We narrowed to 10,436 results for: codon optimized
-
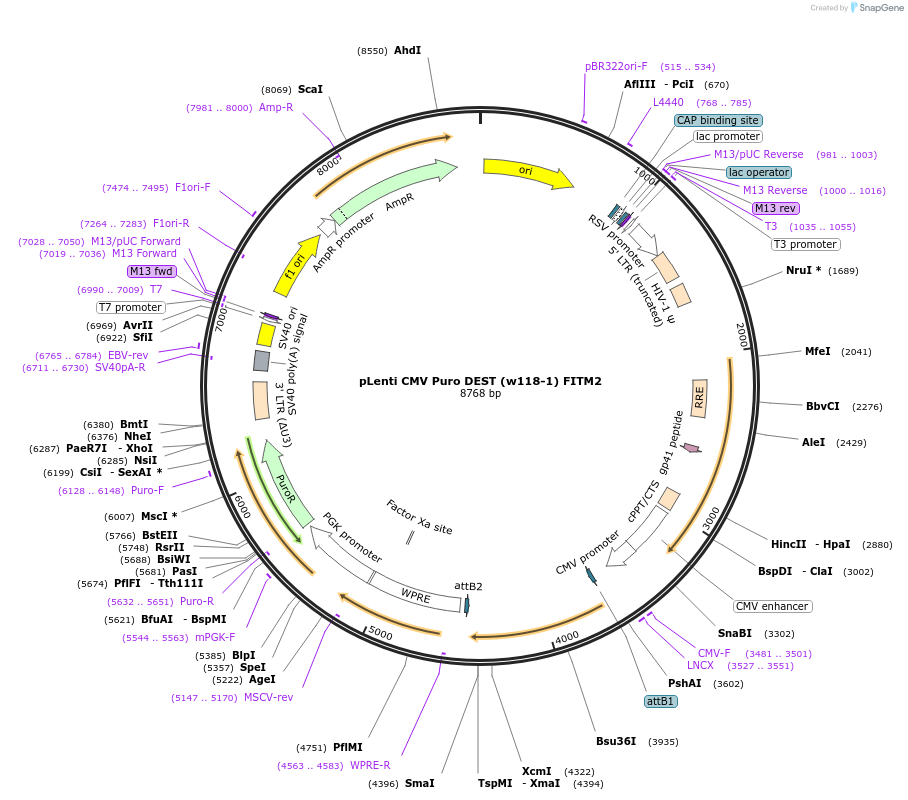
Plasmid#107504PurposeExpresses FITM2, puromycin resistantDepositorInsertFITM2 (codon optimized)
UseLentiviralExpressionMammalianMutationCodon optimized sequencePromoterCMVAvailable SinceNov. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
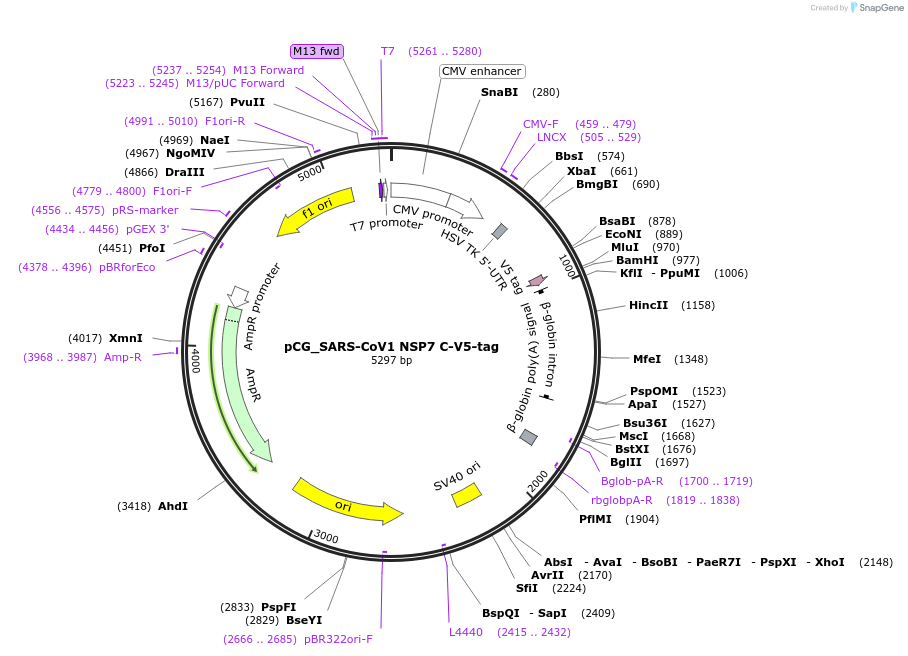
pCG_SARS-CoV1 NSP7 C-V5-tag
Plasmid#179963PurposeMammalian expression vector for SARS-CoV-1 Nsp7, V5-tagged. Sequence codon-optimized.DepositorAvailable SinceOct. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
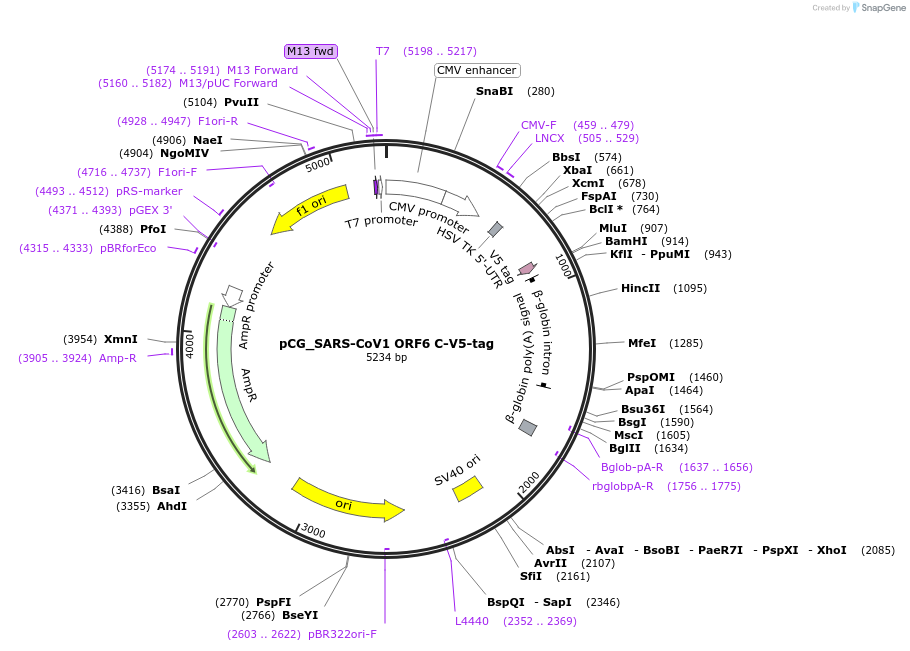
pCG_SARS-CoV1 ORF6 C-V5-tag
Plasmid#179970PurposeMammalian expression vector for SARS-CoV-1 ORF6, V5-tagged. Sequence codon-optimized.DepositorAvailable SinceFeb. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
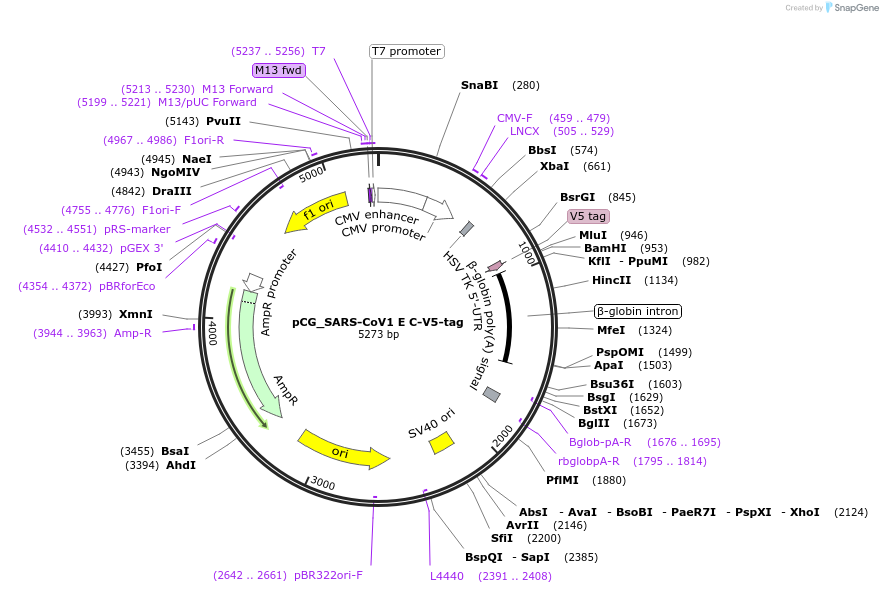
pCG_SARS-CoV1 E C-V5-tag
Plasmid#179966PurposeMammalian expression vector for SARS-CoV-1 E, V5-tagged. Sequence codon-optimized.DepositorAvailable SinceFeb. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
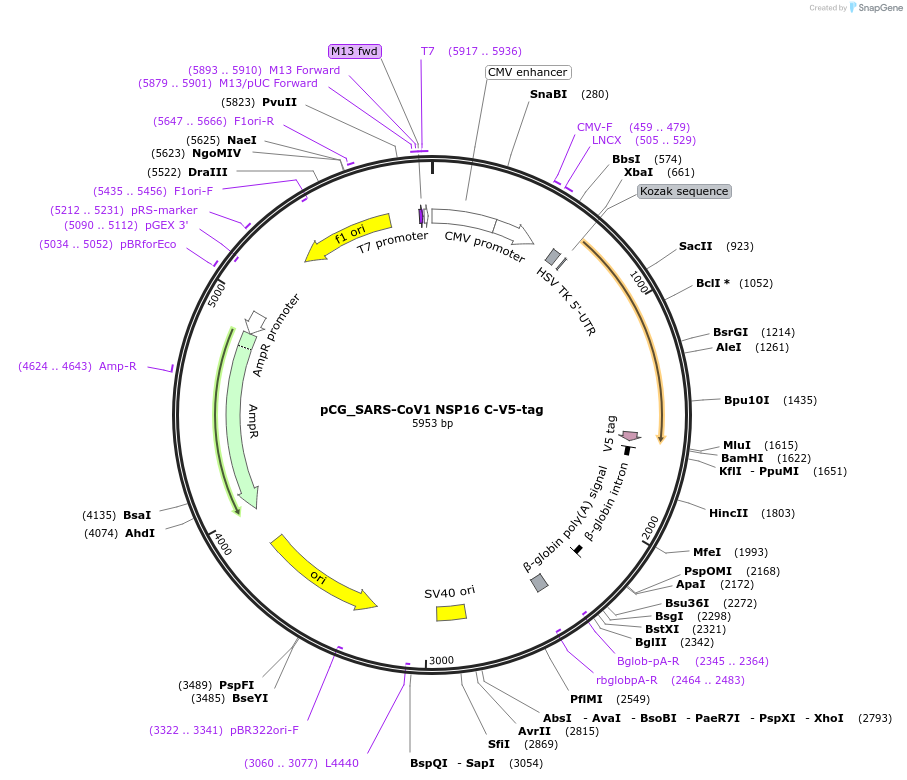
pCG_SARS-CoV1 NSP16 C-V5-tag
Plasmid#179965PurposeMammalian expression vector for SARS-CoV-1 Nsp16, V5-tagged. Sequence codon-optimized.DepositorAvailable SinceFeb. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
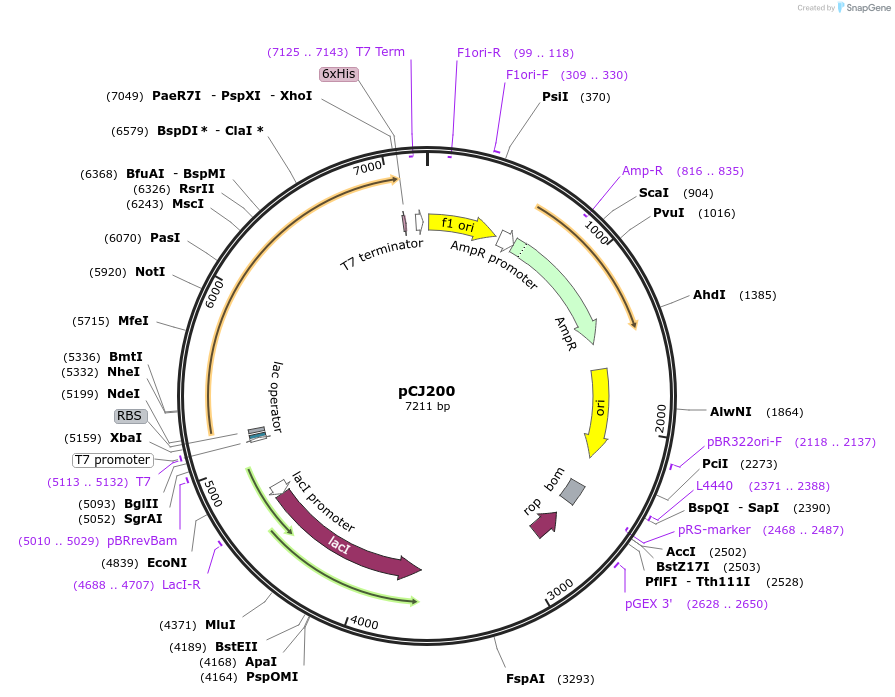
pCJ200
Plasmid#162673PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C and a sequence Tyr75:Phe104 to introduce a 6th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C and a sequence Y75-F104 to introduce a 6th…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
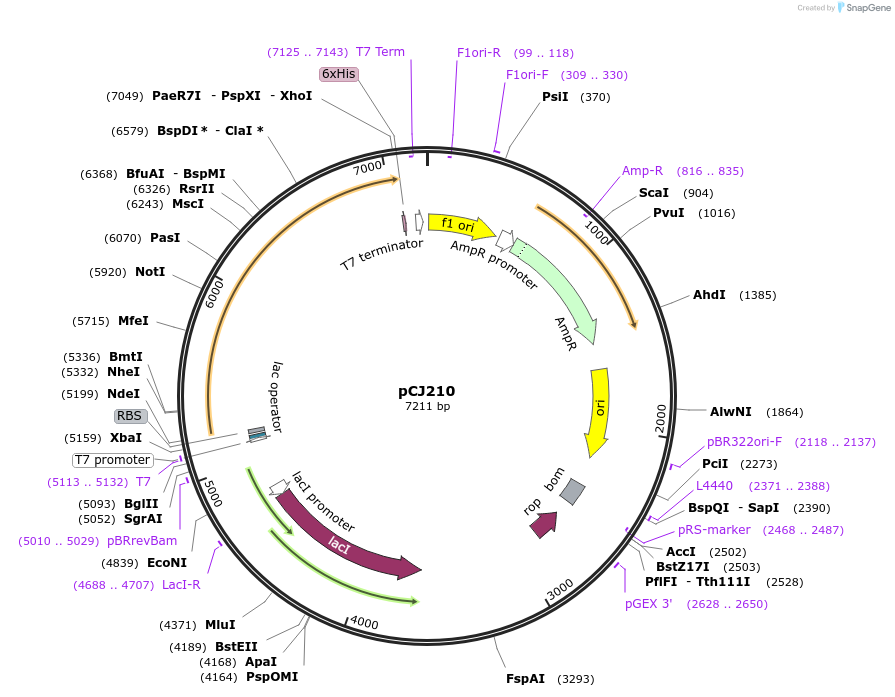
pCJ210
Plasmid#162683PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C, G489C, S530C, and a sequence Tyr75:Phe104 to introduce a 6th and 7th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C, G489C, S530C, and a sequence Y75-F104 to i…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
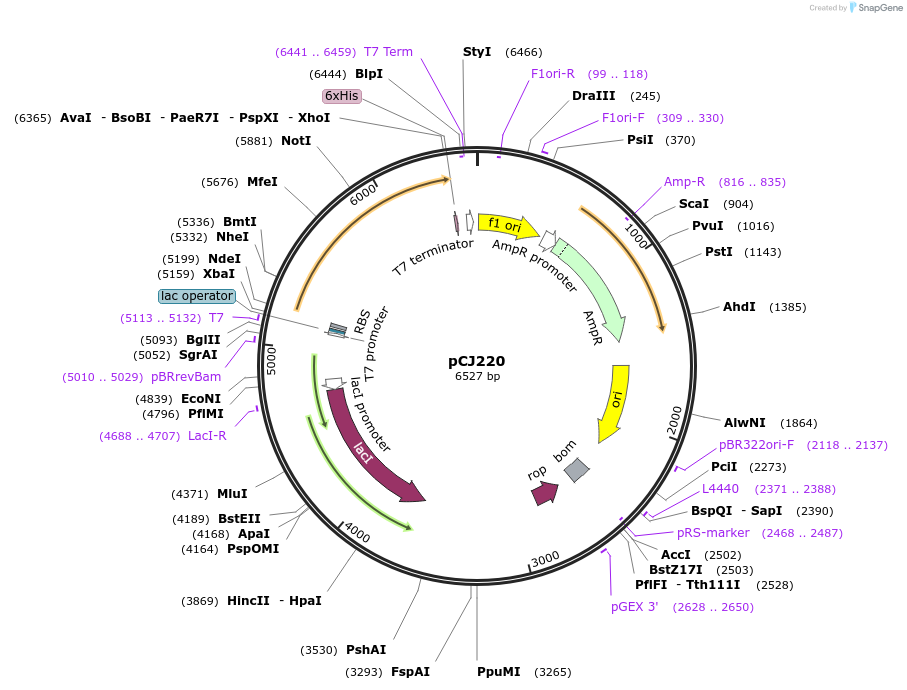
pCJ220
Plasmid#162686PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224W and C529S mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with lid deletion and C224W and C529S mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224W, C529S; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
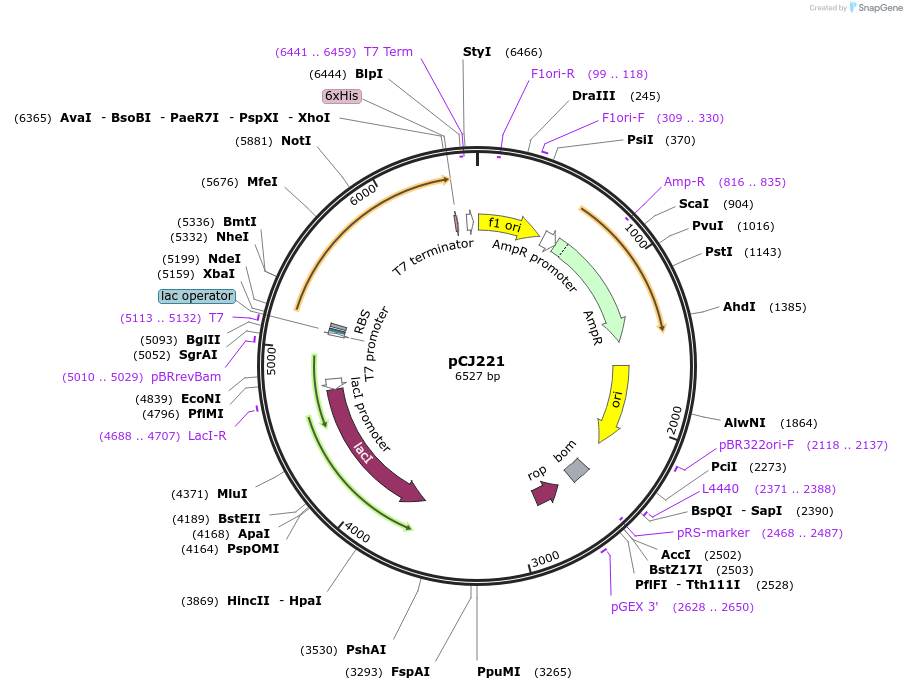
pCJ221
Plasmid#162687PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224H and C529F mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with lid deletion and C224H and C529F mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224H, C529F; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
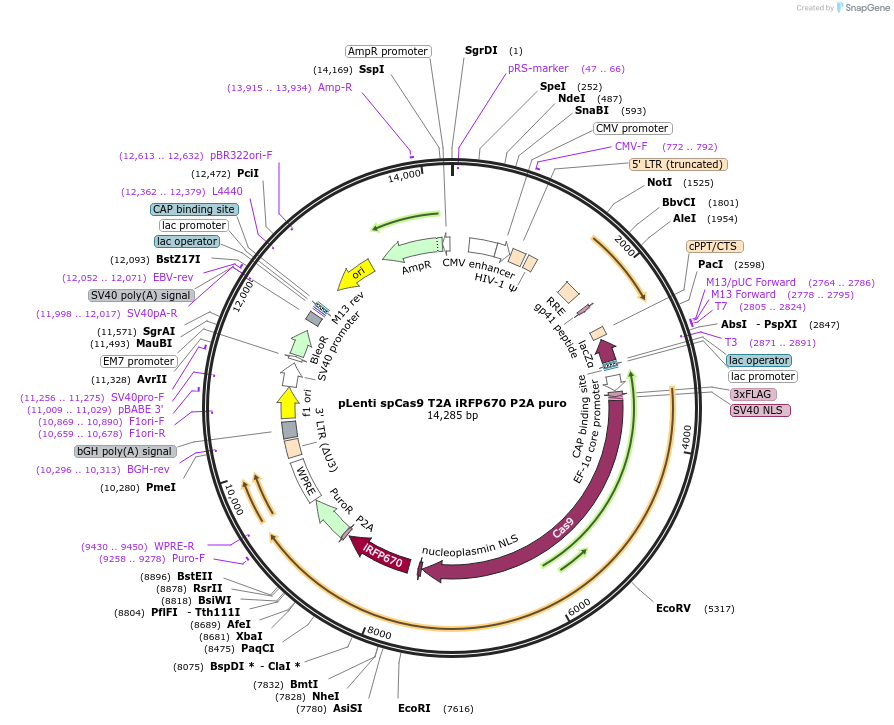
pLenti spCas9 T2A iRFP670 P2A puro
Plasmid#122182PurposeThe plasmid codes for a Flag-spCas9 protein, a iRFP670 fluorescent protein and a puromycin resistance. The plasmid has two BsmBI acceptor sites to insert gRNA expressing sequences.DepositorAvailable SinceFeb. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
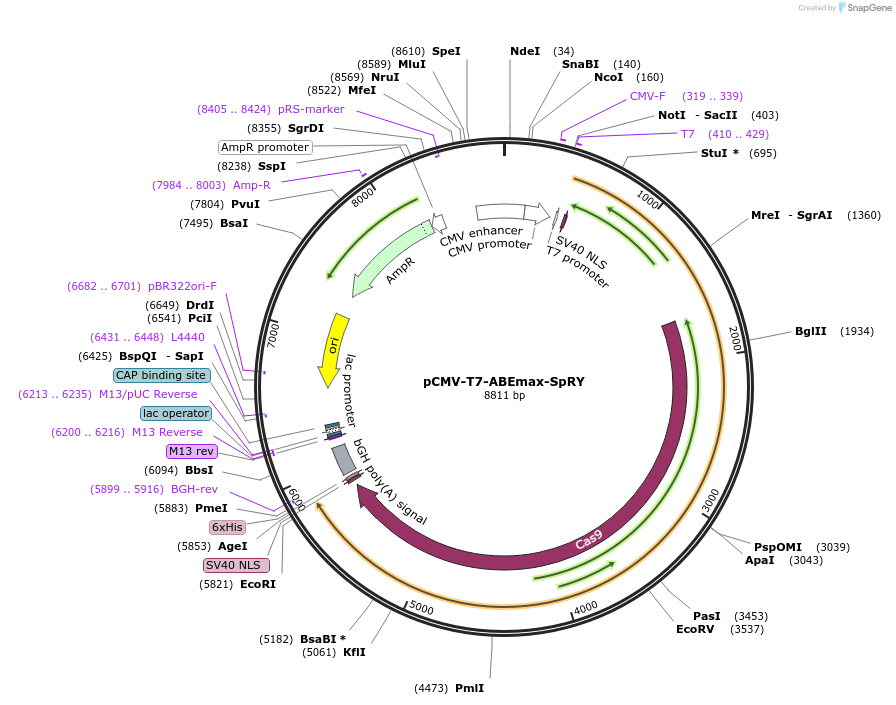
pCMV-T7-ABEmax-SpRY
Plasmid#195279PurposeAllows for in vitro transcription of original pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP (RTW5025) without EGFPDepositorInserthuman codon optimized ABEmax(7.10) SpCas9 variant named SpRY without EGFP
ExpressionMammalianPromoterCMVAvailable SinceJune 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
EP300
Plasmid#39018PurposeBacterial expression for structure determination; may not be full ORFDepositorAvailable SinceMarch 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
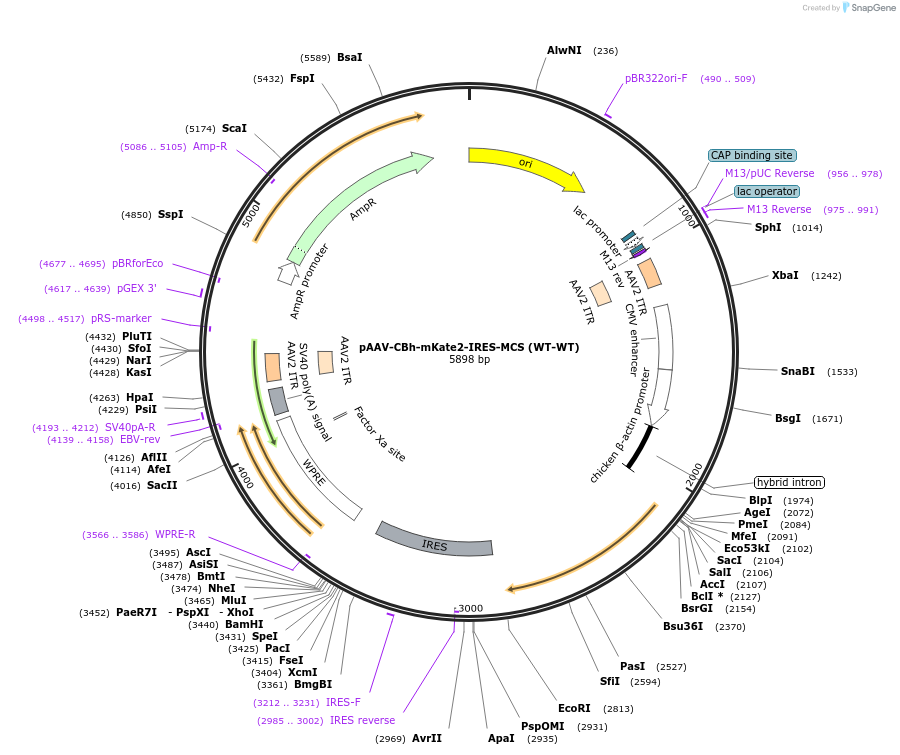
pAAV-CBh-mKate2-IRES-MCS (WT-WT)
Plasmid#105921PurposeAAV-mediated gene expression from CBh promoter. Contains canonical left and right AAV2 ITRsDepositorHas ServiceAAV2InsertsmKate2
IRES-Multiple cloning site
UseAAVMutationcodon optimizedPromoterCBh and same CBh promoter as mKate2Available SinceFeb. 15, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
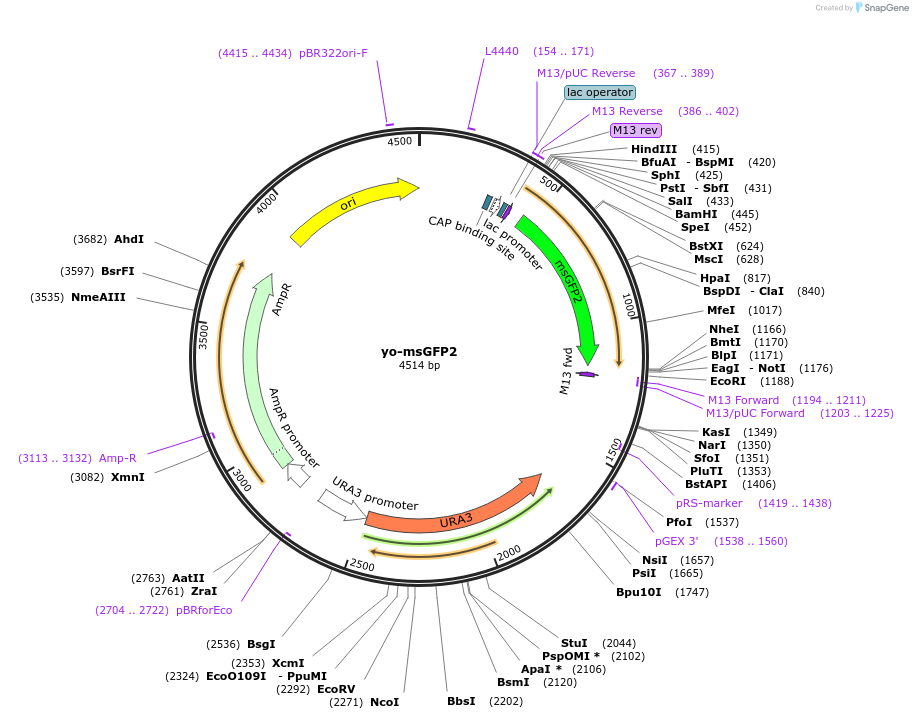
yo-msGFP2
Plasmid#160471PurposeEncodes a yeast codon optimized version of msGFP2DepositorInsertyo-msGFP2
ExpressionYeastAvailable SinceMarch 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
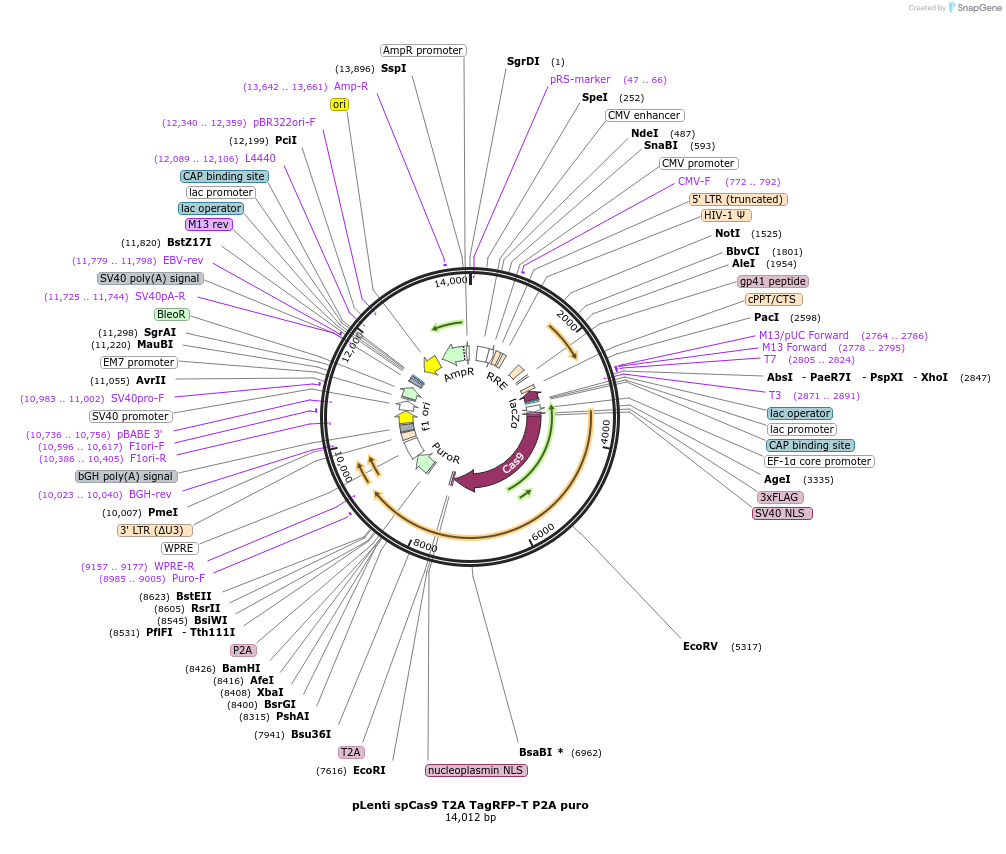
pLenti spCas9 T2A TagRFP-T P2A puro
Plasmid#122200PurposeThe plasmid codes for a Flag-spCas9 protein, a TagRFP-T fluorescent protein and a puromycin resistance. The plasmid has two BsmBI acceptor sites to insert gRNA expressing sequences.DepositorInsertsspCas9
TagRFP-T
UseLentiviralTagsT2AExpressionMammalianAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
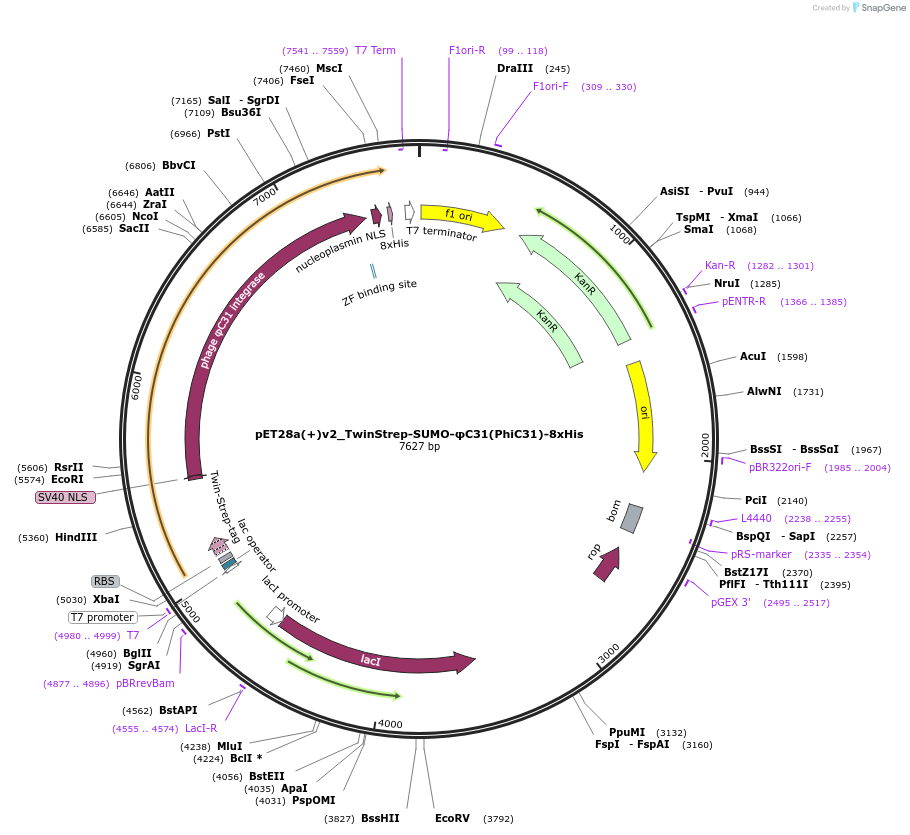
pET28a(+)v2_TwinStrep-SUMO-φC31(PhiC31)-8xHis
Plasmid#246982PurposeExpresses E. coli codon-optimized TwinStrep-SUMO-tagged φC31(PhiC31) integrase with N- and C-terminal NLS, using an improved pET design from Shilling et al. 2020.DepositorInsertφC31(PhiC31)
UseSynthetic BiologyTags8xHis, Nucleoplasmin, SUMO, SV40, and TwinStrepExpressionBacterialPromoterT7Available SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
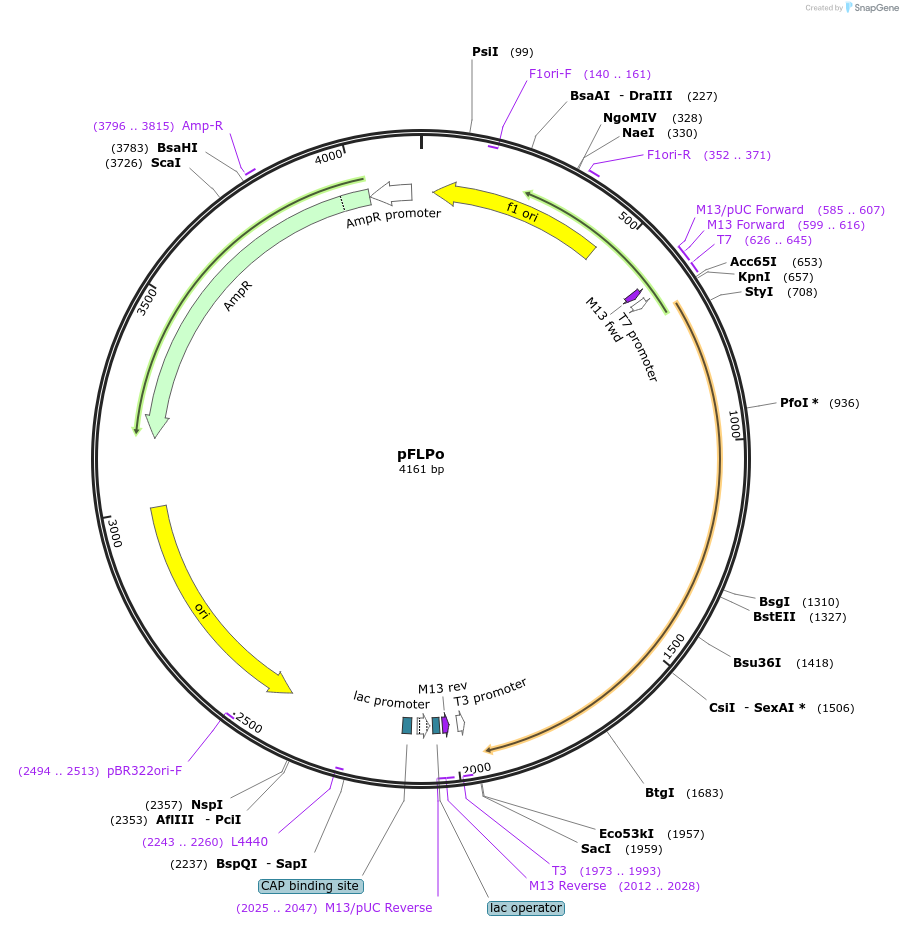
pFLPo
Plasmid#13792PurposeExpression of mouse codon-optimized FLP recombinaseDepositorInsertFLPo
UseCloning vectorExpressionBacterialAvailable SinceJan. 16, 2007AvailabilityAcademic Institutions and Nonprofits only -
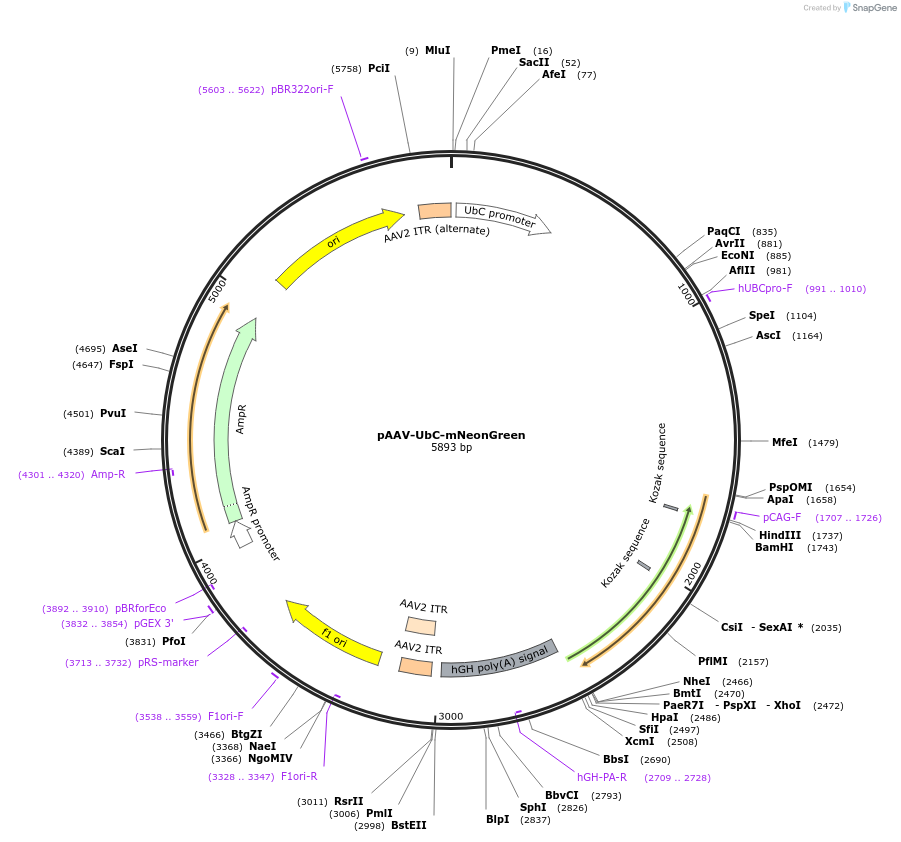
pAAV-UbC-mNeonGreen
Plasmid#124102PurposeExpression of the fluorescent protein mNeonGreen in mammalian cellsDepositorInsertmNeonGreen
UseAAVExpressionMammalianMutationCodon usage was optimized for Homo SapiensPromoterUbCAvailable SinceApril 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
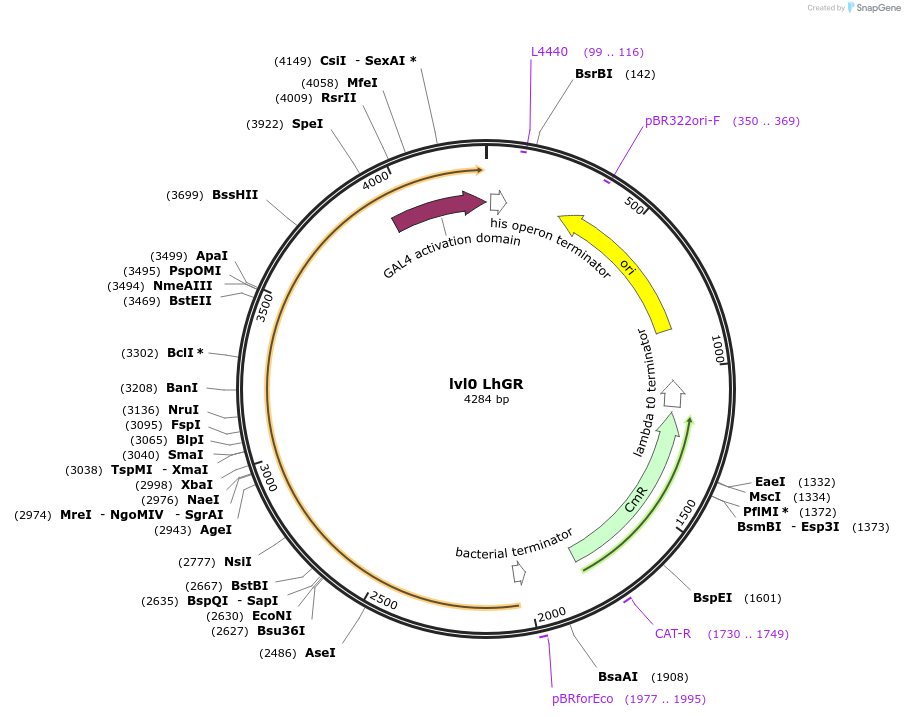
lvl0 LhGR
Plasmid#153408PurposeDex inducible system trans activatorDepositorInsertLhGR CDS
ExpressionPlantMutationCodon optimizedAvailable SinceNov. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
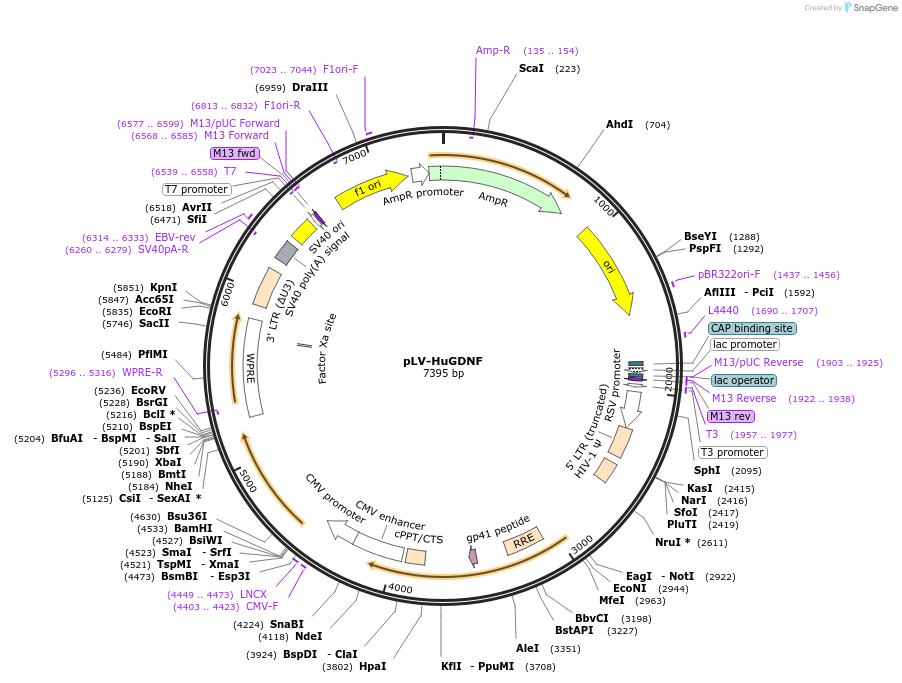
pLV-HuGDNF
Plasmid#182946PurposeTo expres Human GDNF with optimized codon usage for better expressionDepositorAvailable SinceApril 22, 2022AvailabilityAcademic Institutions and Nonprofits only