We narrowed to 3,362 results for: cat.3
-
Plasmid#27505DepositorInsertpmyo-3::yfp::lin-14_3'UTR
ExpressionWormAvailable SinceMarch 10, 2011AvailabilityAcademic Institutions and Nonprofits only -
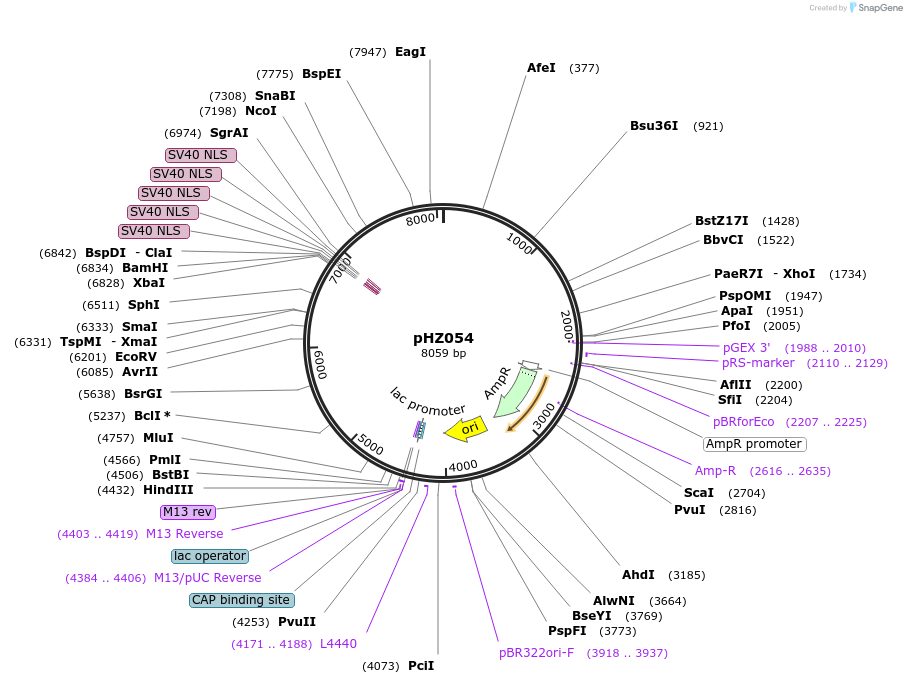
pAAV 3xgRNA KO;Rbfox3
Plasmid#240311PurposeKO:NeuNDepositorInsertKO gRNAs for NeuN
UseAAVMutationNAAvailable SinceJuly 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
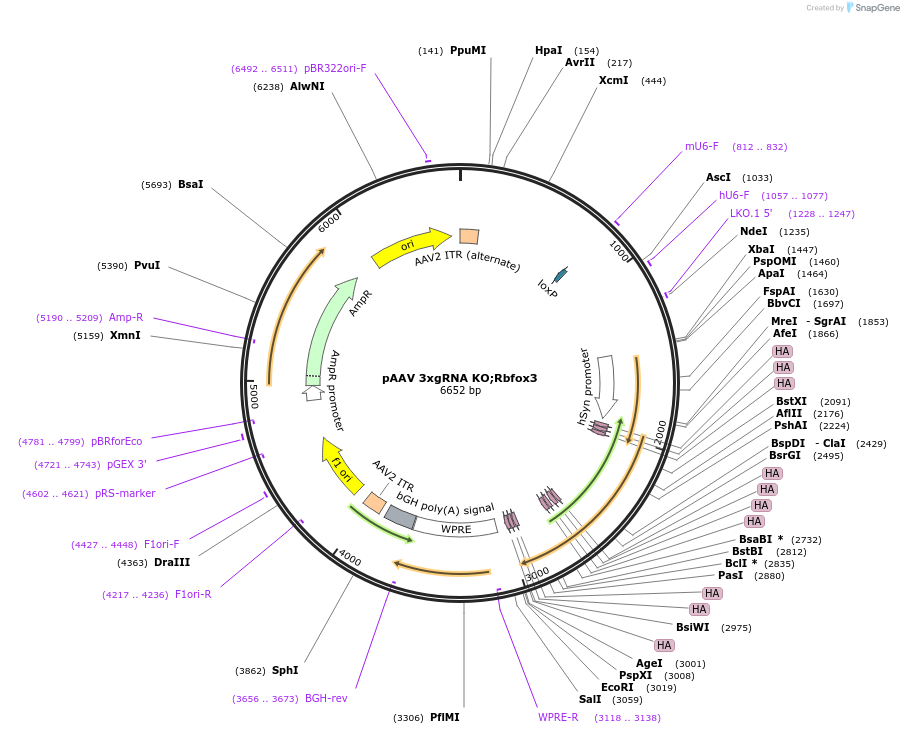
pINER2R
Plasmid#185882PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (11 copy).DepositorInsertPRPL25(Arm 1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm 3)-ARS305-PGAL2>Y.FAST-EVBR1.2A-AcNES1>TRPL41B>RPL25(partial; Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
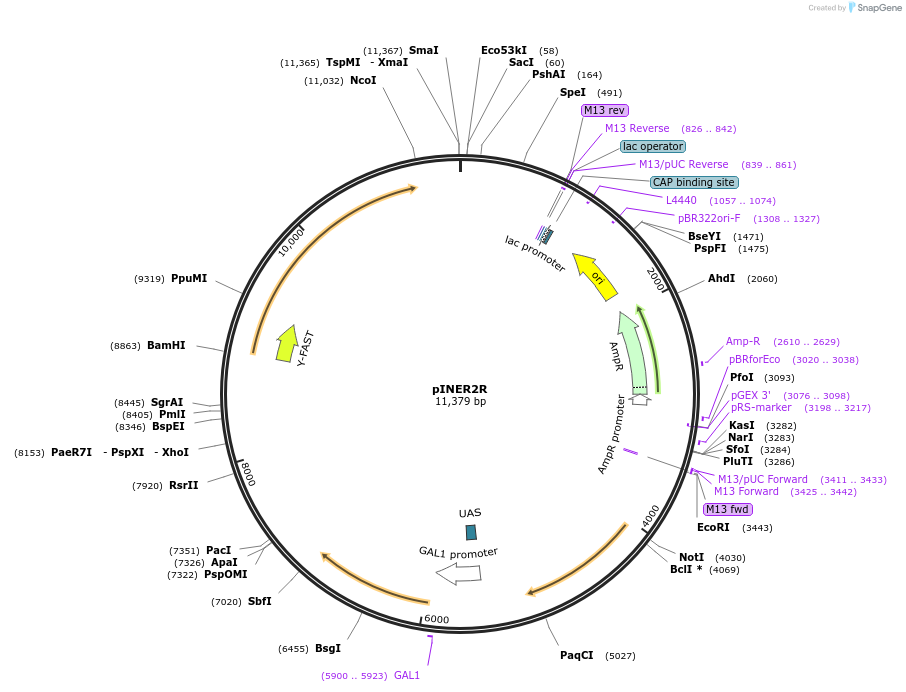
pIT7A6
Plasmid#185896PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (15 copy).DepositorInsertPRPL25(Arm 1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm 3)-ARS305-PSk.GAL2>Y.FAST-EVBR1.2A>TRPL41B-PPDA1>RPL25(partial;Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
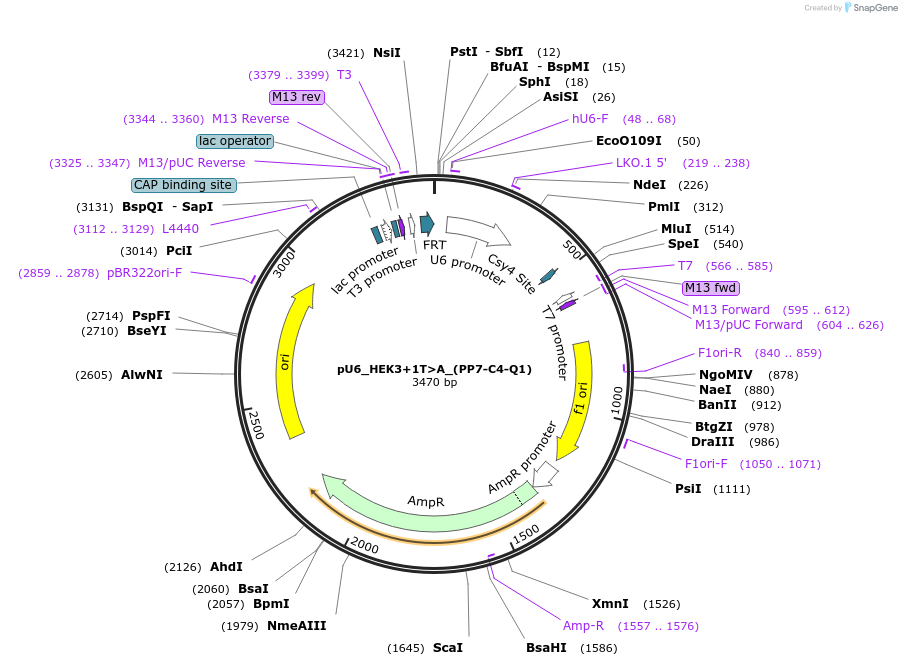
pU6_HEK3+1T>A_(PP7-C4-Q1)
Plasmid#232437PurposepegRNA with optimized 3' modifications to facilitate a +1 T to A prime edit in the HEK3 locusDepositorInsert3'-PP7-tagged pegRNA for rd12 correction driven by human U6 promoter
UseCRISPRPromoterU6Available SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
pBP-T7_RBS-His6-Thrombin
Plasmid#72979PurposeT7 promoter, RBS, His6-tag and thrombin cleave site - derived from pET15b (5' CTAT/3' CATA) fusion)DepositorInsertT7+RBS+His6+thrombin
UseSynthetic BiologyMutationCAT gene - C435G (nucleotide) - silent mutagenesi…Available SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
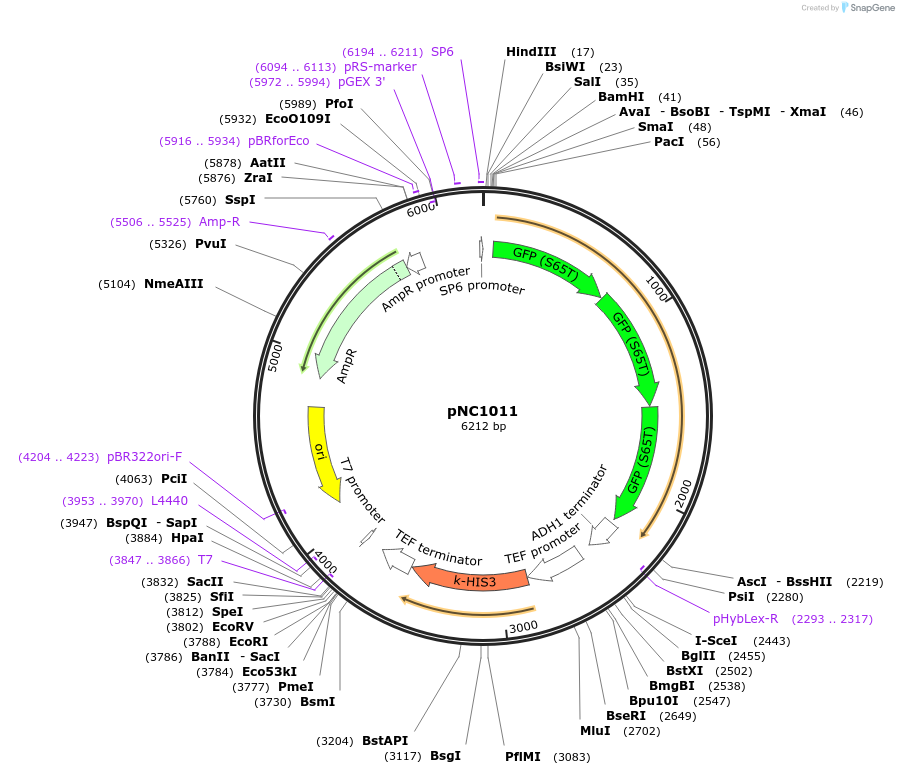
pNC1011
Plasmid#42898DepositorInsert3 tandem copies of GFP*
UseYeast integratingMutationchanged Phenylalanine 64 to Leucine, Serine 65 t…PromoterNoneAvailable SinceMarch 6, 2013AvailabilityAcademic Institutions and Nonprofits only -
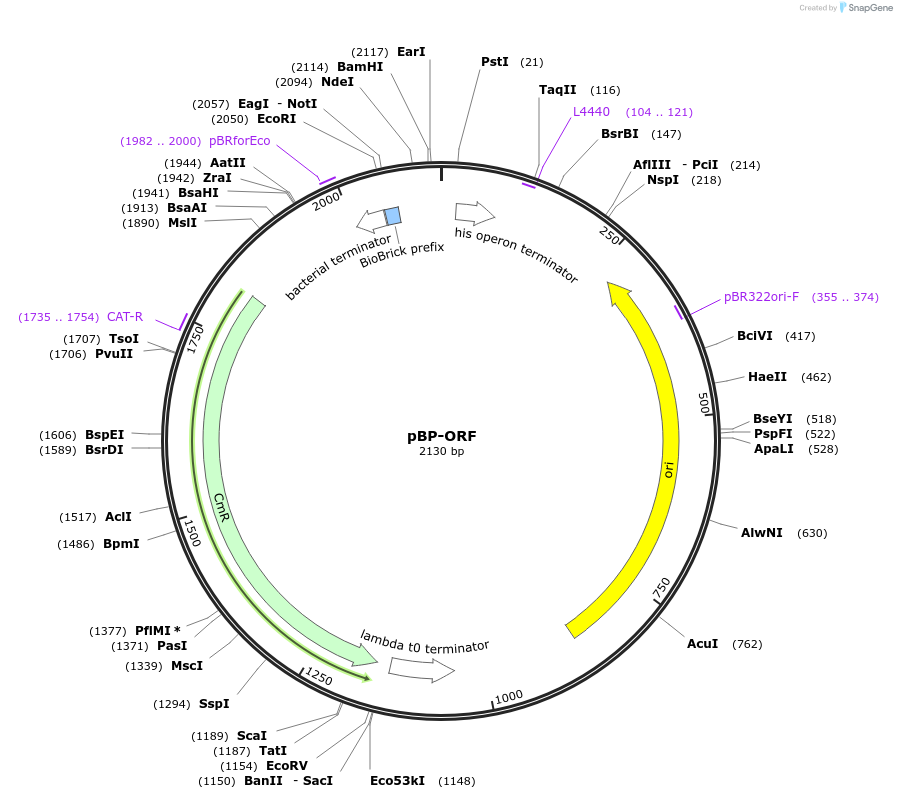
pBP-ORF
Plasmid#72949PurposeEntry vector for ORF's - PCR, gene synthesis or sub-clone using NdeI/BamHI. Alternatively use BsaI with CATA and TCGA fusion sites. Stop codon must be included upstream of chosen 3' restriction siteDepositorInsertLevel 0 - ORFs
UseSynthetic BiologyMutationCAT gene - C435G (nucleotide) - silent mutagenesi…Available SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
pET15b-mISG15(GG)
Plasmid#12448DepositorInsertC-terminal processed form of ISG15
TagsHisExpressionBacterialAvailable SinceJan. 4, 2007AvailabilityAcademic Institutions and Nonprofits only -
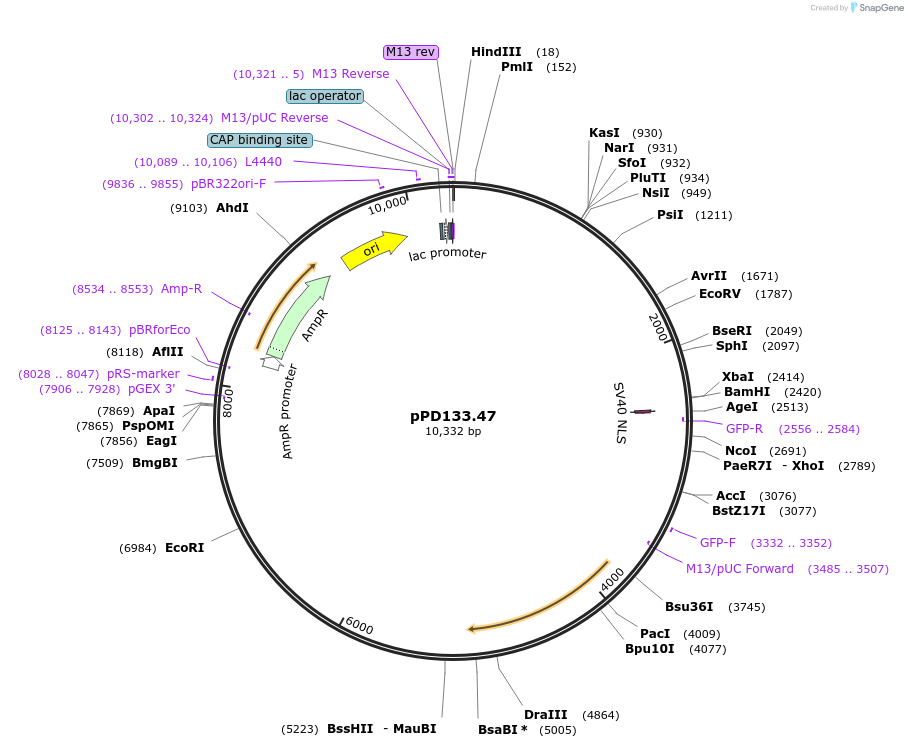
pPD133.47
Plasmid#27510DepositorInsertpmyo-3::cfp::lacZ::unc-54_3'UTR
ExpressionWormAvailable SinceMarch 15, 2011AvailabilityAcademic Institutions and Nonprofits only -
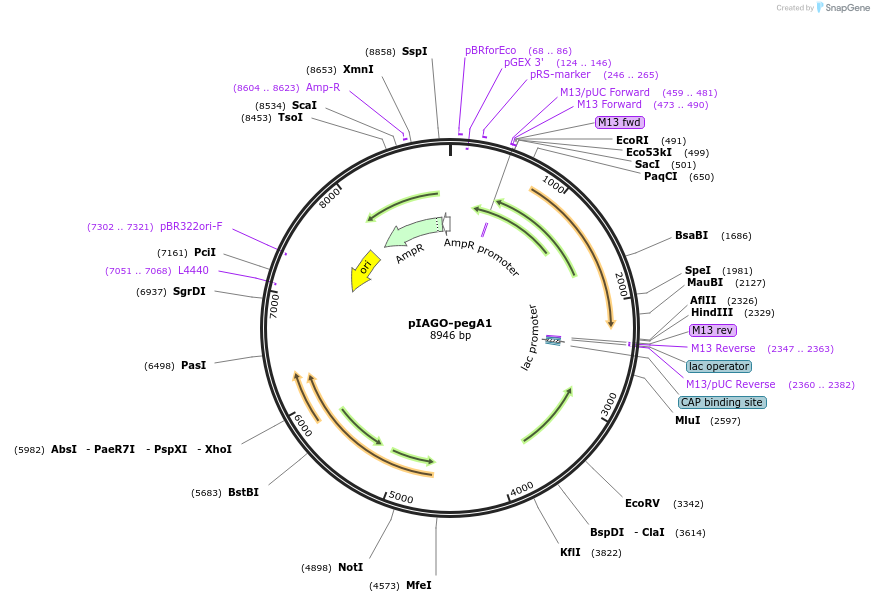
pIAGO-pegA1
Plasmid#114455PurposeExpression of PegA glycosyltansferase in streptomycetesDepositorInsertpegA
ExpressionBacterialPromoterermEAvailable SinceFeb. 21, 2019AvailabilityAcademic Institutions and Nonprofits only -
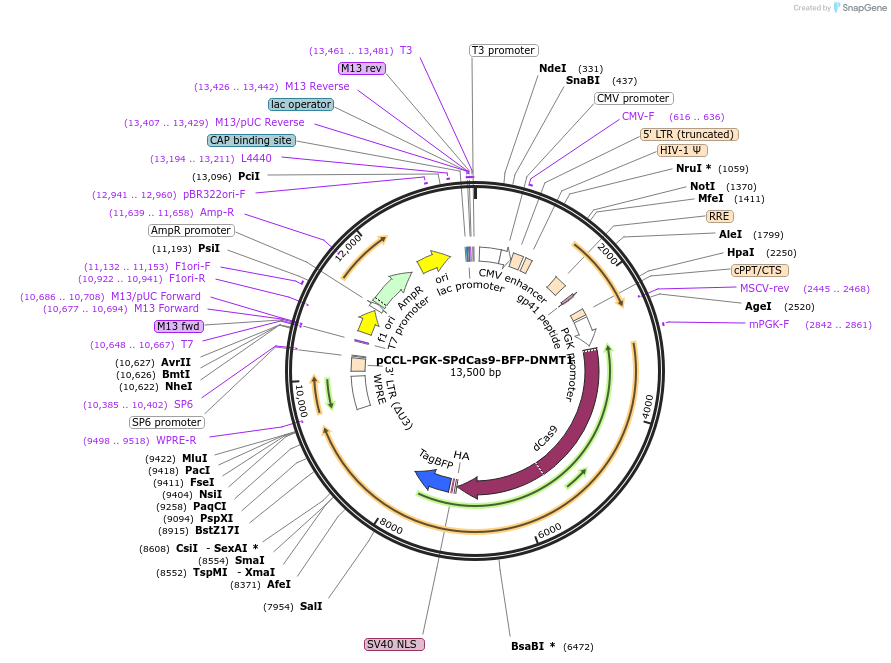
pCCL-PGK-SPdCas9-BFP-DNMT1
Plasmid#66818PurposedCas9 fused to BFP and the human DNMT1 catalytic domainDepositorInsertdCas9-BFP-DNMT1, catalytic domain (DNMT1 )
ExpressionMammalianAvailable SinceJune 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
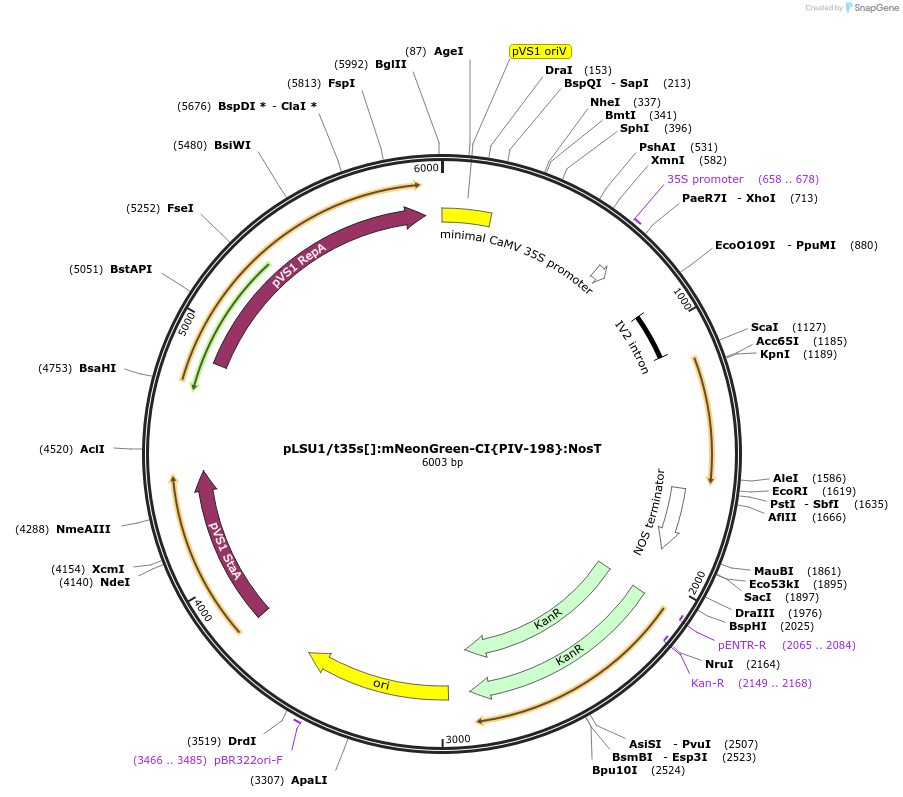
pLSU1/t35s[]:mNeonGreen-CI{PIV-198}:NosT
Plasmid#212171PurposeThis binary vector expresses mNeonGreen containing the PIV intron (198bp into the gene) with extra 3' splice site removed (corrected intron) with the truncated 35sCAMV promoterDepositorInsertTomato codon optimized version of mNeonGreen-I{PIV-198}
ExpressionPlantMutationmNeonGreen codon-optimized for tomato and PIV int…Promotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
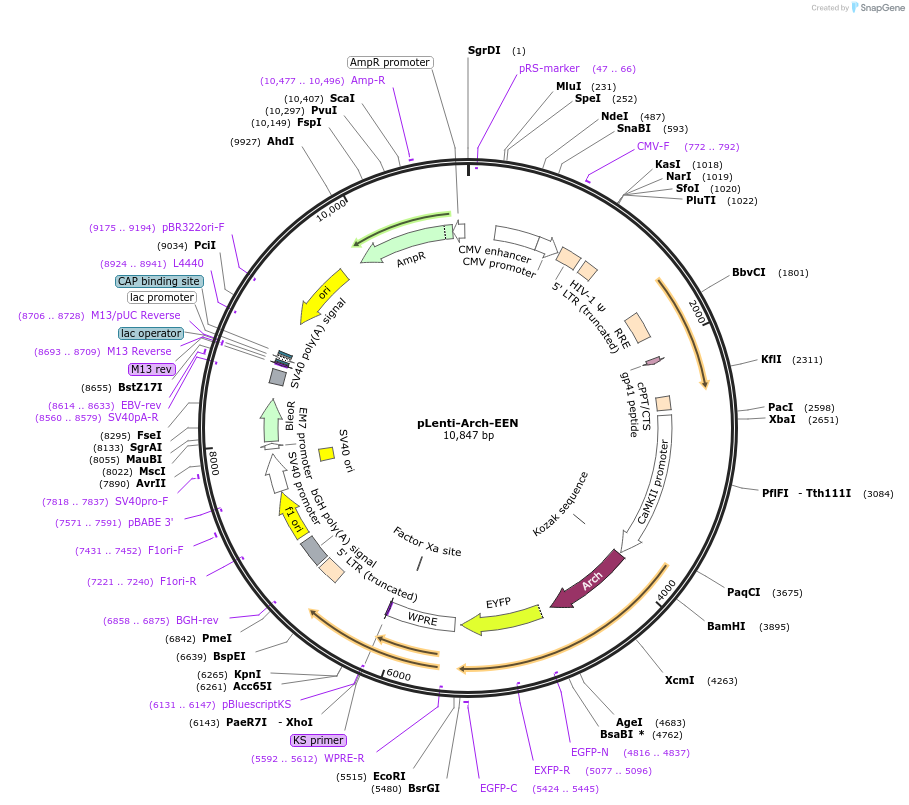
pLenti-Arch-EEN
Plasmid#45189PurposeArch voltage indicator with double mutations D95N-D106E (Arch-EEN), with faster kinetics and greater fluorescence dynamic range than Arch-D95NDepositorInsertenhanced Archaerhodopsin-3
UseLentiviralTagseYFPExpressionMammalianMutationD95N, D106EPromoterCamKIIaAvailable SinceJuly 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
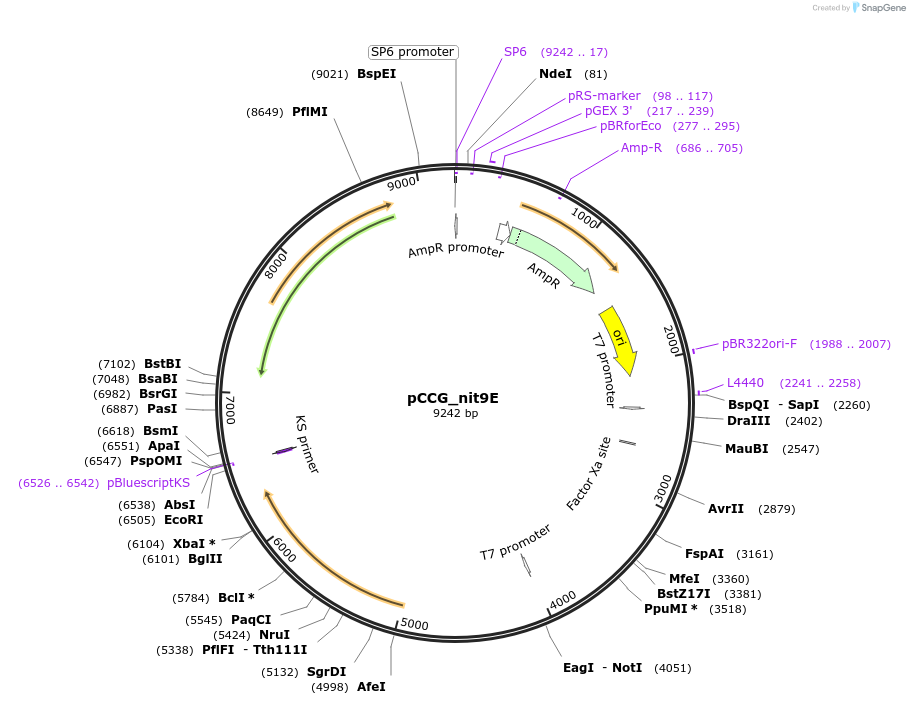
pCCG_nit9E
Plasmid#228419PurposeExpression of NIT-9E domain in N. crassa at the his-3 locusDepositorInsertNIT-9
UseProtein expression in n. crassaMutationtruncation of the N terminal G domain and linkage…Available SinceFeb. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
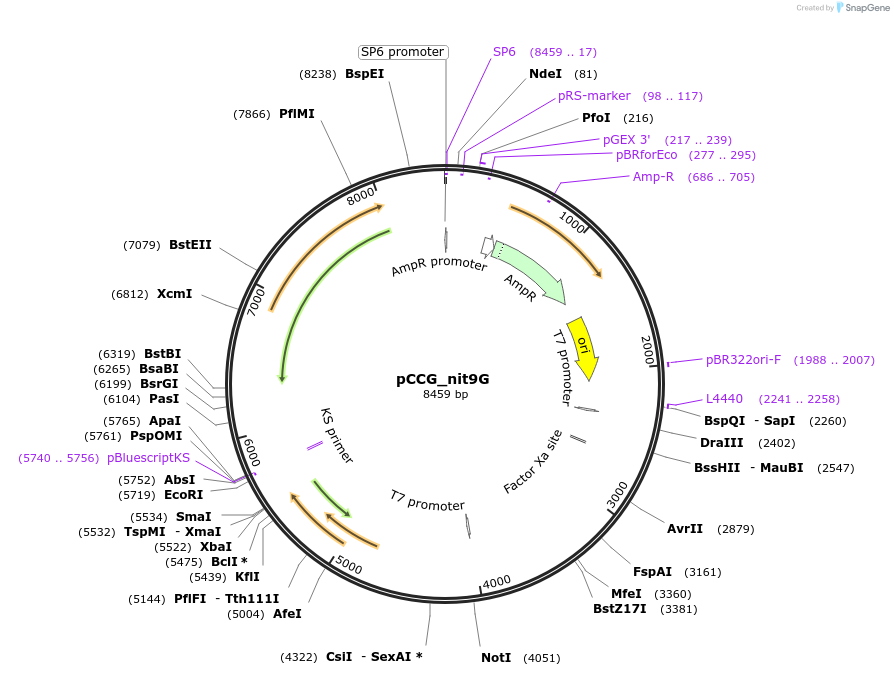
pCCG_nit9G
Plasmid#228420PurposeExpression of NIT-9G domain in N. crassa at the his-3 locusDepositorInsertNIT-9
UseProtein expression in n. crassaMutationtruncation of the C terminal E domain and linkage…Available SinceFeb. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
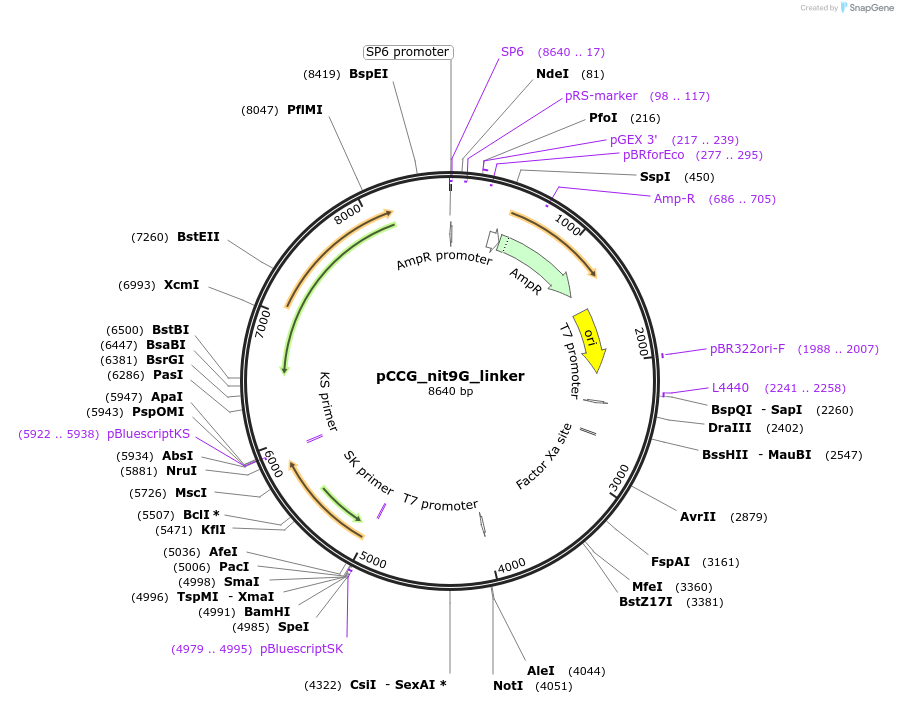
pCCG_nit9G_linker
Plasmid#228413PurposeExpression of NIT-9G with linkage region at the C terminus in N. crassa at the his-3 locusDepositorInsertNIT-9
UseProtein expression in n. crassaMutationtruncation of the C terminal E domainAvailable SinceJan. 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
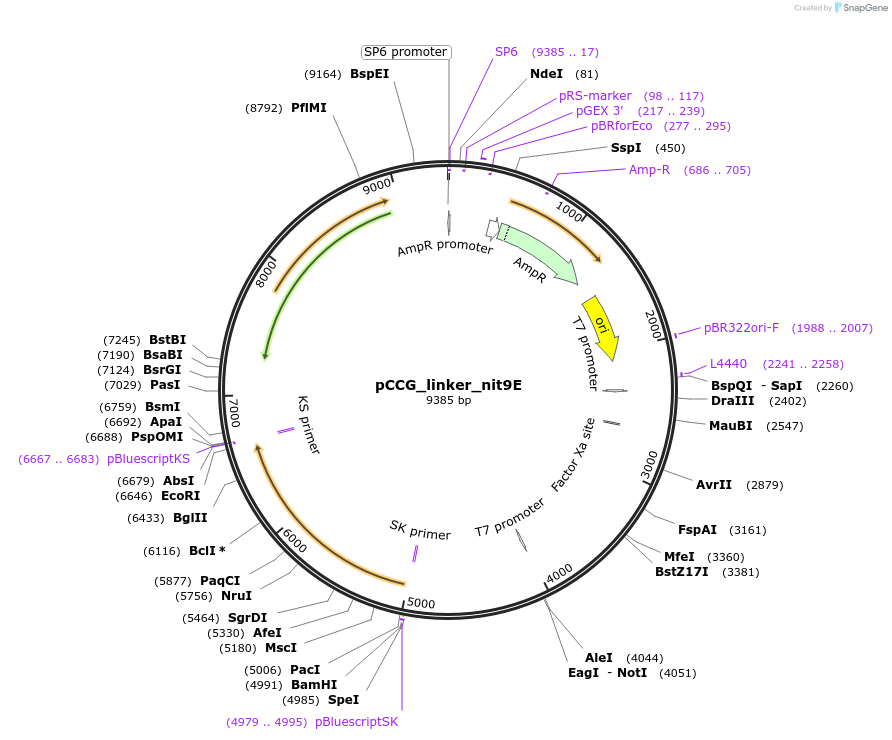
pCCG_linker_nit9E
Plasmid#228412PurposeExpression of NIT-9E with linkage region at the N terminus in N. crassa at the his-3 locusDepositorInsertNIT-9
UseProtein expression in n. crassaMutationtruncation of the N terminal G domainAvailable SinceJan. 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
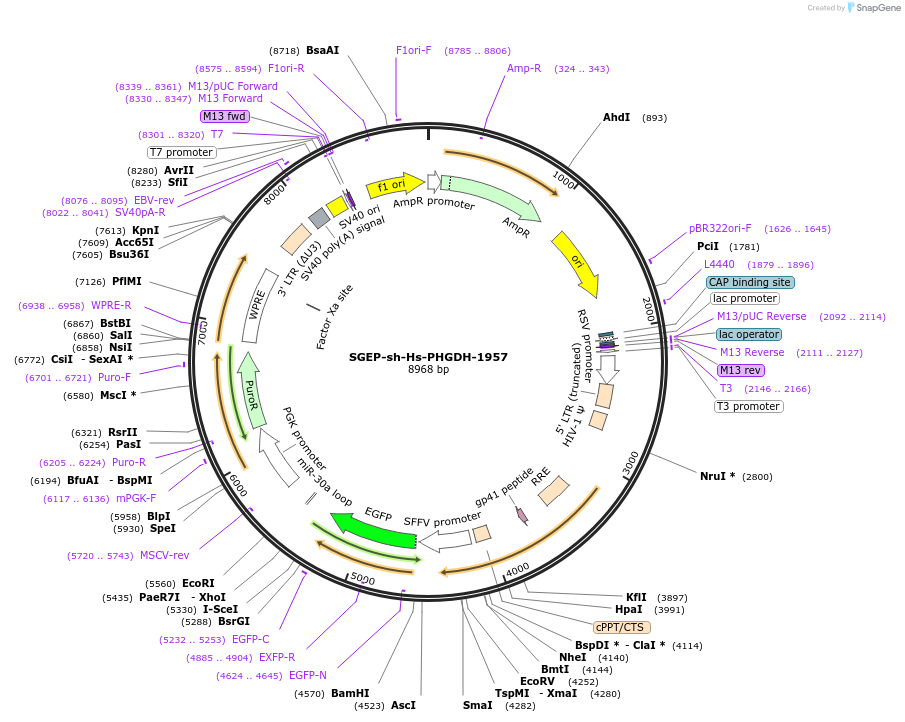
SGEP-sh-Hs-PHGDH-1957
Plasmid#188674PurposeshRNADepositorAvailable SinceOct. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
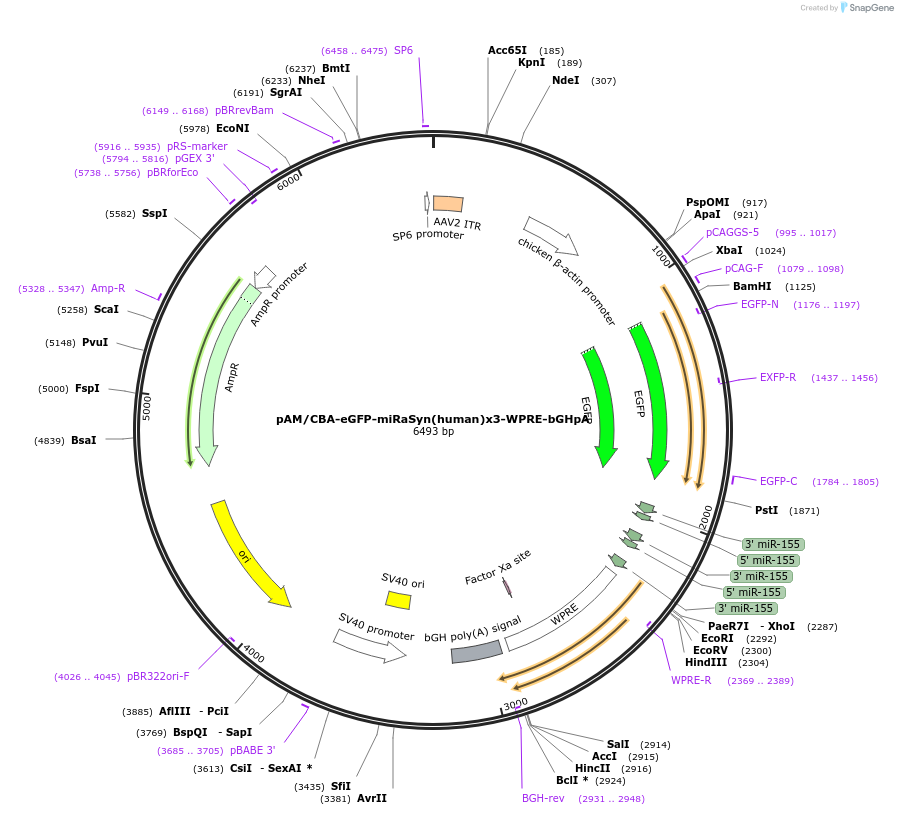
pAM/CBA-eGFP-miRaSyn(human)x3-WPRE-bGHpA
Plasmid#194247PurposeExpresses 3 miRNAs targeting human alpha-SynucleinDepositorAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only