We narrowed to 2,494 results for: pcas9
-
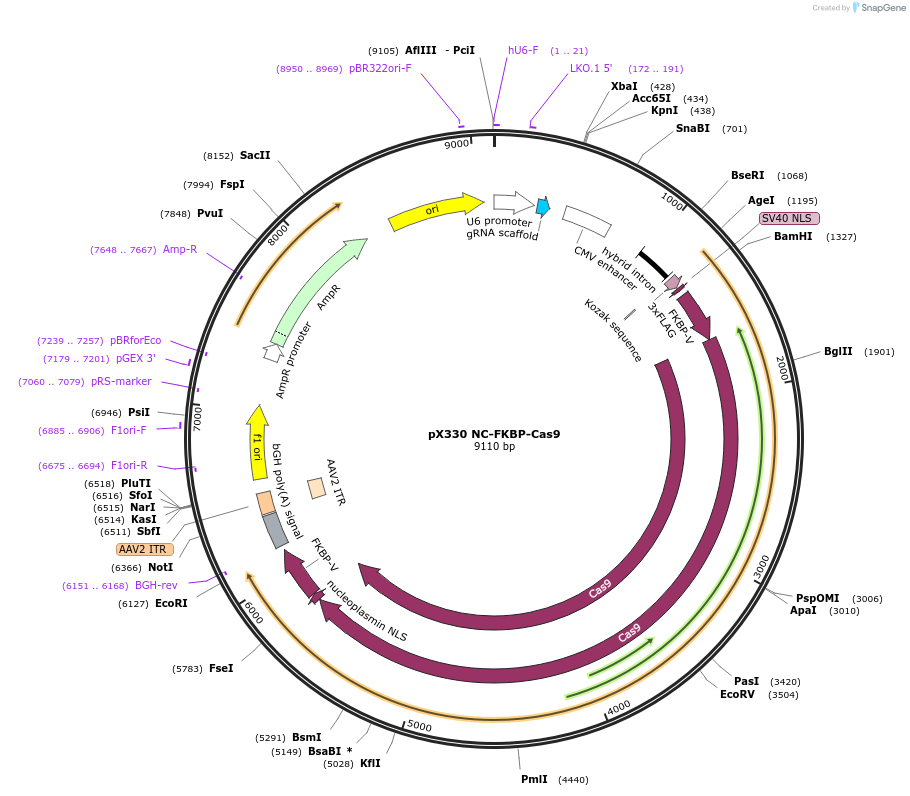
Plasmid#162725PurposeMammalian expressionDepositorInsertFKBP F36V
UseCRISPRTags3xFLAGExpressionMammalianPromoterCBhAvailable SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
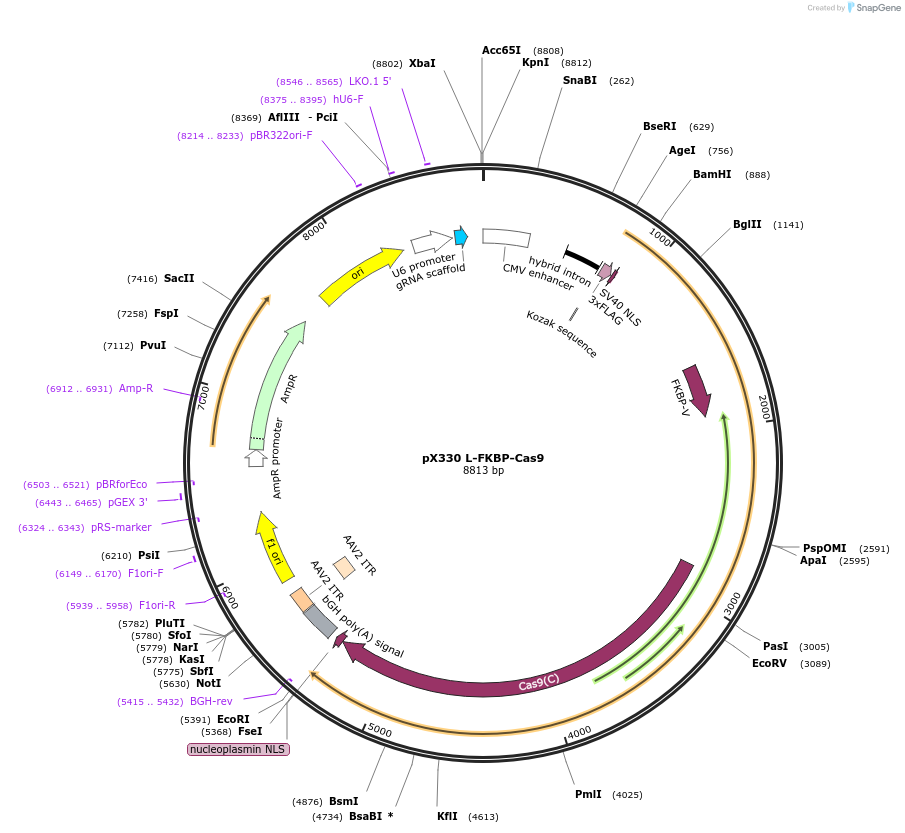
pX330 L-FKBP-Cas9
Plasmid#162728PurposeMammalian expressionDepositorInsertFKBP F36V
UseCRISPRTags3xFLAGExpressionMammalianPromoterCBhAvailable SinceJan. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
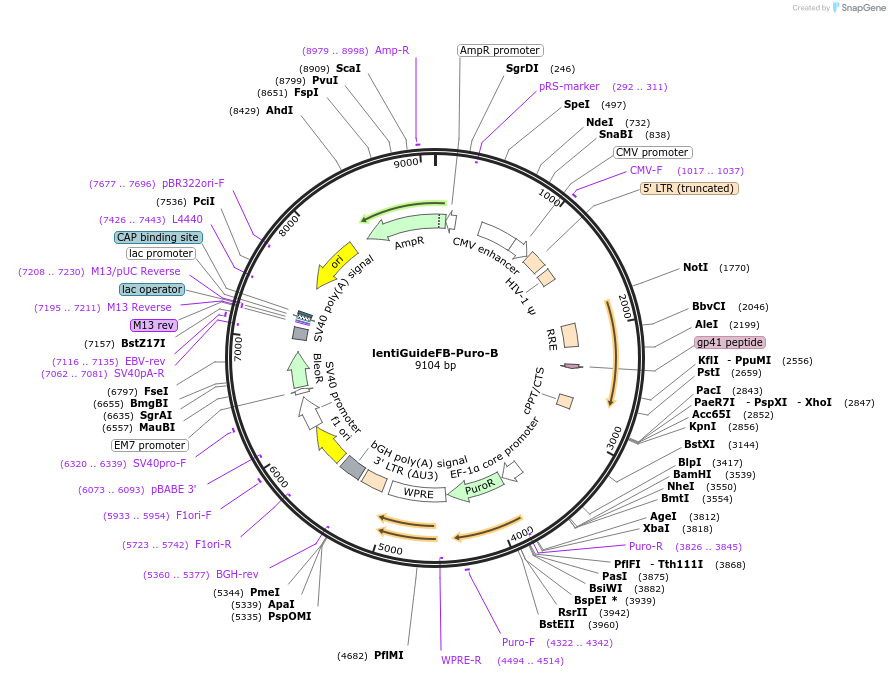
lentiGuideFB-Puro-B
Plasmid#192507PurposeFor cloning of sgRNAs compatible with PspCas9. Contains a 5' direct repeat and a unique sgRNA scaffold variant with a capture sequence. Cloning guide RNAs using BsmBI.DepositorInsertbU6-sgRNACR2-CS-BsmBI-EFS-Puro-WPRE
UseLentiviralPromoterbU6Available SinceOct. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
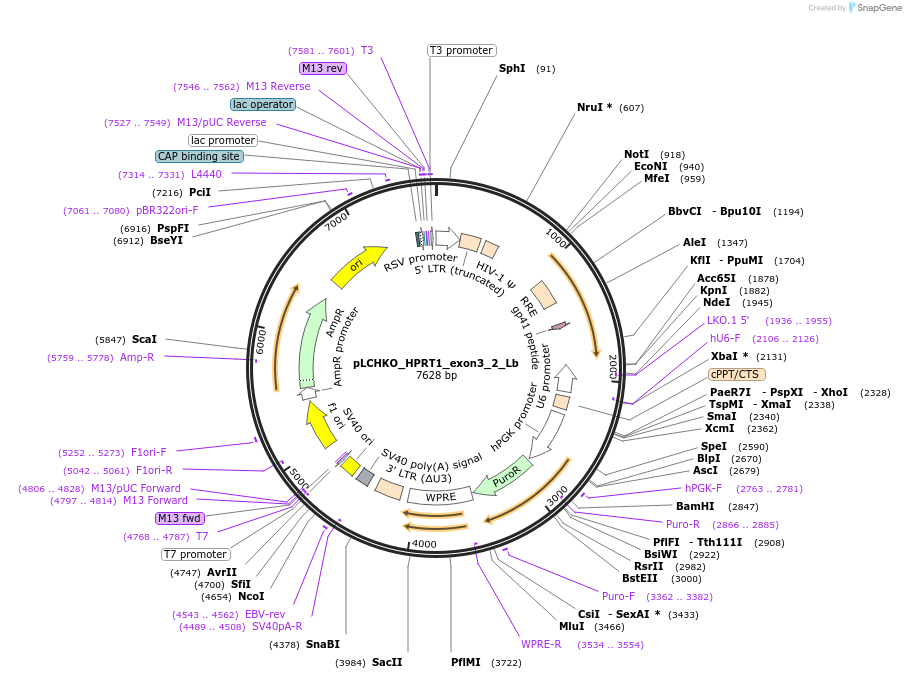
pLCHKO_HPRT1_exon3_2_Lb
Plasmid#155054PurposeLentiviral plasmid for expressing U6 driven hybrid guide (hg)RNA for deletion of HPRT1 exon 3 using SpCas9 and LbCas12a nucleases (CHyMErA system)DepositorInsert(hg)RNA for deletion of HPRT1 exon 3
UseCRISPR and LentiviralExpressionMammalianPromoterU6Available SinceAug. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
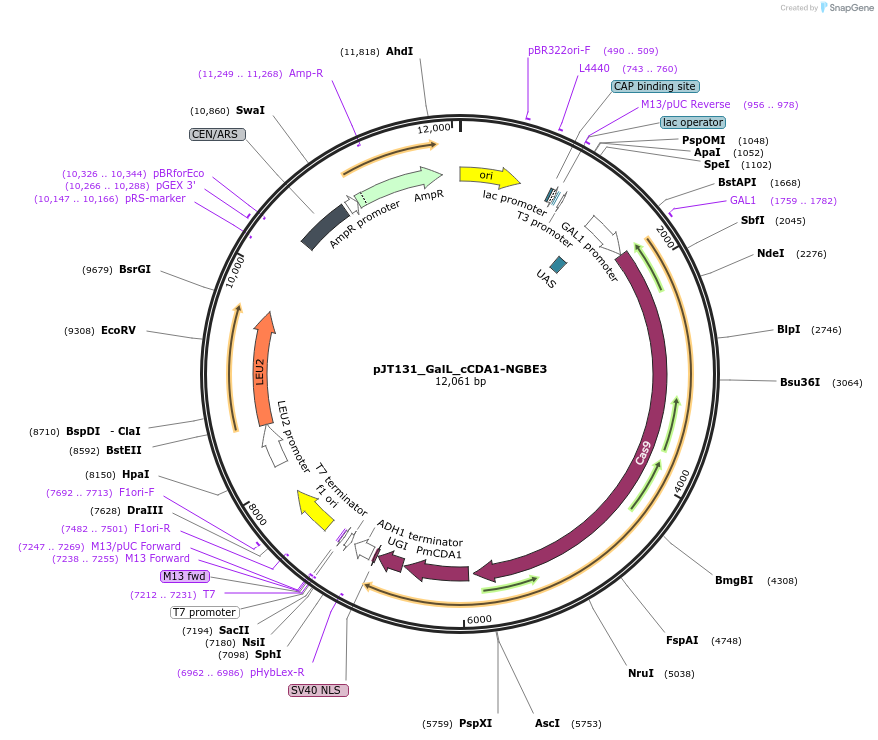
pJT131_GalL_cCDA1-NGBE3
Plasmid#145095PurposeExpressing base editor cCDA1-NGBE3 in yeast cellsDepositorInsertcCDA1-NGBE3
UseCRISPRExpressionYeastMutationspCas9(D10A/L1111R/D1135V/G1218R/E1219F/A1322R/R1…PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
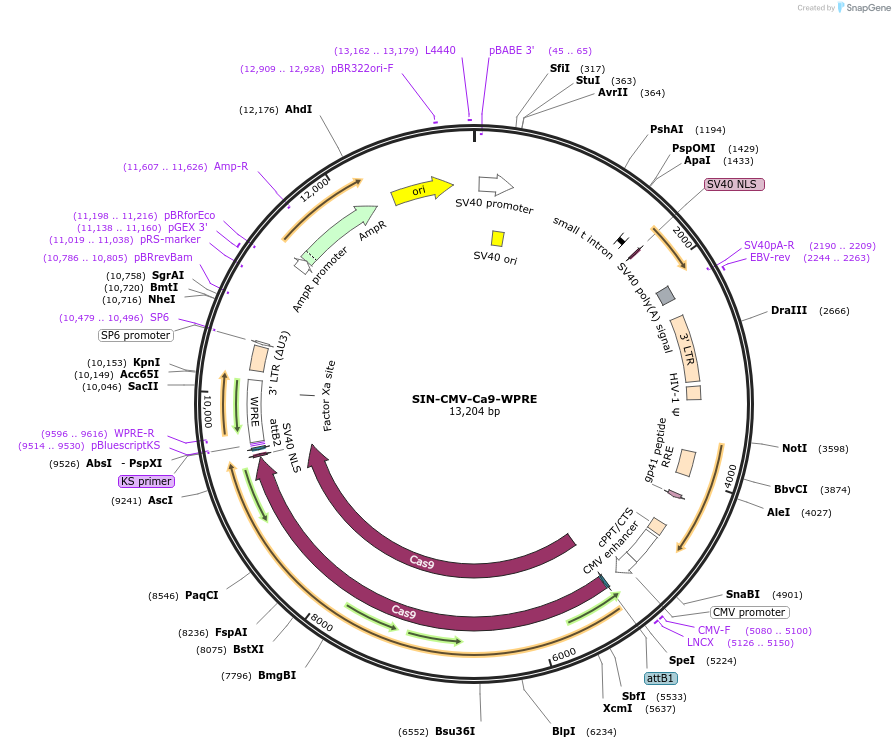
SIN-CMV-Ca9-WPRE
Plasmid#87874PurposeTransfer plasmid for the production of lentiviral vectorsDepositorInsertspCas9
UseLentiviralTagsnlsMutationHuman codon-optimizedPromoterCMVAvailable SinceSept. 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
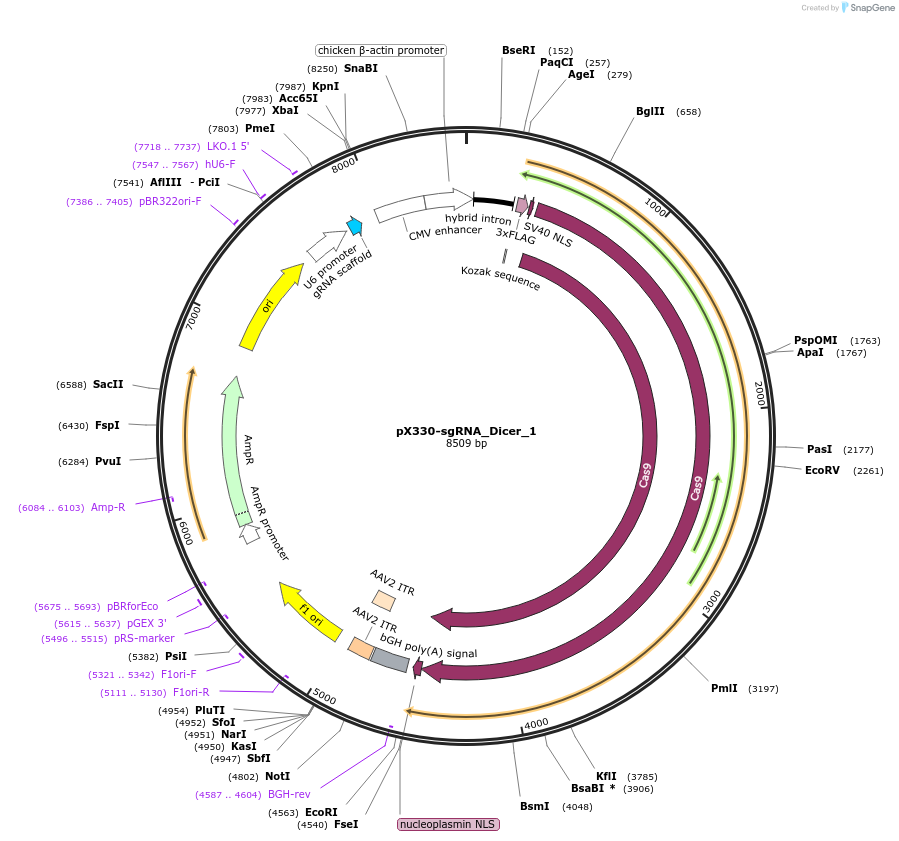
pX330-sgRNA_Dicer_1
Plasmid#68807Purposespecific sgRNA against mouse Dicer1 gene cloned in the pX330 backbone (Addgene Number 42230). Generation of Dicer1 knockout mESCs.DepositorInsertsgRNA mouse Dicer1
UseCRISPR and Mouse TargetingExpressionBacterial and MammalianAvailable SinceFeb. 13, 2017AvailabilityAcademic Institutions and Nonprofits only -
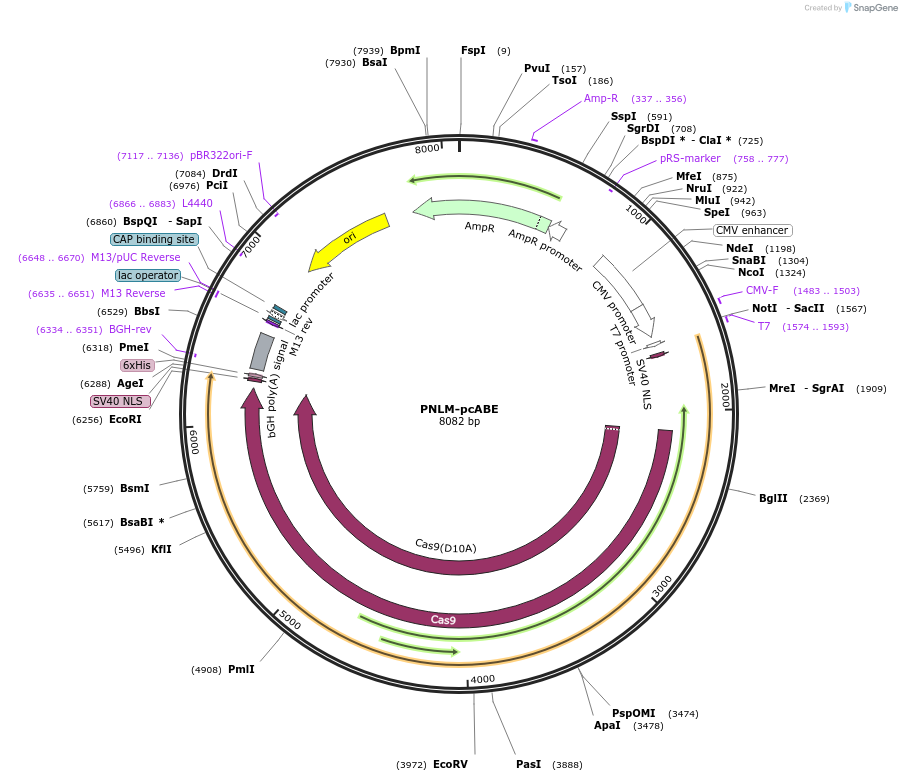
PNLM-pcABE
Plasmid#237618PurposePNLM-pcABE generated by pre-trained Protein-Nucleic Acid constrained Language Model exhibits high efficiency, a condensed editing window, and a shortened size.DepositorInsertecTadA(8e)-nSpCas9
UseCRISPRExpressionMammalianMutationdeleted amino acids 2-8 and 147-152PromoterpCMVAvailable SinceFeb. 6, 2026AvailabilityAcademic Institutions and Nonprofits only -
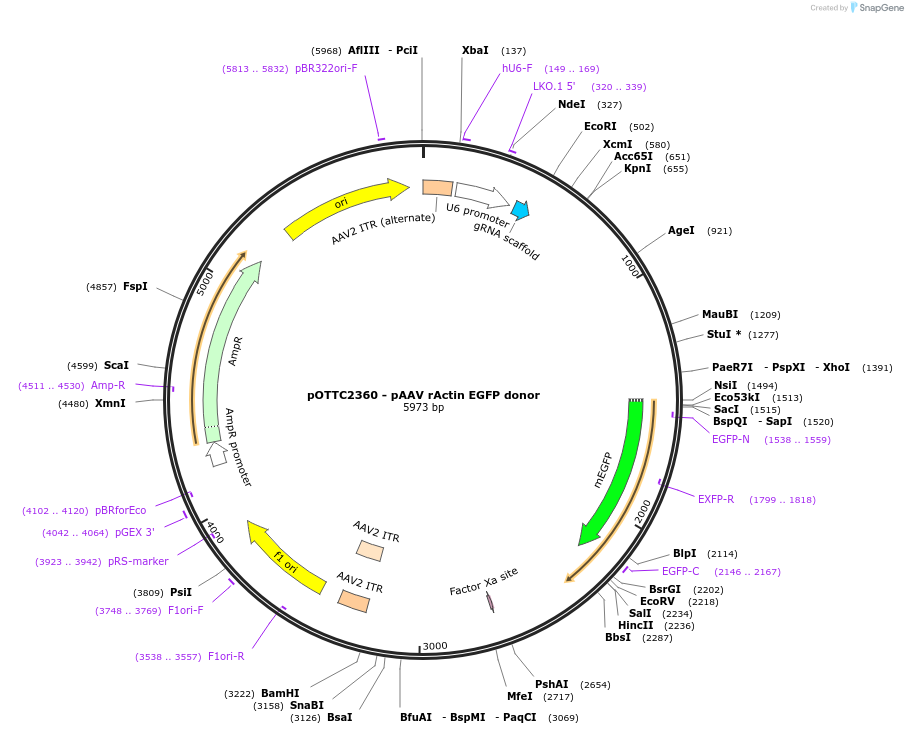
pOTTC2360 - pAAV rActin EGFP donor
Plasmid#228444PurposeAn adeno-associated viral vector to express a SpCas9 guide RNA targeting rat beta actin also carrying an HDR template for insertion of mEGFP to the break site.DepositorInsertmEGFP
UseAAV and CRISPRExpressionMammalianAvailable SinceJan. 26, 2026AvailabilityAcademic Institutions and Nonprofits only -
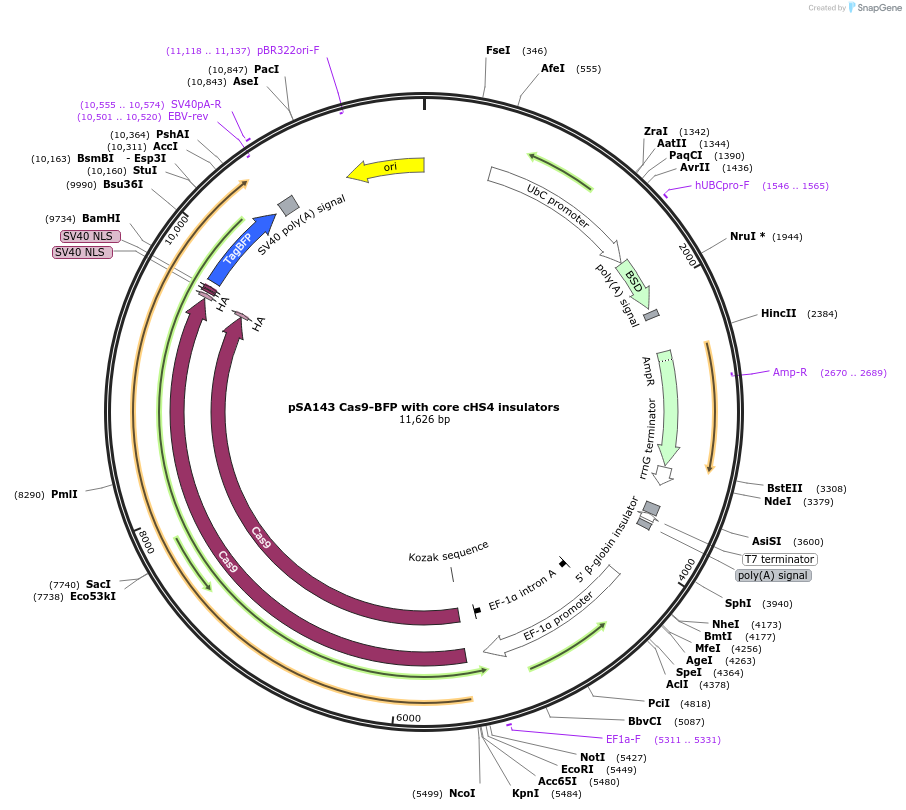
pSA143 Cas9-BFP with core cHS4 insulators
Plasmid#249153PurposeOverexpression of SpCas9-BFP and blasticidin resistance gene. Flanked by cHS4 core insulators. Contains Cp36 recombination site.DepositorInsertCas9-TagBFP with core cHS4 insulators
UseCRISPR; Cp36 donor dnaExpressionMammalianPromoterEF1aAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
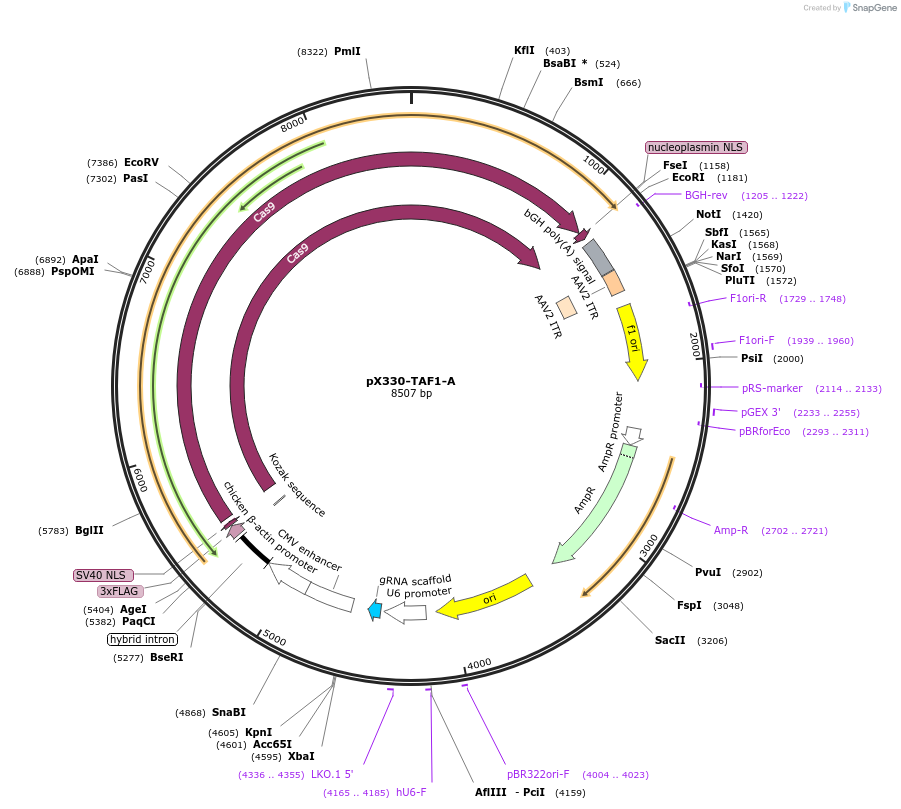
pX330-TAF1-A
Plasmid#247346PurposeExpresses SpCas9 and a sgRNA targeting the human TAF1-A loci for knock-in.DepositorAvailable SinceNov. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
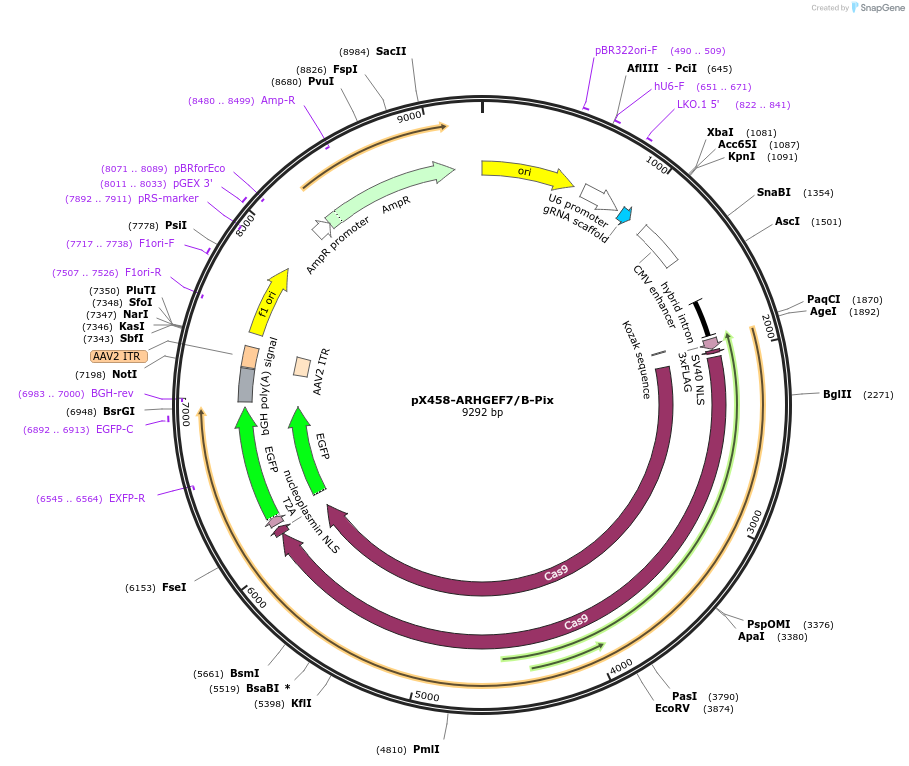
pX458-ARHGEF7/B-Pix
Plasmid#241368PurposeMammalian expression of SpCas9 and gRNA targeting ARHGEF7 (β-Pix)DepositorAvailable SinceAug. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
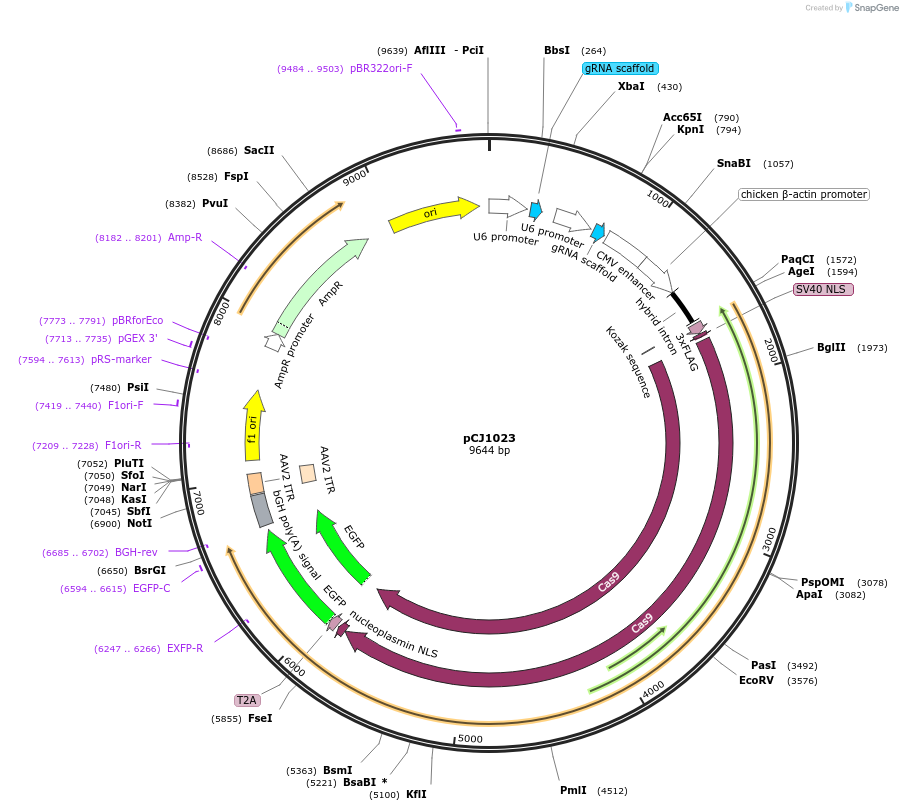
pCJ1023
Plasmid#228763PurposePlasmid expressing Cas9 and gRNAs for mouse Ift88 and Pkd2. Use for disruption of mouse Ift88 and Pkd2 in cultured cells.DepositorUseCRISPRTags3XFLAG and GFPExpressionMammalianPromoterhuman U6Available SinceDec. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
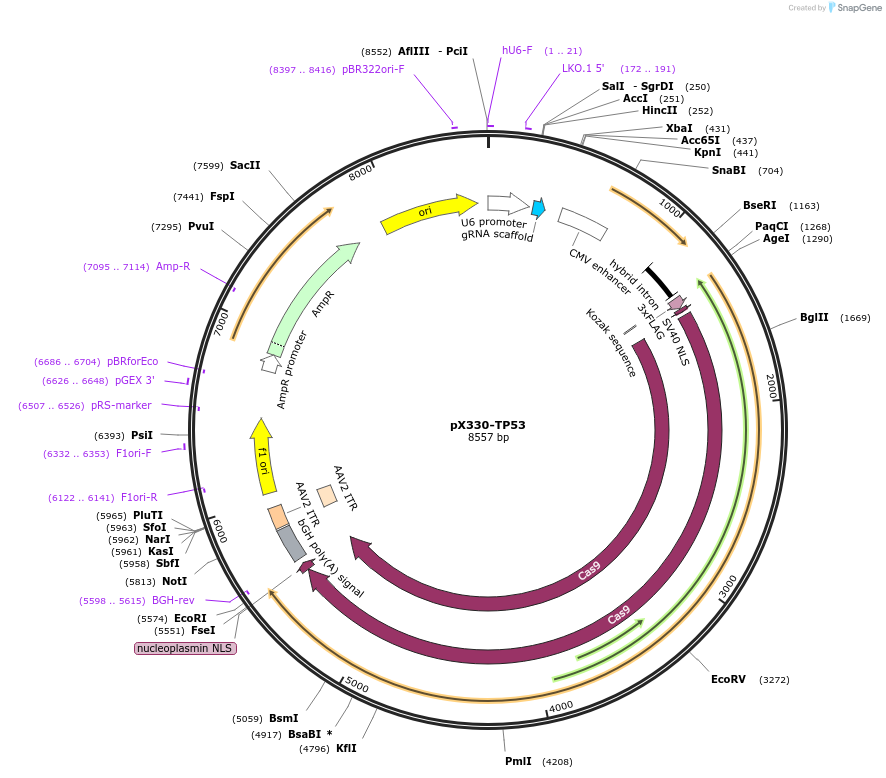
pX330-TP53
Plasmid#227318PurposeExpresses SpCas9 and a sgRNA targeting the N-terminus of TP53 for knock-in.DepositorAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
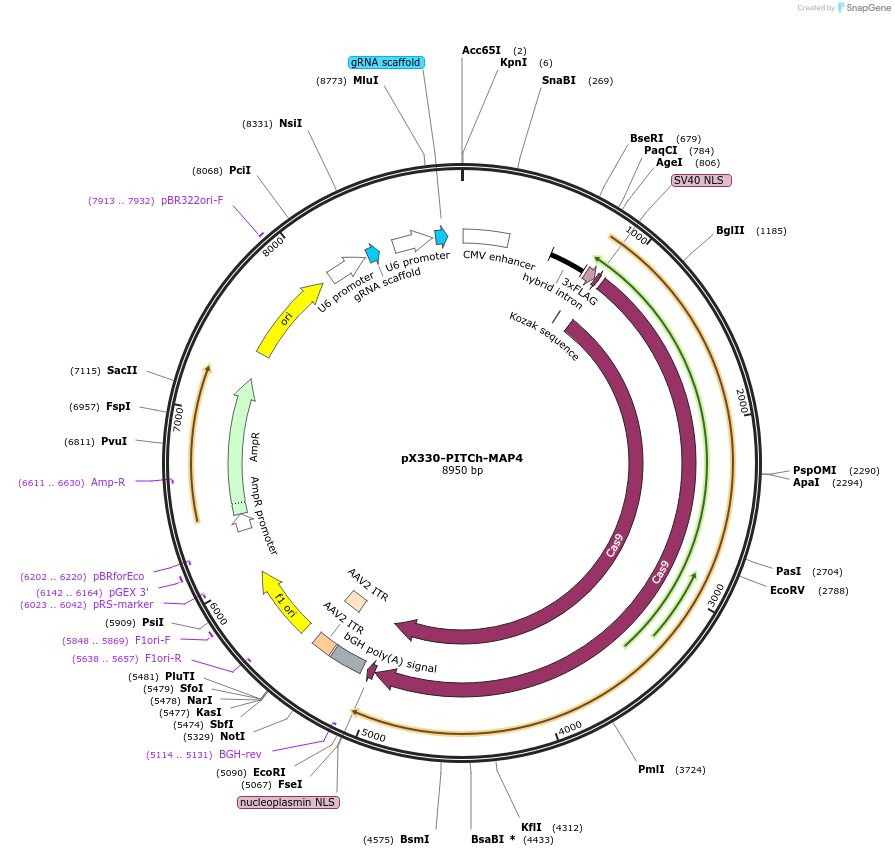
pX330-PITCh-MAP4
Plasmid#227295PurposeExpresses SpCas9, the PITCh gRNA, and a sgRNA targeting the N-terminus of MAP4 for knock-in.DepositorAvailable SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
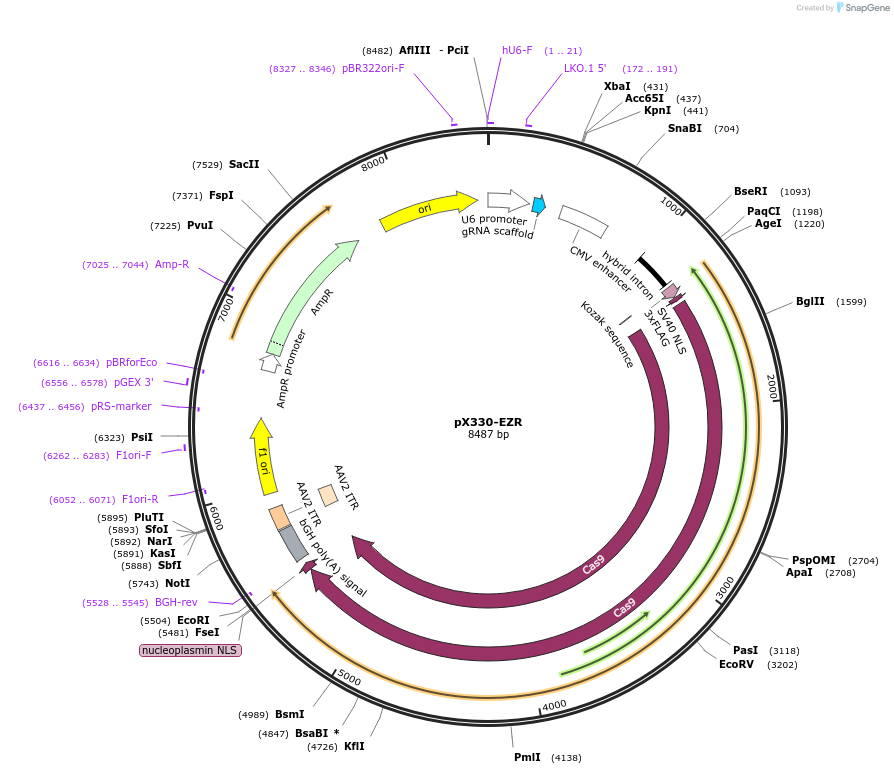
pX330-EZR
Plasmid#227293PurposeExpresses SpCas9 and a sgRNA targeting the C-terminus of EZR for knock-in.DepositorAvailable SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
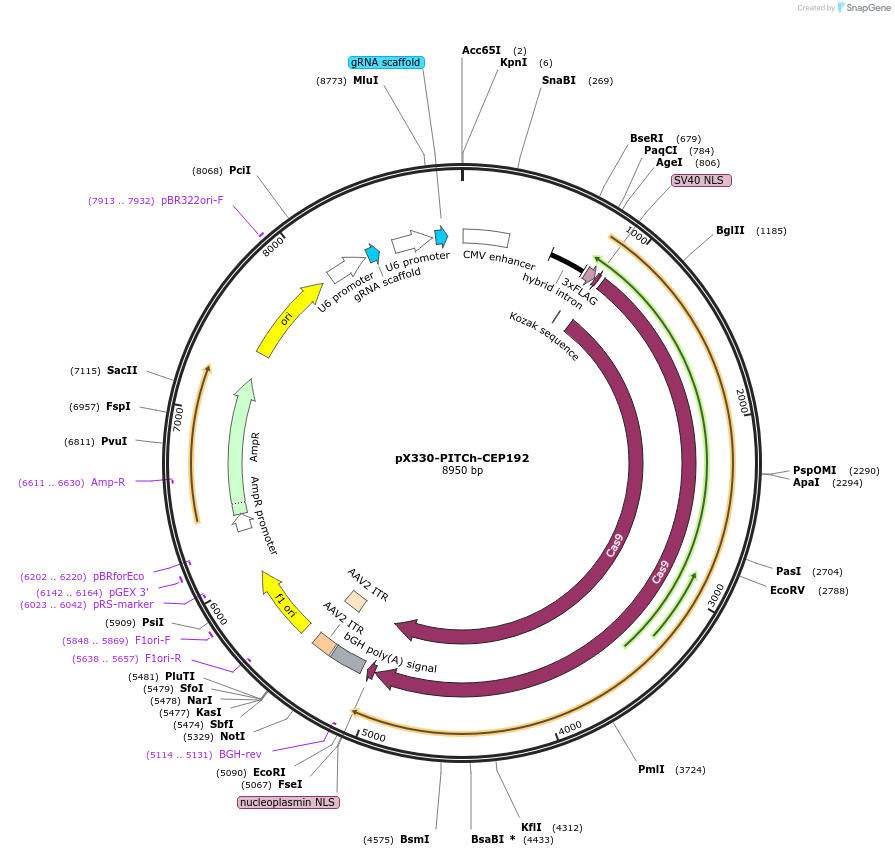
pX330-PITCh-CEP192
Plasmid#227288PurposeExpresses SpCas9, the PITCh gRNA, and a sgRNA targeting the C-terminus of CEP192 for knock-in.DepositorInsertsgRNA Targeting C-terminus of CEP192 (CEP192 Human)
UseCRISPRExpressionMammalianPromoterU6Available SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
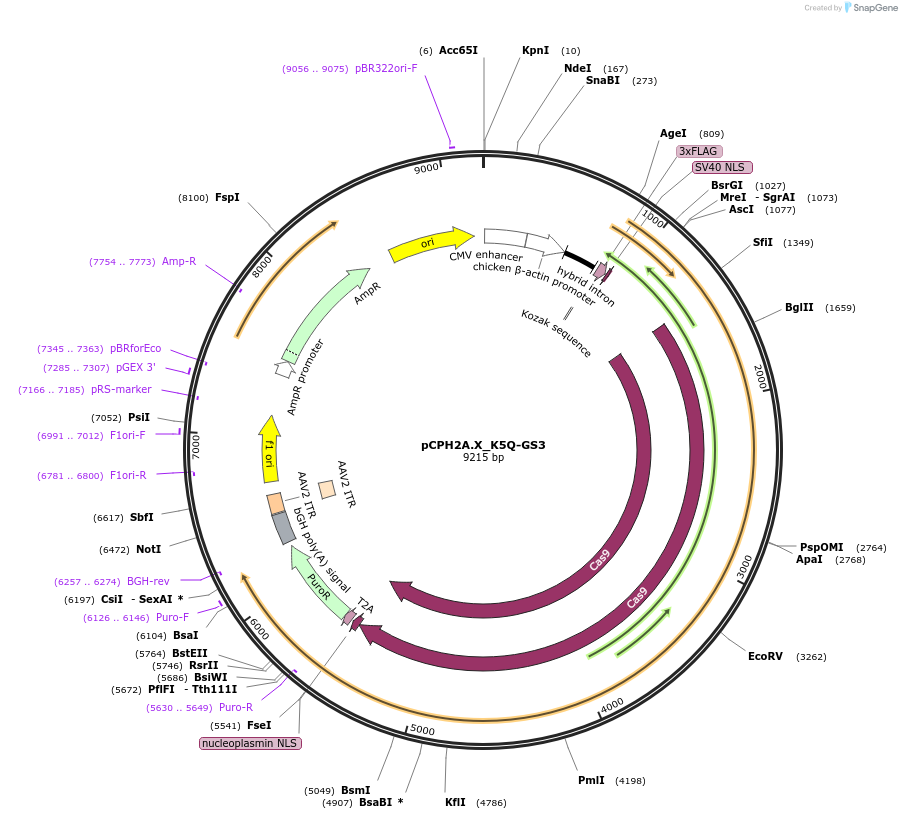
pCPH2A.X_K5Q-GS3
Plasmid#204746PurposeExpression of Cas9 and human H2AX with the K5Q mutationDepositorAvailable SinceJune 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
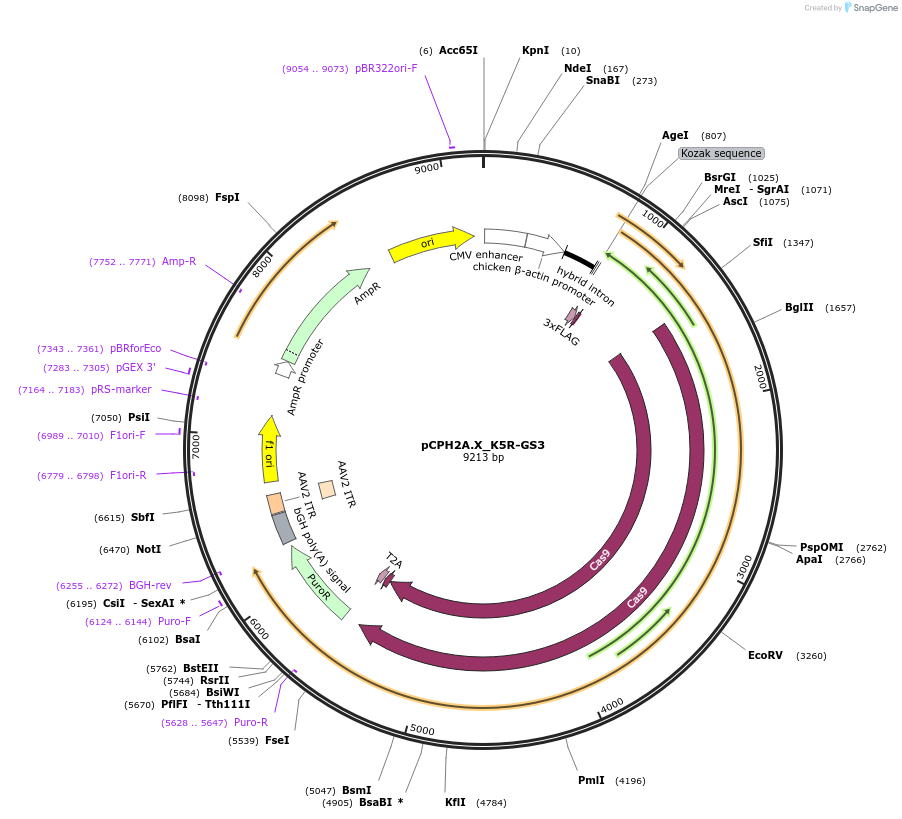
pCPH2A.X_K5R-GS3
Plasmid#204747PurposeExpression of Cas9 and human H2AX with the K5R mutationDepositorAvailable SinceJune 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
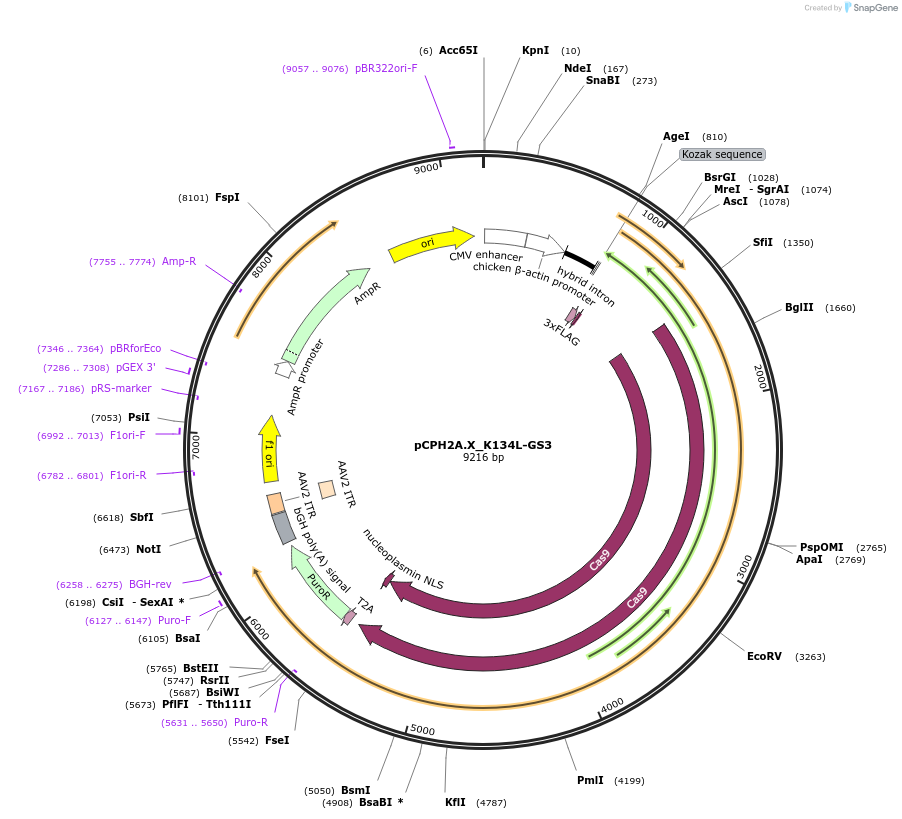
pCPH2A.X_K134L-GS3
Plasmid#204750PurposeExpression of Cas9 and human H2AX with the K134L mutationDepositorAvailable SinceJune 13, 2024AvailabilityAcademic Institutions and Nonprofits only