We narrowed to 4,755 results for: Drosophila
-
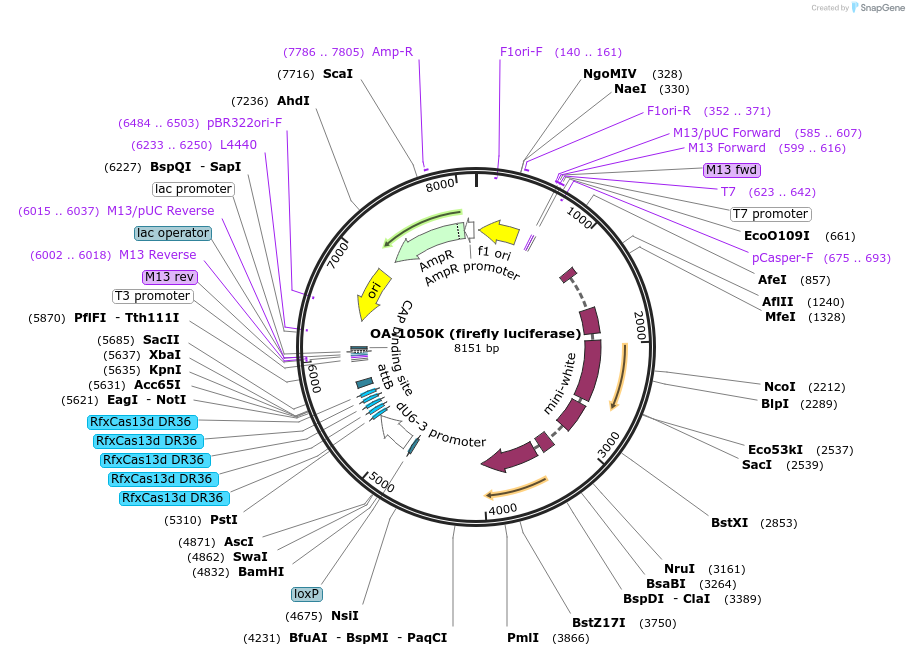
Plasmid#132422Purposeexpress arrays of gRNA targeting Firefly Luciferase under dU6-3 promoterDepositorInsertfirefly Luciferase gRNA array
ExpressionInsectPromoterU6-3Available SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
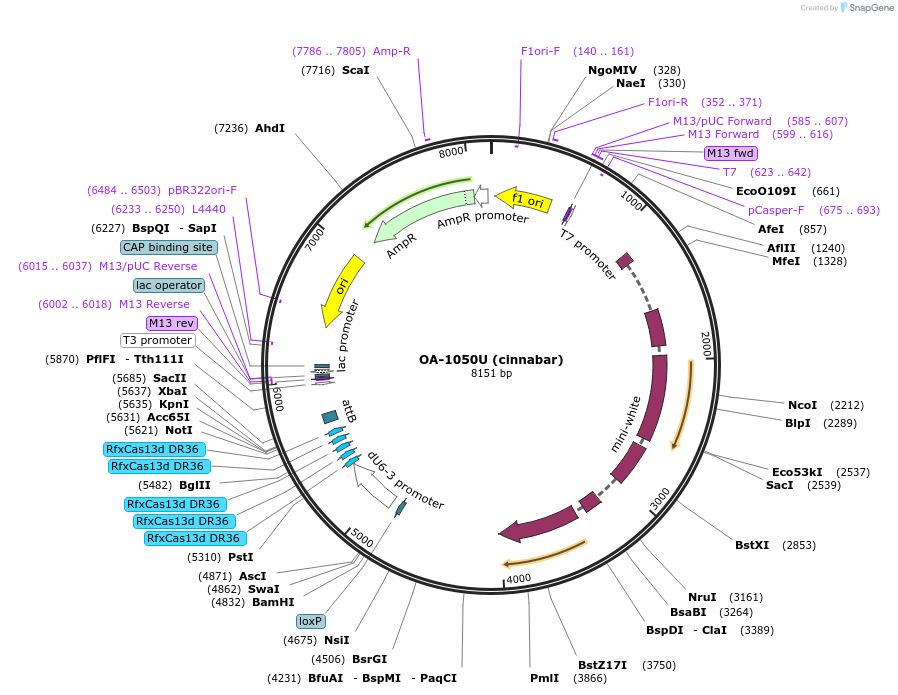
OA-1050U (cinnabar)
Plasmid#132423Purposeexpress arrays of gRNA targeting Cinnabar under dU6-3 promoterDepositorInsertcinnabar gRNA array
ExpressionInsectPromoterU6-3Available SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
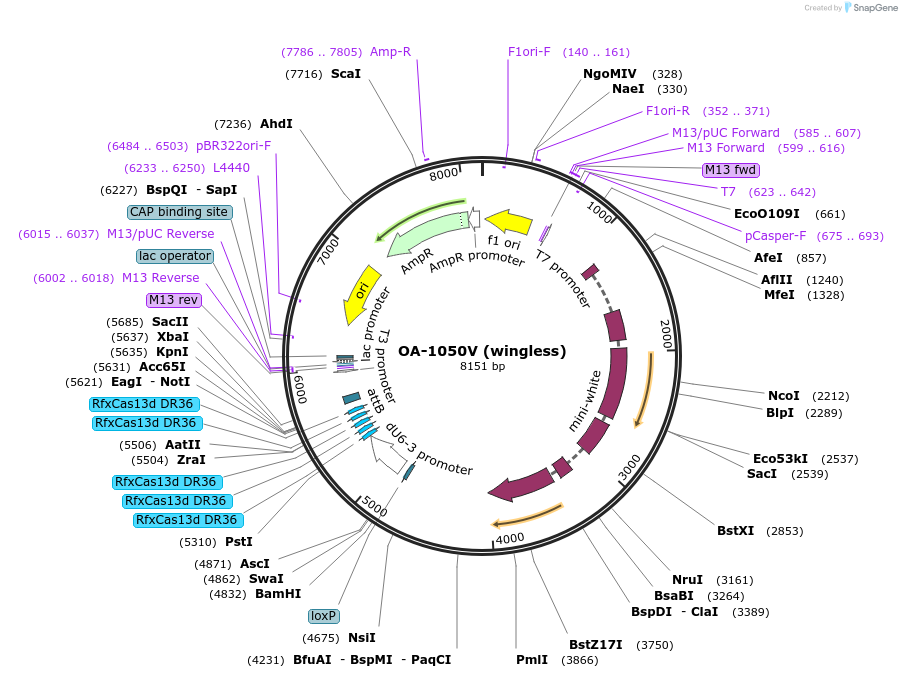
OA-1050V (wingless)
Plasmid#132424Purposeexpress arrays of gRNA targeting Wingless under dU6-3 promoterDepositorInsertwingless gRNA array
ExpressionInsectPromoterU6-3Available SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
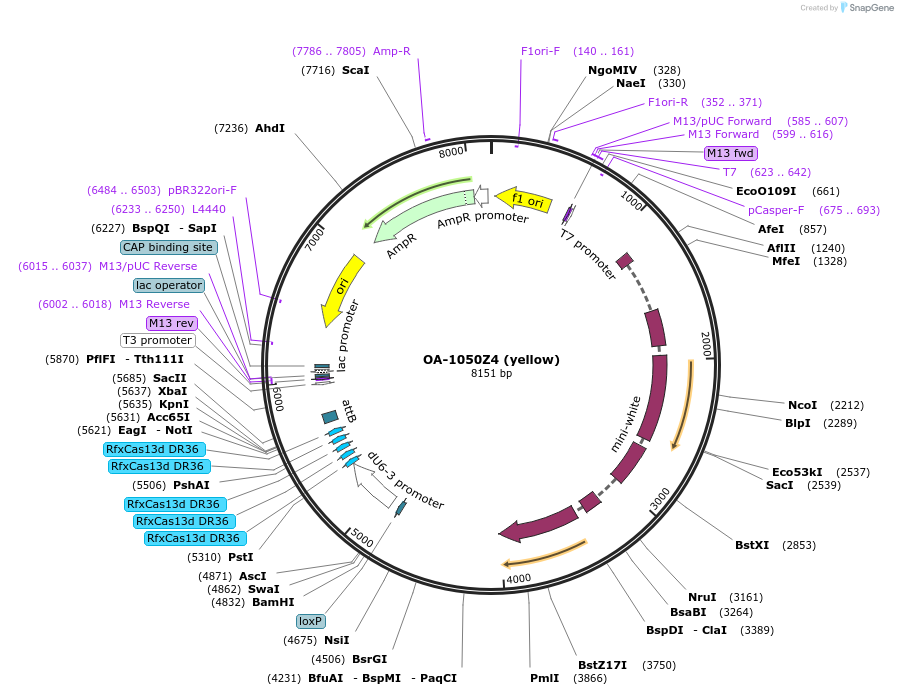
OA-1050Z4 (yellow)
Plasmid#132425Purposeexpress arrays of gRNA targeting Yellow under dU6-3 promoterDepositorInsertyellow gRNA array
ExpressionInsectPromoterU6-3Available SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
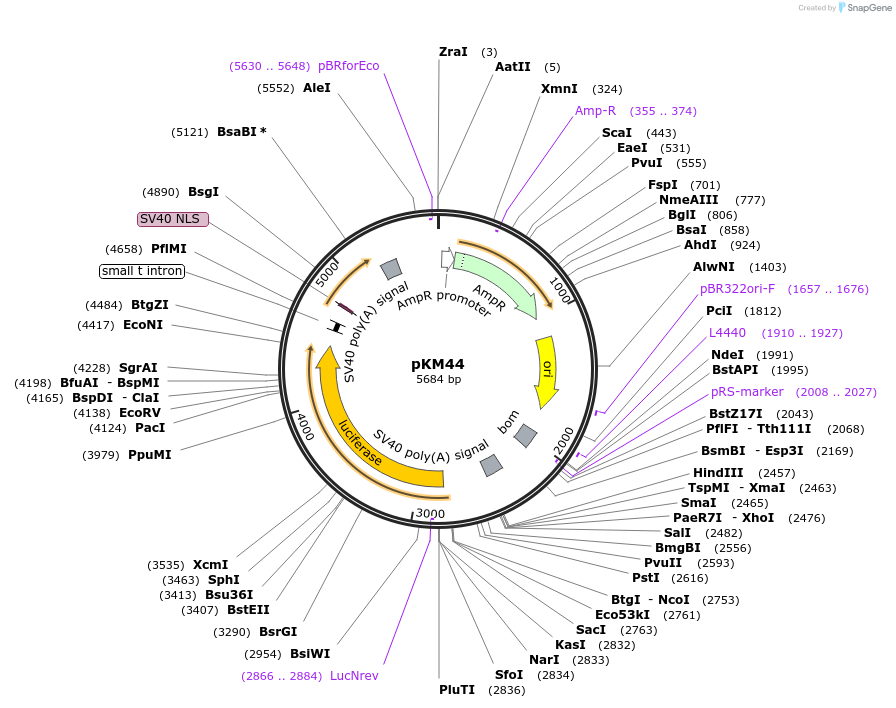
pKM44
Plasmid#154153PurposeFirefly luciferase under the control of the Drosophila melanogaster minimal Hsp70 promoterDepositorInsertFirefly luciferase
UseLuciferasePromoterDrosophila melanogaster minimal Hsp70 promoterAvailable SinceAug. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
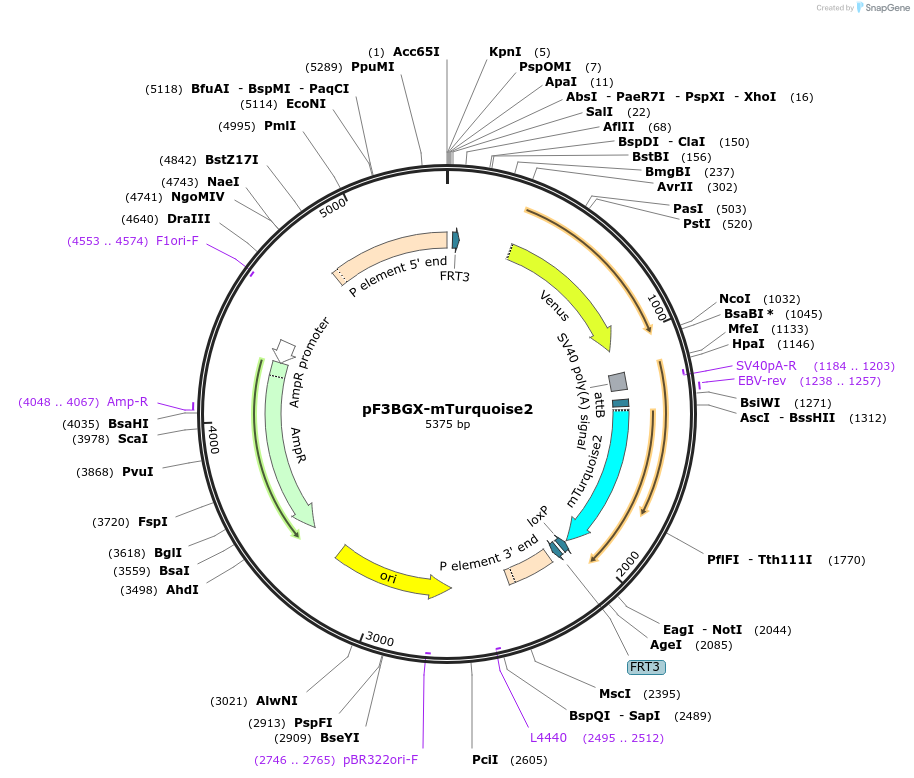
pF3BGX-mTurquoise2
Plasmid#138390PurposeRMCE vector for generating C-terminal mTurqouise2 fusionDepositorInsertmTurquoise2
ExpressionInsectAvailable SinceMarch 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
pGL3_zfh1_CP-candidate_luc+
Plasmid#86391PurposeLuciferase validation vector, where candidates are inserted at the position of the core promoter. The candidates responsiveness to the upstream enhancer is reflected by luciferase activityDepositorInsertD. melanogaster zfh1 enhancer
UseLuciferaseAvailable SinceMarch 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
pRK5-FLAG-dSestrin (CG11299-PD)
Plasmid#73675PurposeoverexpressionDepositorAvailable SinceMarch 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_CUS1E07
Plasmid#71086PurposeGateway entry cloneDepositorInsertTaf13 (Taf13 Fly)
UseGateway entry vectorAvailable SinceFeb. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_DUM1B01
Plasmid#71089PurposeGateway entry cloneDepositorInsertClk (Clk Fly)
UseGateway entry vectorAvailable SinceFeb. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF1B11
Plasmid#70848PurposeGateway entry cloneDepositorInsertpont (pont Fly)
UseGateway entry vectorAvailable SinceDec. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF1C11
Plasmid#70849PurposeGateway entry cloneDepositorInsertOrc5 (Orc5 Fly)
UseGateway entry vectorAvailable SinceDec. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF1D12
Plasmid#70857PurposeGateway entry cloneDepositorInsertRpn3 (Rpn3 Fly)
UseGateway entry vectorAvailable SinceDec. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF1E12
Plasmid#70858PurposeGateway entry cloneDepositorInsertnot (not Fly)
UseGateway entry vectorAvailable SinceDec. 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF2F11
Plasmid#71052PurposeGateway entry cloneDepositorInsertRbf (Rbf Fly)
UseGateway entry vectorAvailable SinceDec. 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF4H02
Plasmid#71026PurposeGateway entry cloneDepositorInsertBre1 (Bre1 Fly)
UseGateway entry vectorAvailable SinceDec. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF4A03
Plasmid#71027PurposeGateway entry cloneDepositorInsertlds (lds Fly)
UseGateway entry vectorAvailable SinceDec. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF4E02
Plasmid#71024PurposeGateway entry cloneDepositorInsertsimj (simj Fly)
UseGateway entry vectorAvailable SinceDec. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF1H07
Plasmid#71031PurposeGateway entry cloneDepositorInsertexd (exd Fly)
UseGateway entry vectorAvailable SinceDec. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF3D01
Plasmid#70999PurposeGateway entry cloneDepositorInsertChro (Chro Fly)
UseGateway entry vectorAvailable SinceDec. 1, 2015AvailabilityAcademic Institutions and Nonprofits only