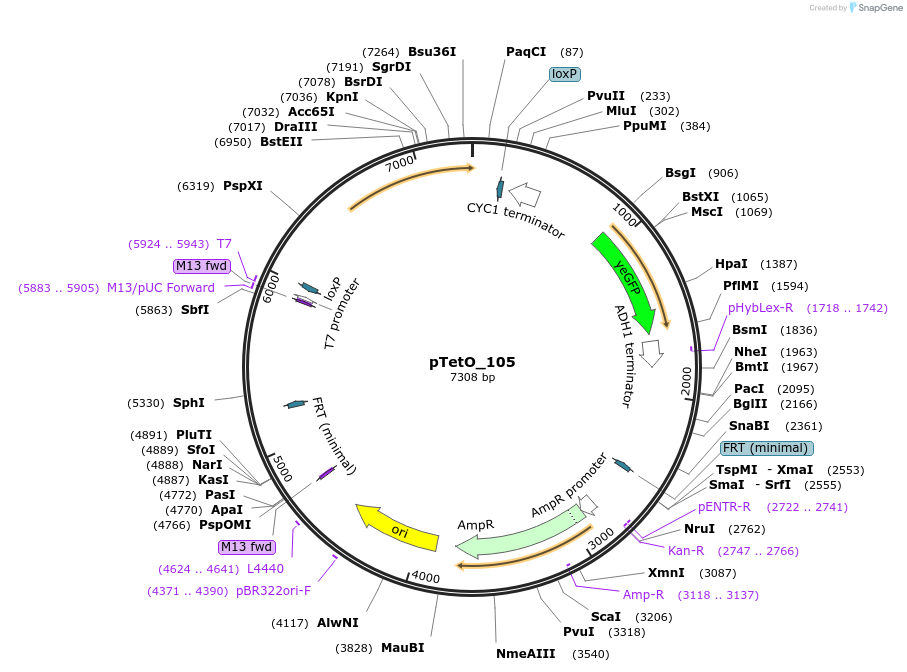
pTetO_105
Plasmid#149599PurposepXI - 3 -LoxP -KlURA3 -pTEF1 - TetO_105 -yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastPromoterTEF1-TetO_105Available SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
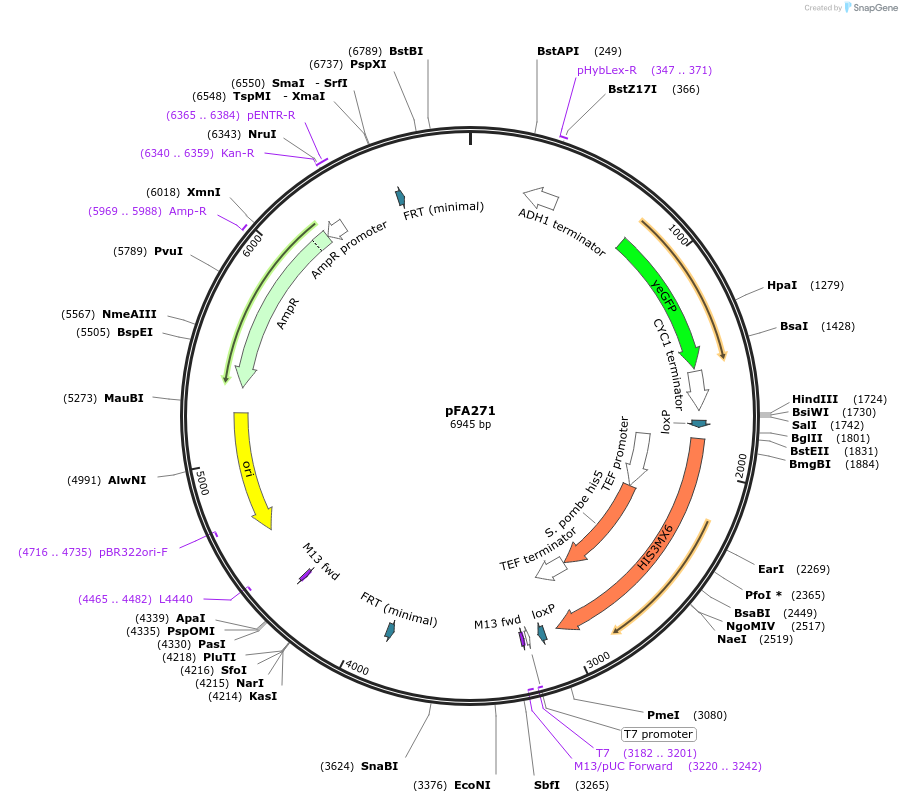
pFA271
Plasmid#149600Purpose(parent plasmid pCFB262) pXII - 4 -LoxP -SpHIS5 -209bp - pCYC1_BenO_183 -yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastAvailable SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
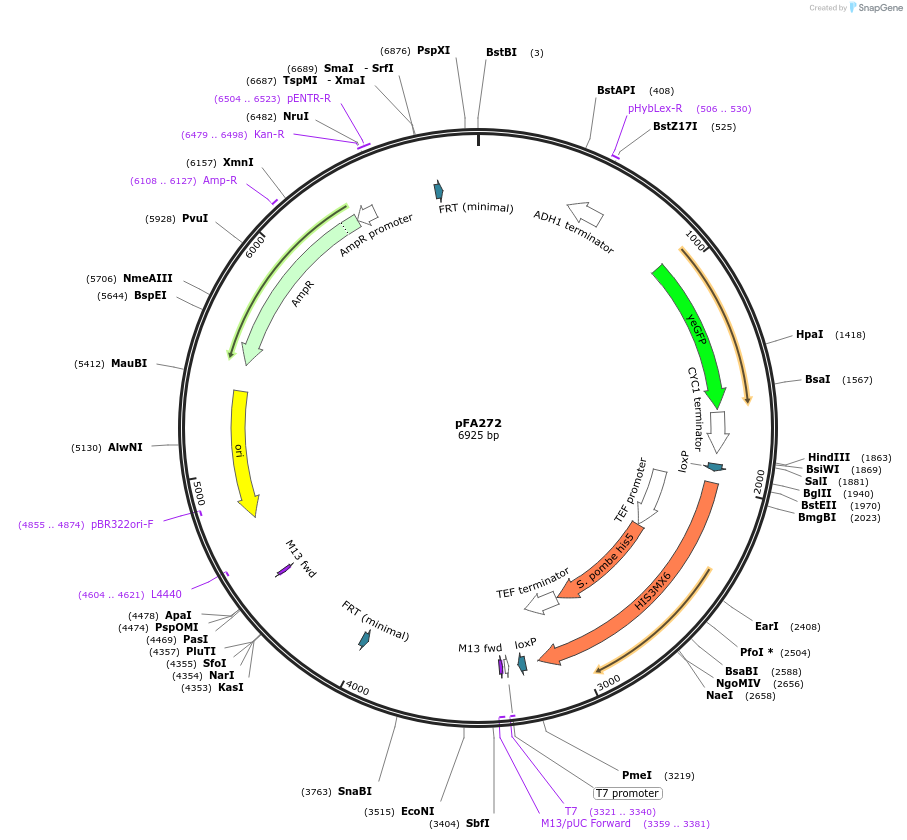
pFA272
Plasmid#149601Purpose(parent plasmid pCFB262) pXII - 4 -LoxP -SpHIS5 -209bp - pCYC1_BenO_182 -yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastAvailable SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
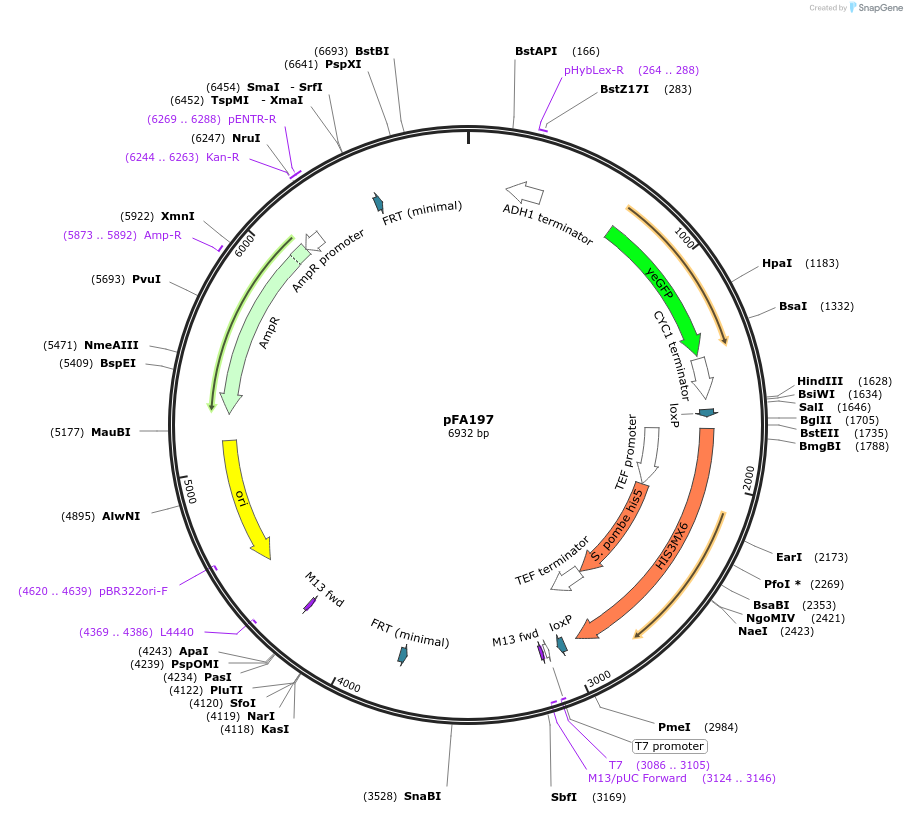
pFA197
Plasmid#149602Purpose(parent plasmid pCFB262) pXII - 4 -LoxP -SpHIS5 -209bp - pCYC1 -PcaO -108 -yeGFPDepositorInsertyeGFP
UseSynthetic BiologyExpressionYeastPromoterPcaO_1O8Available SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
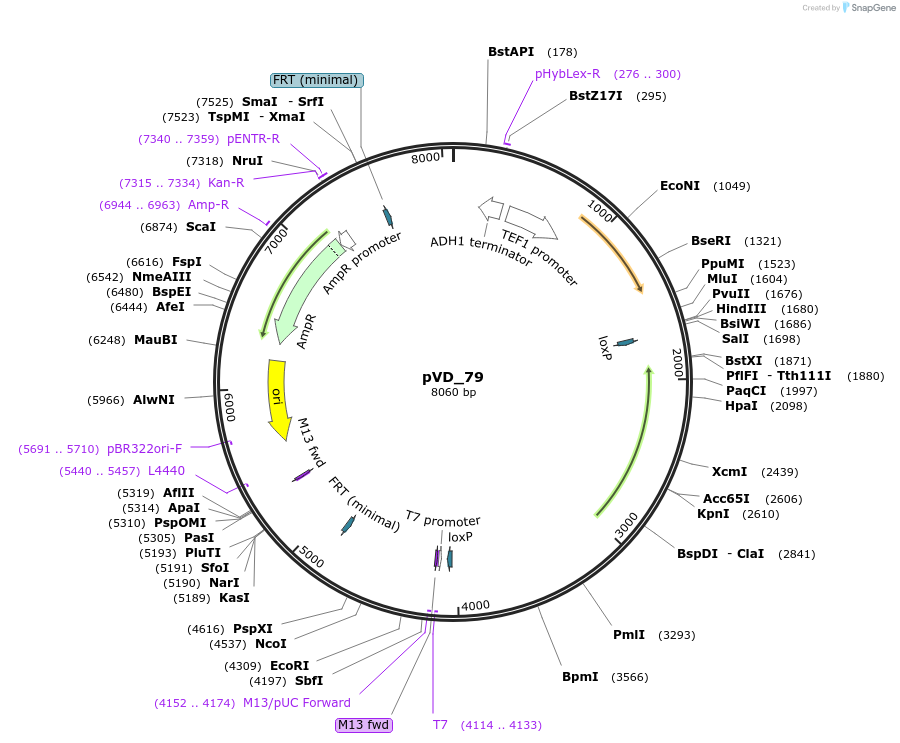
pVD_79
Plasmid#149603Purpose(parent plasmid pCFB257) pX - 3 -LoxP -KlLEU2 -pTEF1 - TetRDepositorInsertTetR
UseSynthetic BiologyExpressionYeastPromoterTEF1Available SinceDec. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
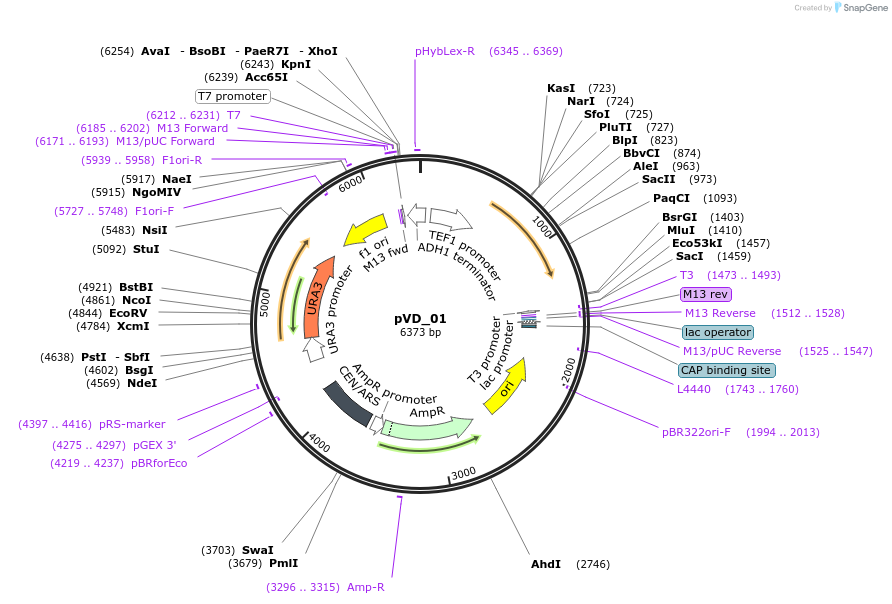
pVD_01
Plasmid#149606PurposepTEF1-VanR inserted in pRS416 backboneDepositorInsertVanR
UseSynthetic BiologyExpressionYeastPromoterTEF1Available SinceDec. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
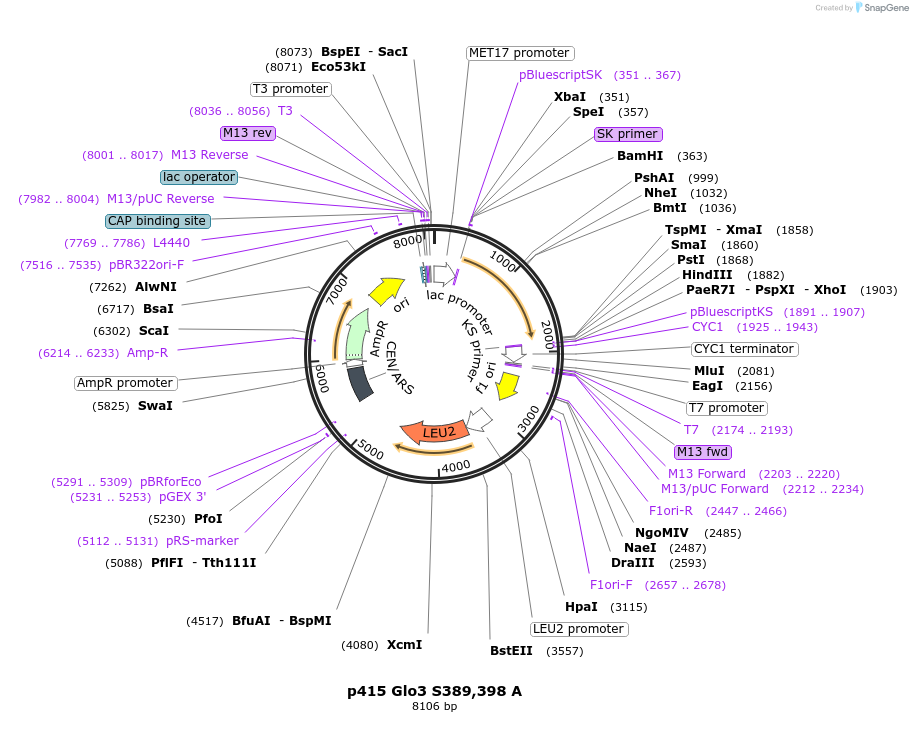
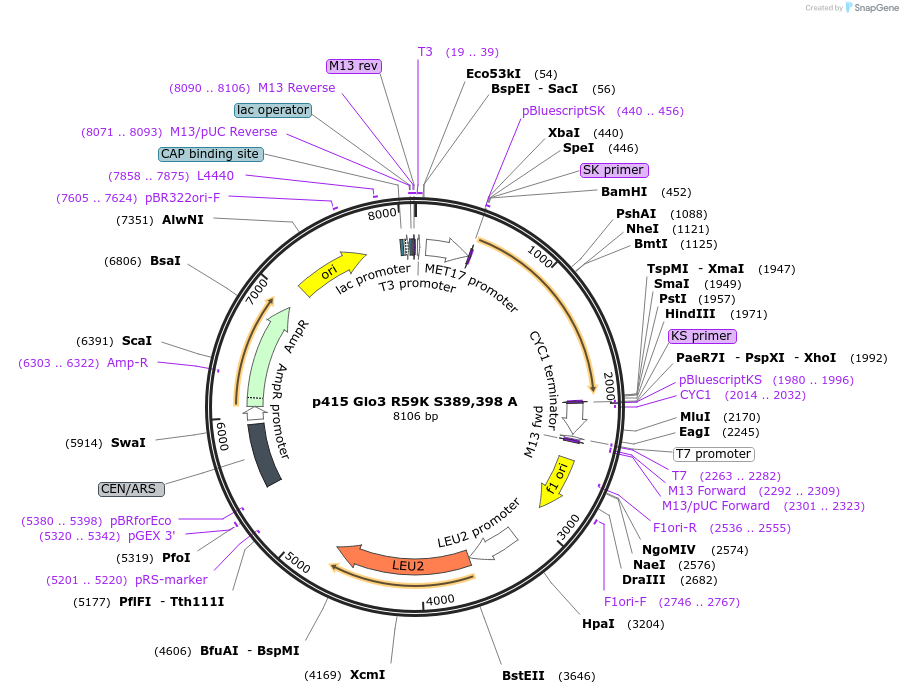
p415 Glo3 S389,398 A
Plasmid#129485PurposeExpression in S. cerevisiaeDepositorInsertGlo3 S389,398 A (GLO3 Budding Yeast)
ExpressionYeastMutationMutation of serines 389 and 398 to alanineAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
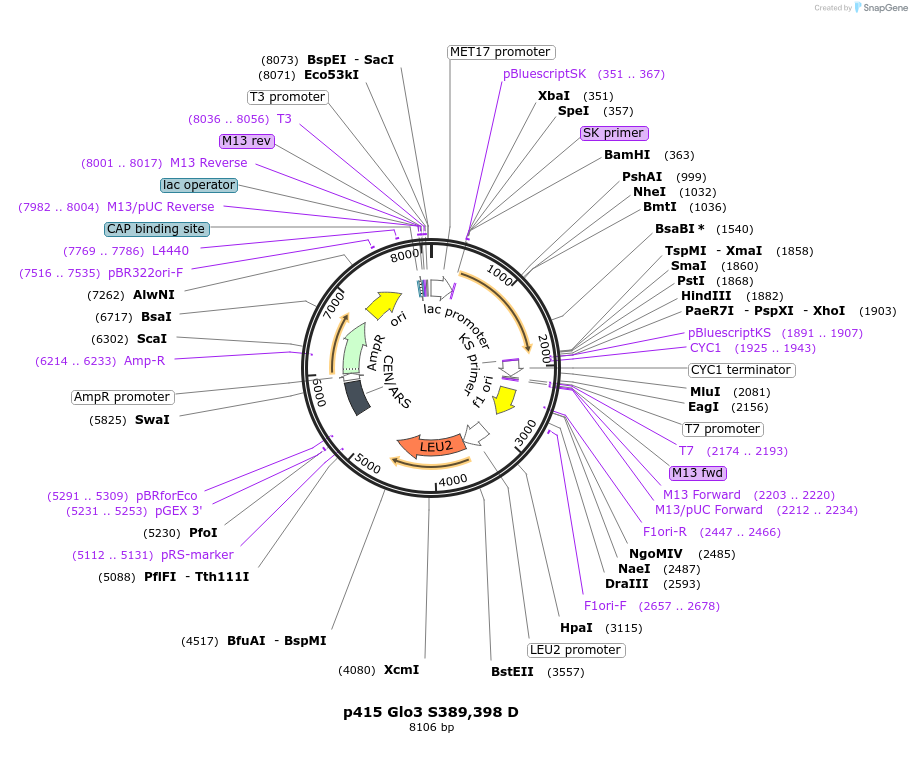
p415 Glo3 S389,398 D
Plasmid#129486PurposeExpression in S. cerevisiaeDepositorInsertGlo3 S389,398 D (GLO3 Budding Yeast)
ExpressionYeastMutationMutation of serines 389 and 398 to AspAvailable SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
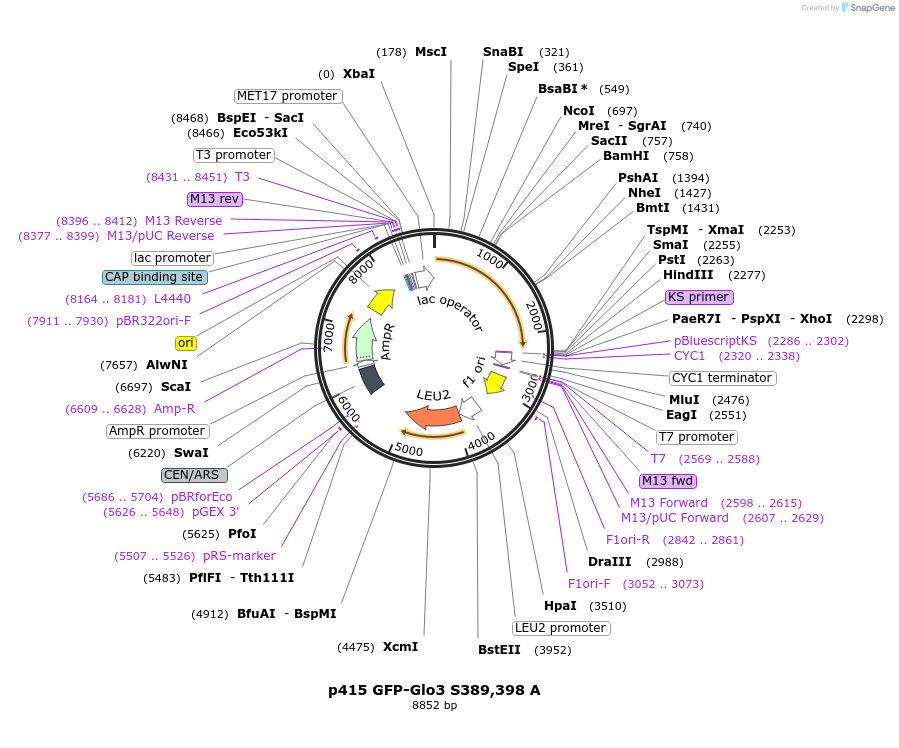
p415 GFP-Glo3 S389,398 A
Plasmid#112660PurposeExpression S.cerevisiaeDepositorInsertGFP- Glo3 S389,398A
TagssfGFPExpressionYeastMutationmutations (S389A, S398A) in Glo3 motif, non-phosp…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
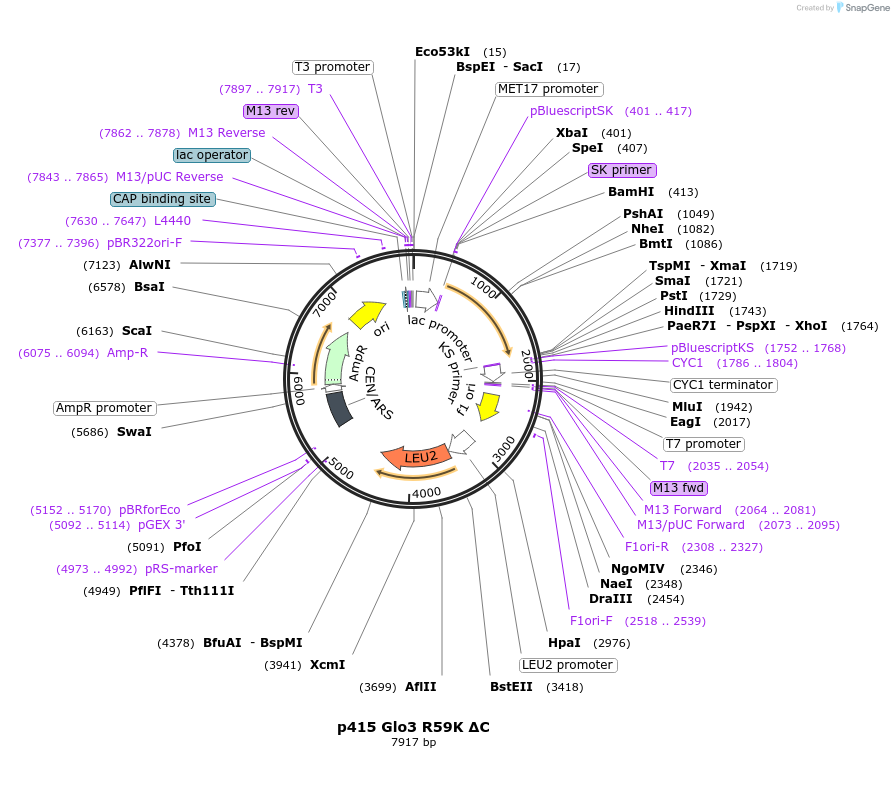
p415 Glo3 R59K ΔC
Plasmid#112653PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K ΔC
ExpressionYeastMutationR59K, Δ431-493 (deletion of C-terminal amphipathi…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
p415 Glo3 R59K S389,398 A
Plasmid#112656PurposeExpression S.cerevisiaeDepositorInsertGlo3 R59K S389,398A
ExpressionYeastMutationR59K, mutations (S389A, S398A) in Glo3 motif, non…PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
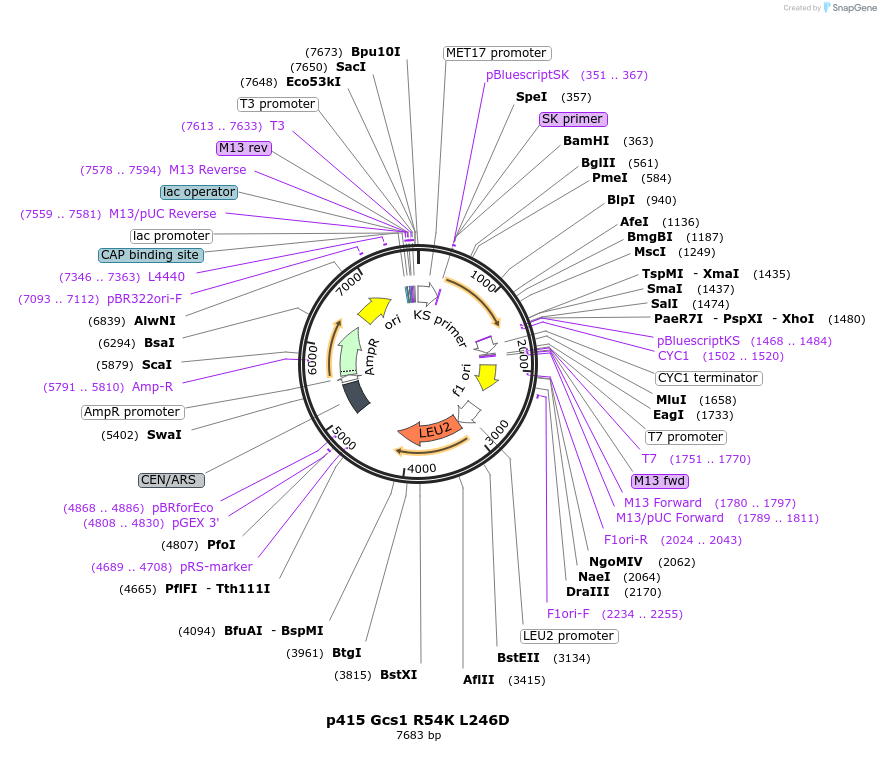
p415 Gcs1 R54K L246D
Plasmid#112648PurposeExpression S.cerevisiaeDepositorInsertGcs1 R54K L246D
ExpressionYeastMutationR54K & L246D (in ALPS motif)PromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
p415 Gcs1 Δ3xF AxxA
Plasmid#112650PurposeExpression S.cerevisiaeDepositorInsertGcs1 Δ3xF AxxA
ExpressionYeastMutationR54K, W304A, W307A, W310A, W349A, F353APromoterMet25Available SinceSept. 25, 2020AvailabilityAcademic Institutions and Nonprofits only