We narrowed to 4,703 results for: gla
-
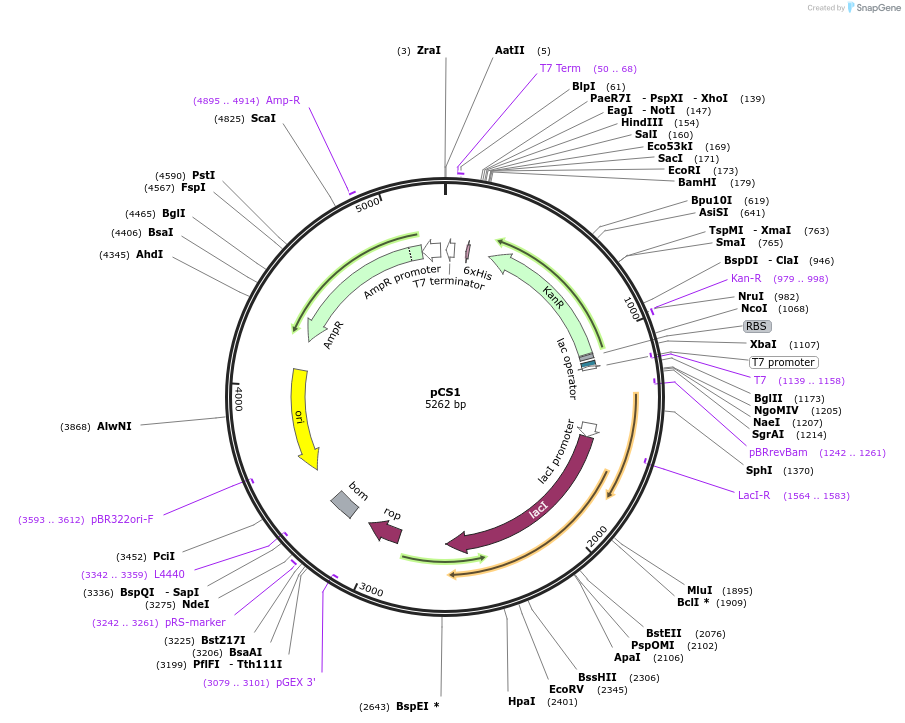
Plasmid#55749PurposeProduces aminoglycoside phosphotransferase (3′)-IIa when in the presence of T7 RNA Polymerase.DepositorInsertKanR
ExpressionBacterialPromoterT7-lacAvailable SinceJuly 15, 2014AvailabilityAcademic Institutions and Nonprofits only -
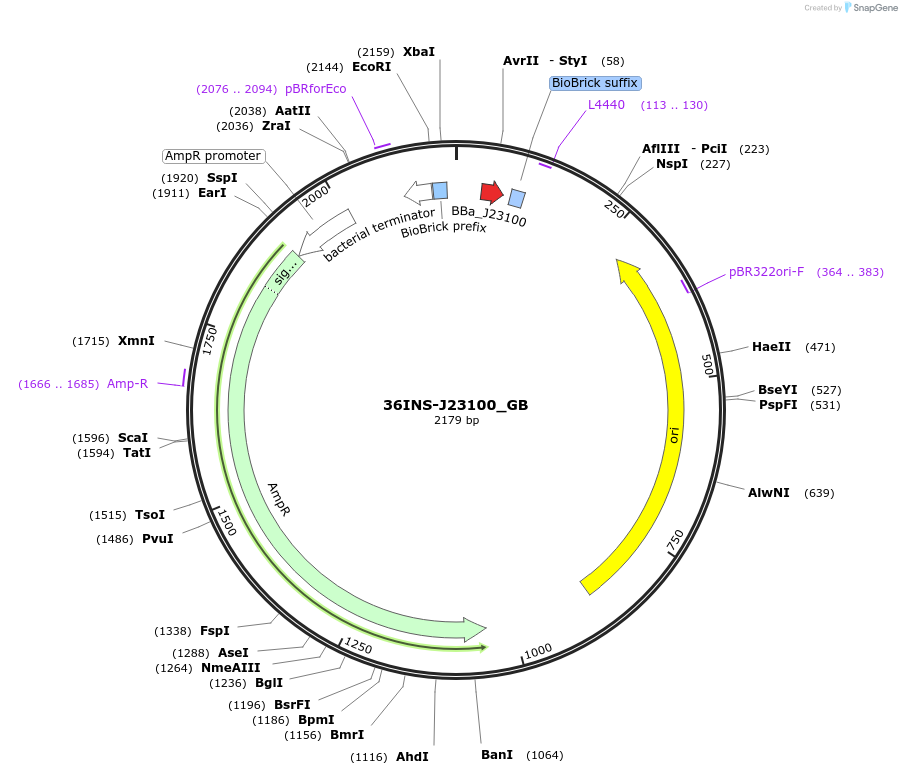
36INS-J23100_GB
Plasmid#78669Purpose36INS-J23100_GB Insulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_J23100. [G:36INS-J23100:B]DepositorInsertInsulated J23100 Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
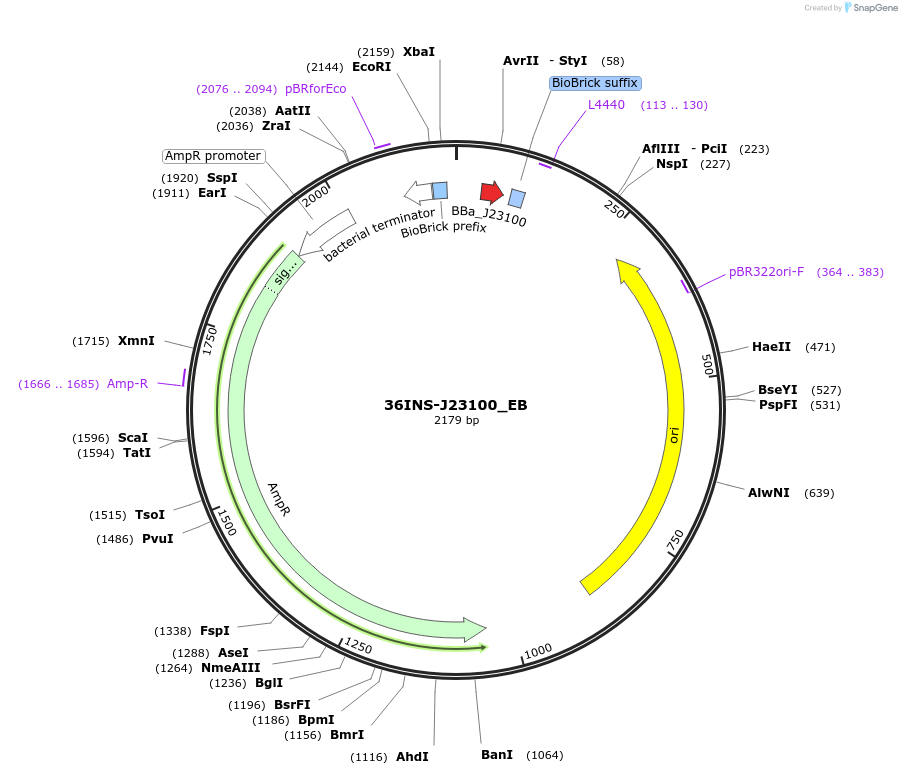
36INS-J23100_EB
Plasmid#78667Purpose36INS-J23100_EB Insulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_J23100. [E:36INS-J23100:B]DepositorInsertInsulated J23100 Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
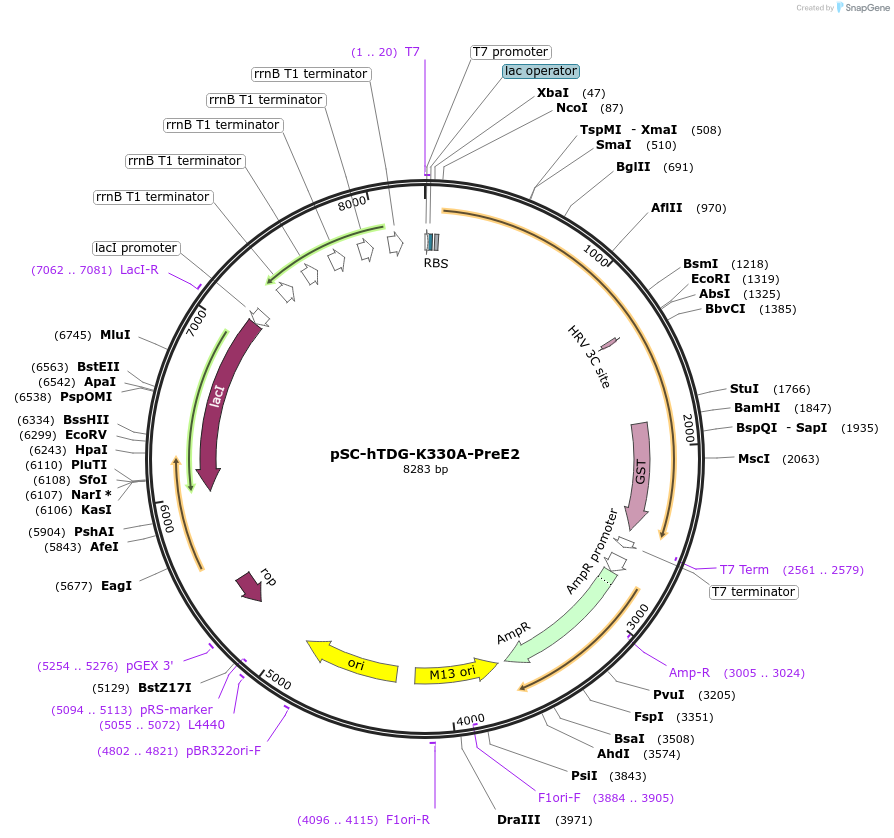
pSC-hTDG-K330A-PreE2
Plasmid#52273PurposeBacterial expression of human TDG-K330A C-terminally fused to SUMO-E2 and GSTDepositorInserthTDG (TDG Human)
TagsUbc9-GSTExpressionBacterialMutationK330A, SUMO acceptor site mutationPromoterT7Available SinceJuly 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
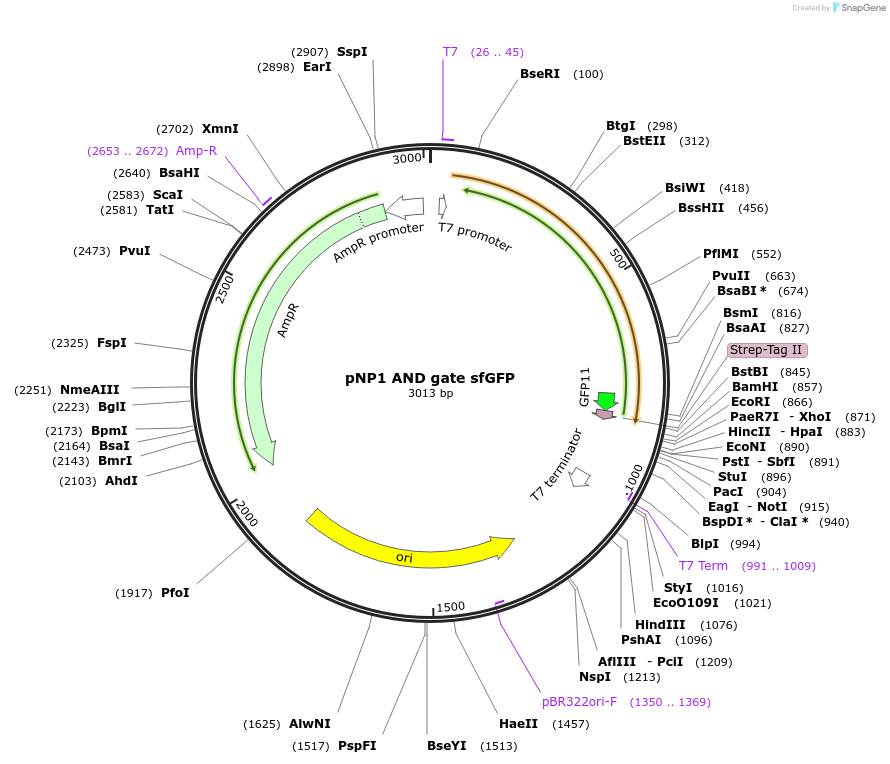
pNP1 AND gate sfGFP
Plasmid#107361PurposeExpresses sfGFP (a GFP) in Ecoli off a T7 promoter when pNP1 AND input A and pNP1 AND input B are co-expressedDepositorInsertAND gate + sfGFP
ExpressionBacterialPromoterT7Available SinceJan. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
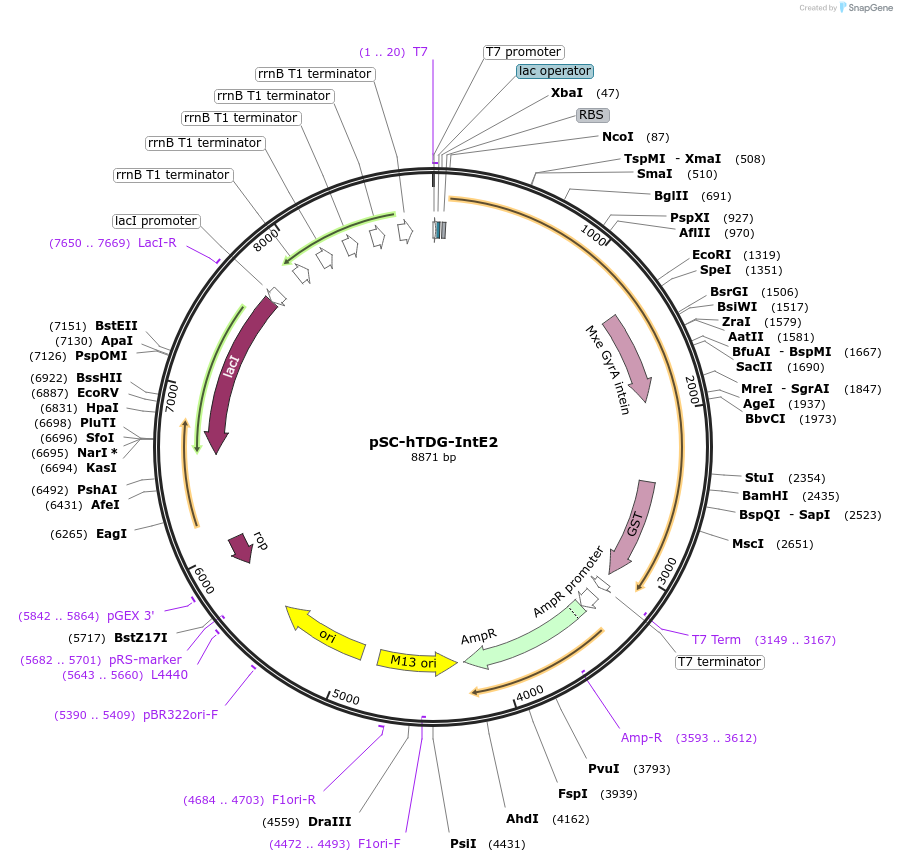
pSC-hTDG-IntE2
Plasmid#52270PurposeBacterial expression of human TDG C-terminally fused to GyrA intein, SUMO-E2 and GSTDepositorAvailable SinceJuly 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
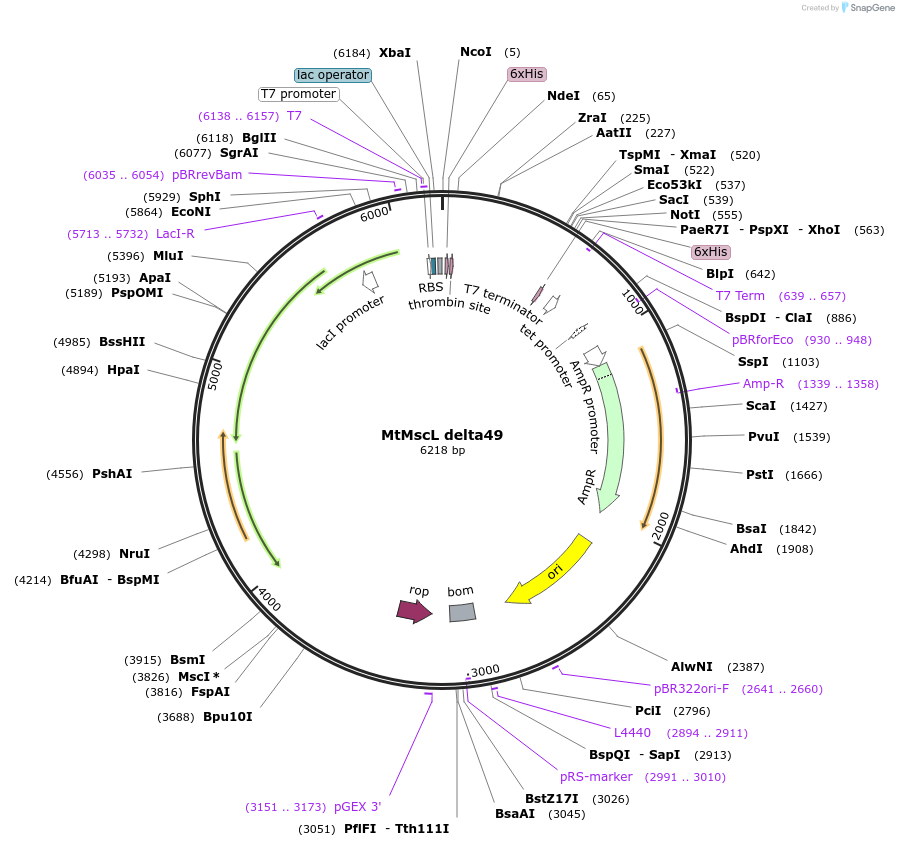
MtMscL delta49
Plasmid#118342PurposeM. tuberculosis MscL (delta49) in pET19bDepositorInsertMscL (mscL Mycobacterium tuberculosis)
Tags6x His Tag and Thrombin Cleavage siteExpressionBacterialMutationChanged Glutamic Acid (Glu) 102 to Stop CodonPromoterT7 PromoterAvailable SinceNov. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
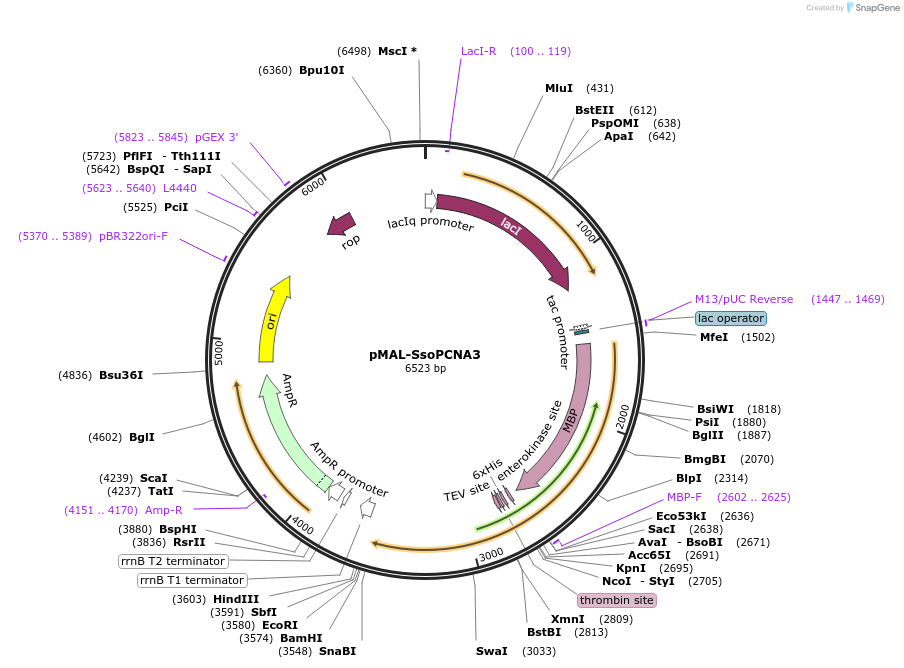
pMAL-SsoPCNA3
Plasmid#107069PurposeExpresses S. solfataricus PCNA3 fused to MBP in E. coliDepositorInsertS. solfataricus PCNA3
Tags6xHis-thrombin cleavage site and MBPExpressionBacterialPromotertac promoterAvailable SinceMarch 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
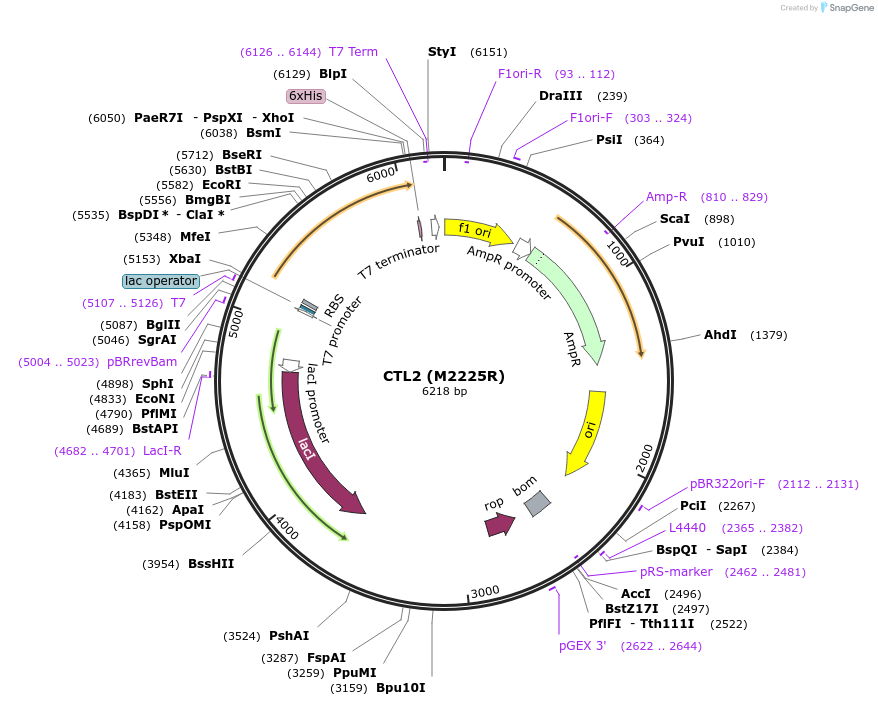
CTL2 (M2225R)
Plasmid#96952PurposeExpresses C-Terminal segment of C. elegans Piezo1 with M2225R mutation in E. coliDepositorInsertCTL2 (M222R) of PIEZO1
Tags6x Histidine TagExpressionBacterialPromoterT7 PromoterAvailable SinceOct. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
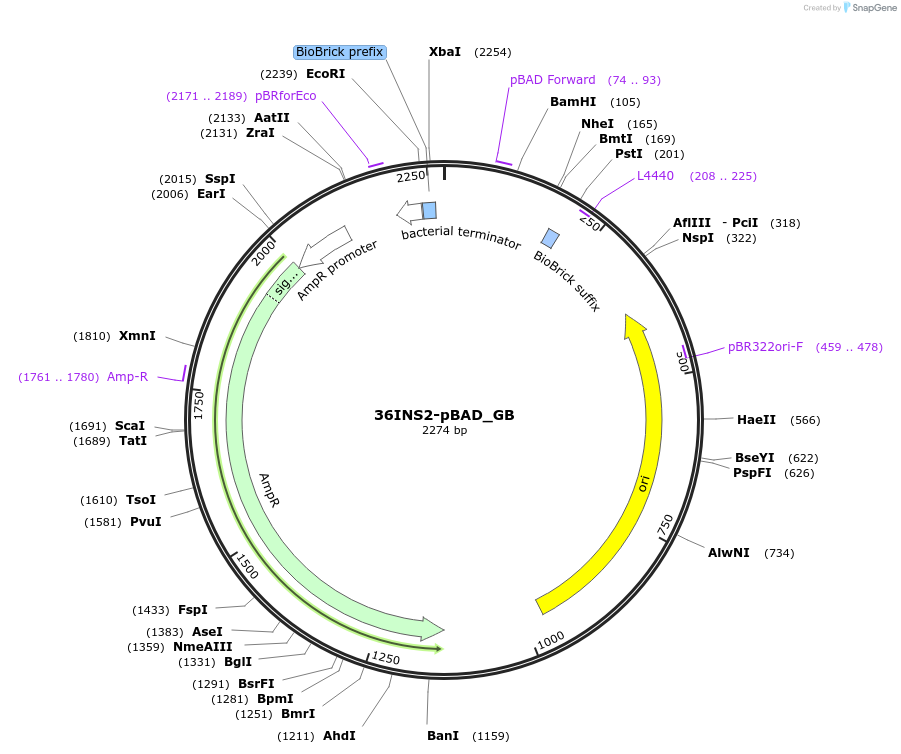
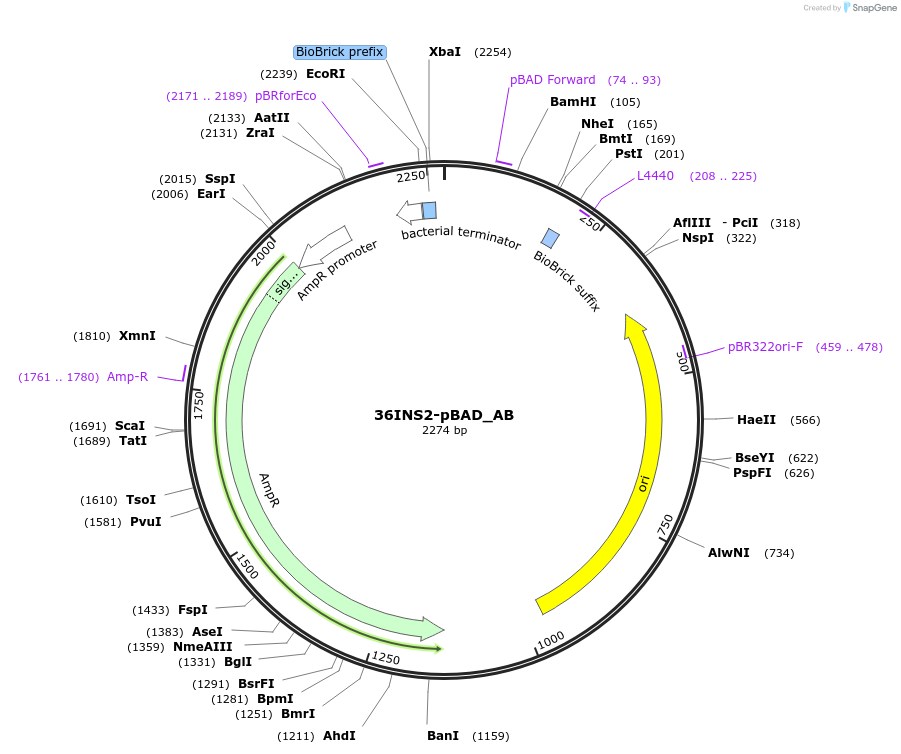
36INS2-pBAD_GB
Plasmid#78677PurposeInsulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_I13453. Has near identical expression as 36INS1-pBAD_AB [G:36INS2-pBAD:B]DepositorInsertInsulated pBAD Promoter
UseSynthetic BiologyAvailable SinceAug. 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
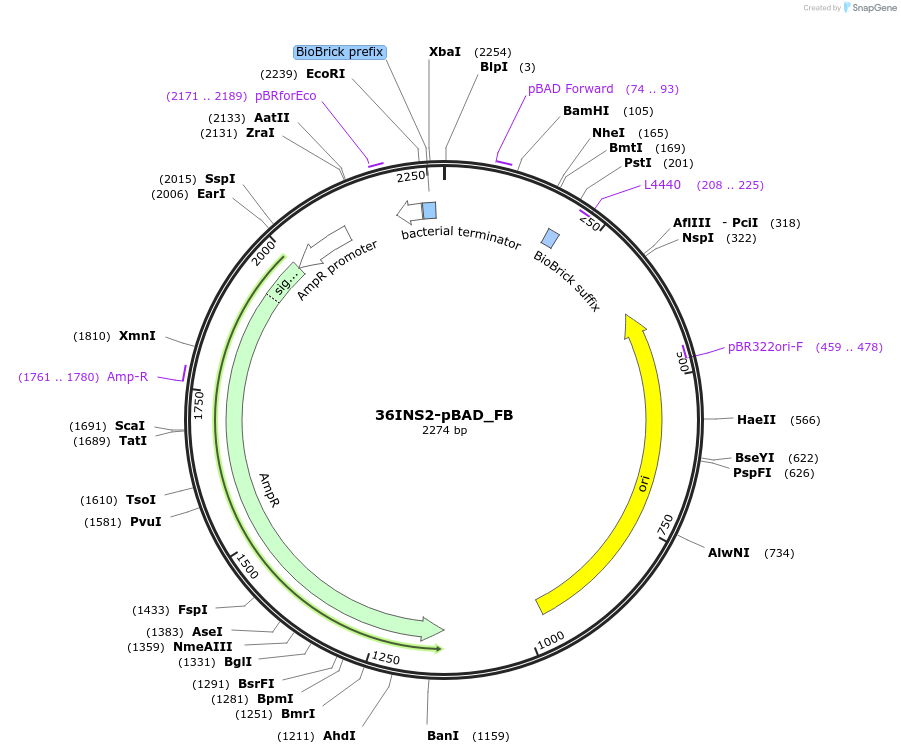
36INS2-pBAD_FB
Plasmid#78676PurposeInsulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_I13453. Has near identical expression as 36INS1-pBAD_AB [F:36INS2-pBAD:B]DepositorInsertInsulated pBAD Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
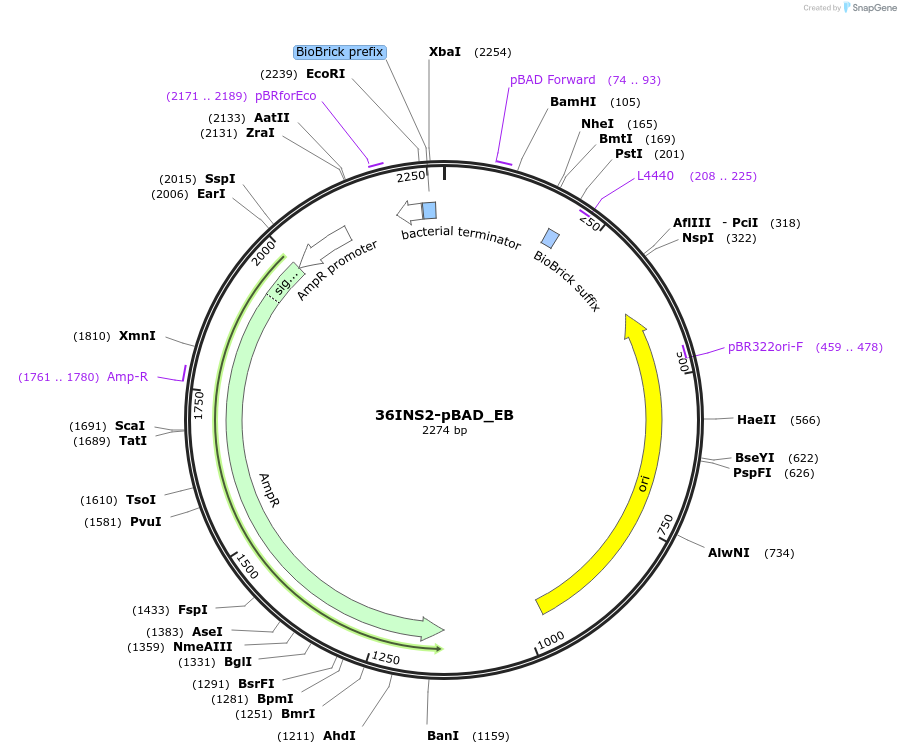
36INS2-pBAD_EB
Plasmid#78675PurposeInsulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_I13453. Has near identical expression as 36INS1-pBAD_AB [E:36INS2-pBAD:B]DepositorInsertInsulated pBAD Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
36INS2-pBAD_AB
Plasmid#78674PurposeInsulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected. Modified from BBa_I13453. Has near identical expression as 36INS1-pBAD_AB [A:36INS2-pBAD:B]DepositorInsertInsulated pBAD Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
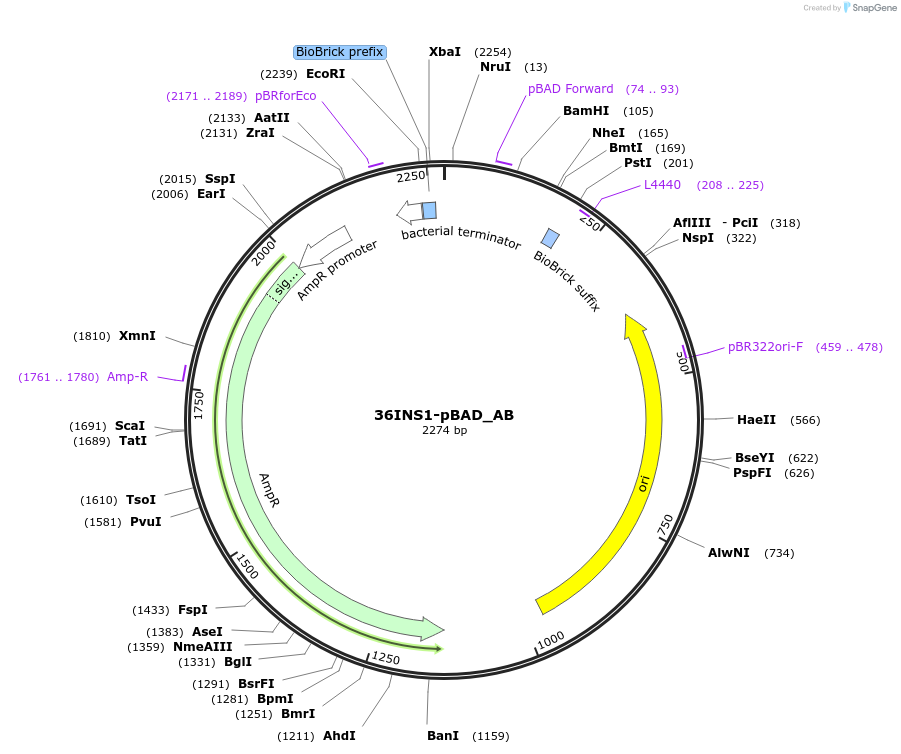
36INS1-pBAD_AB
Plasmid#78670PurposeInsulated MoClo promoter for E. coli. 36nt DNA spacer empirically selected.Modified from BBa_I13453 to adjust spacing in MoClo system. [A:36INS1-pBAD:B]DepositorInsertInsulated pBAD Promoter
UseSynthetic BiologyAvailable SinceAug. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
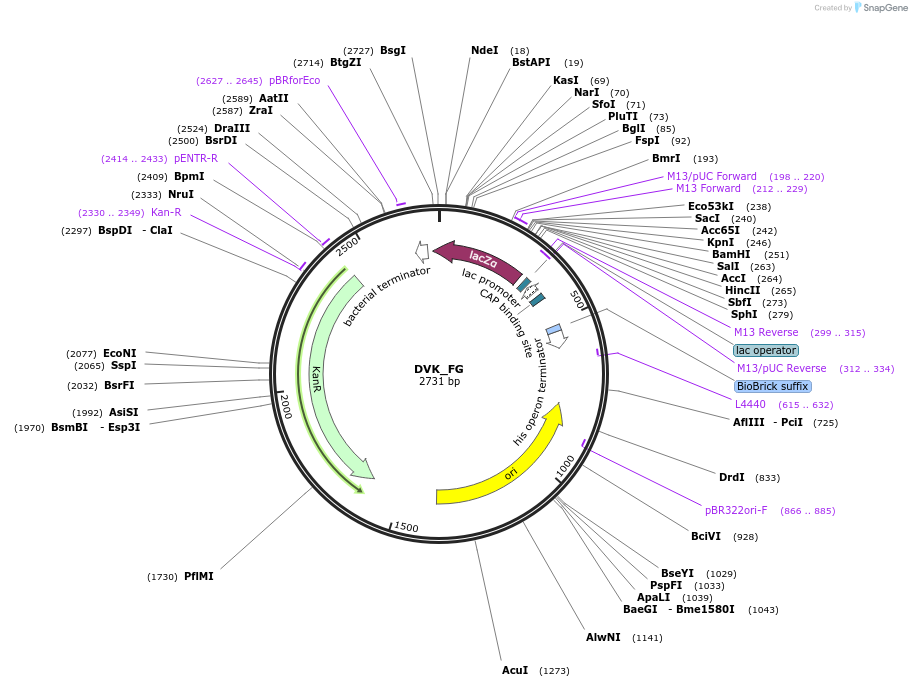
DVK_FG
Plasmid#66070PurposeMoClo Destination Vector: (based on pSB1K3) [F:LacZa:G]DepositorTypeEmpty backboneUseSynthetic Biology; Destination vectorAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
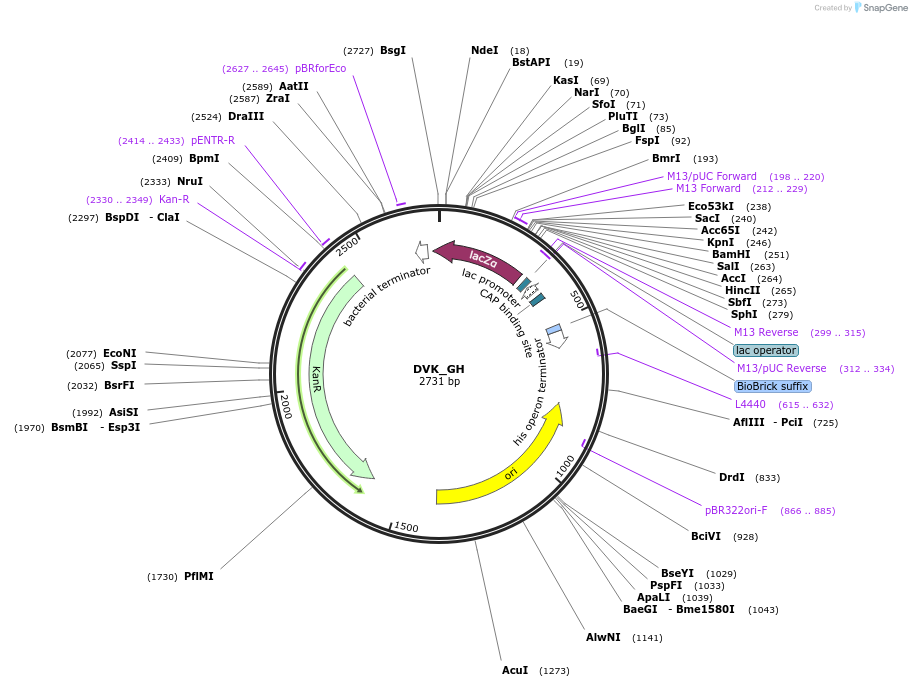
DVK_GH
Plasmid#66071PurposeMoClo Destination Vector: (based on pSB1K3) [G:LacZa:H]DepositorTypeEmpty backboneUseSynthetic Biology; Destination vectorAvailable SinceJan. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
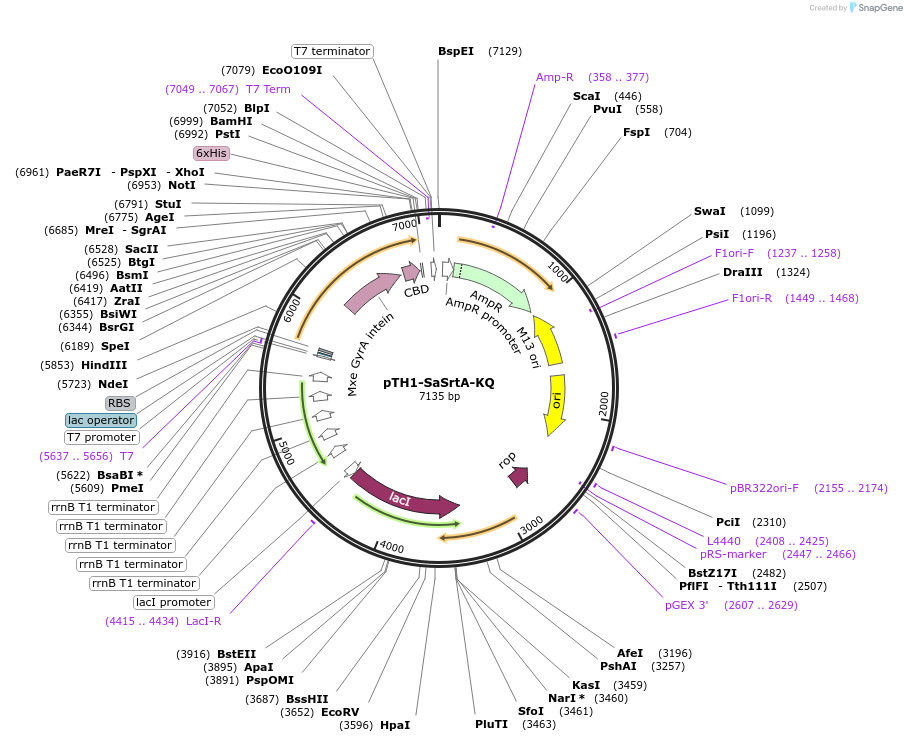
pTH1-SaSrtA-KQ
Plasmid#64982PurposeExpresses the E105K/E108Q mutant of Staphylococcus aureus sortase A in E. coliDepositorInsertsortase A
TagsHis6 tag, Mxe GyrA intein, and chitin-binding pro…ExpressionBacterialMutationdeleted amino acids 1-59, E105K, E108QAvailable SinceJune 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
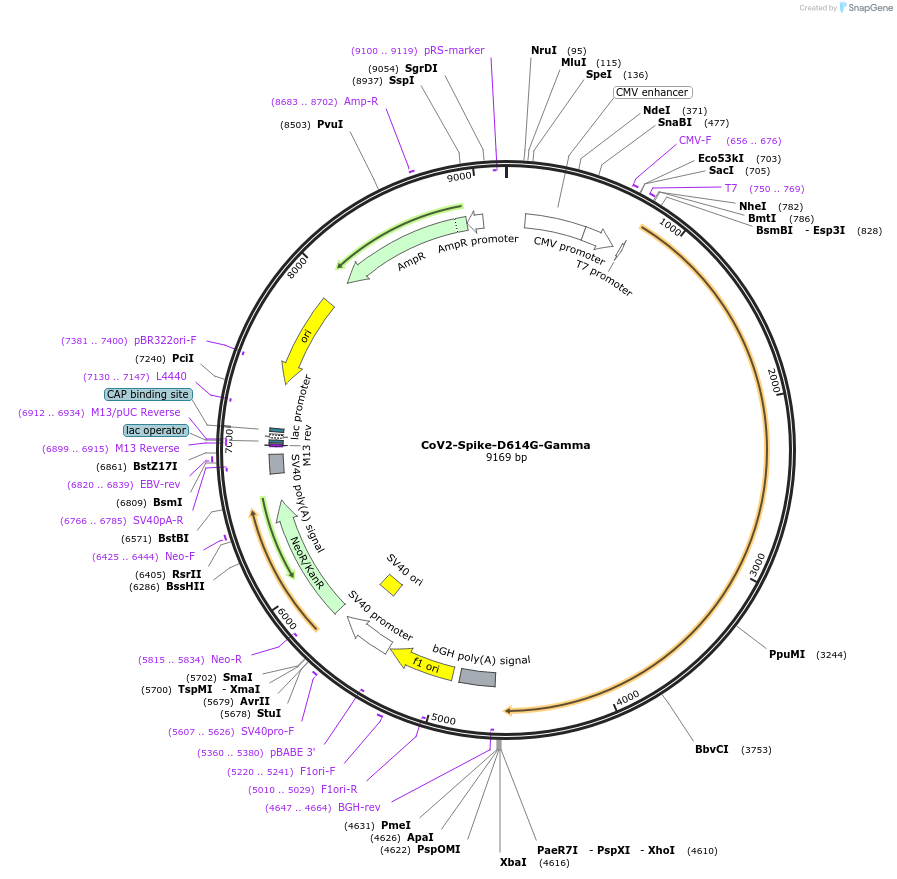
CoV2-Spike-D614G-Gamma
Plasmid#177963PurposeTransient mammalian expression of codon optimized SARS-CoV-2 Spike protein (Gamma: L13F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F)DepositorInsertSARS-CoV-2 Spike protein (Gamma: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F) (S )
ExpressionMammalianMutationL18F, T20N, P26S, D138Y, R190S, K417T, E484K, N50…Available SinceNov. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
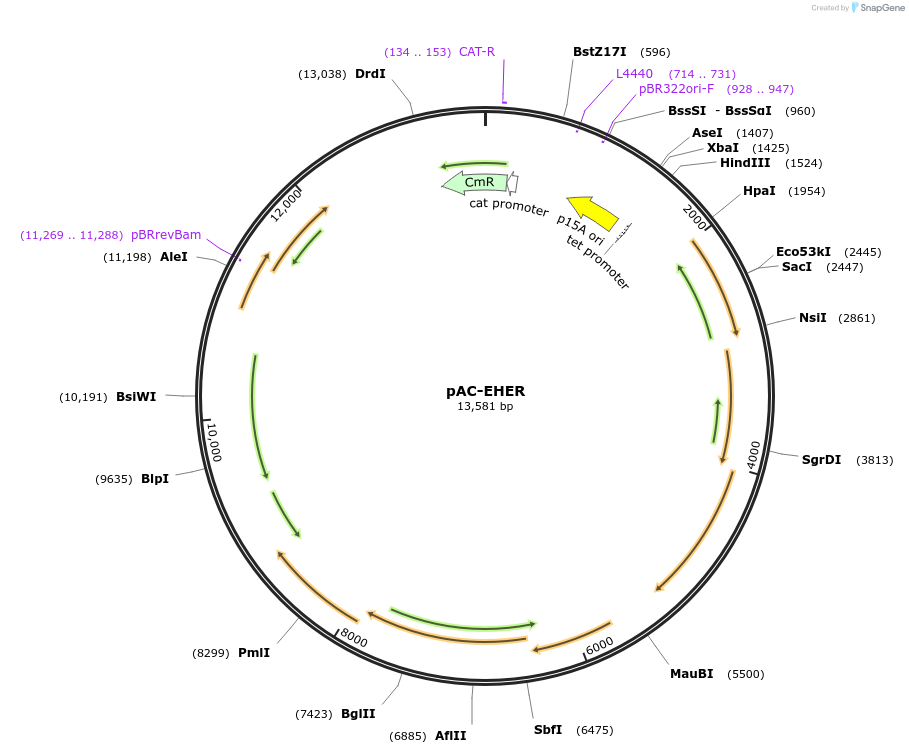
pAC-EHER
Plasmid#53262PurposeContains a 7 gene carotenoid pathway gene cluster of Erwinia herbicola (Pantoea agglomerans) Eho10 and thereby produces zeaxanthin diglucoside in Escherichia coliDepositorInsertcrtE, idi, crtX, crtY, crtI, crtB, crtZ
UseLow copy number bacterial cloning vectorPromoterendogenous promotersAvailable SinceJan. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
pSmac-GFP
Plasmid#40881DepositorAvailable SinceNov. 9, 2012AvailabilityAcademic Institutions and Nonprofits only