We narrowed to 421 results for: pet-28
-
Plasmid#30025DepositorInsertpLtetO-1:MicC, pLtetO-1:fhlA::mCherry, pLlacO-1:ompC::gfp
UseSynthetic BiologyTagsmCherry, GFPExpressionBacterialAvailable SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
HL 2625 (=HL 716 + pHL 908)
Plasmid#30028DepositorInsertpLlacO-1:RhyB::mCherry, pLtetO-1m9:sodB::gfp
UseSynthetic BiologyTagsmCherry, GFPExpressionBacterialAvailable SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
HL 3612 (=HL 716 + pHL 841)
Plasmid#30082DepositorInsertpLtetO-1:RhyB-mCherry, pLlacO-1:ompC::gfp
UseSynthetic BiologyTagsmCherry, GFPExpressionBacterialAvailable SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
HL 3619 (=HL 716 + pHL 1228)
Plasmid#30083DepositorInsertpLtetO-1:DsrA-mCherry, pLlacO-1:ompC::gfp
UseSynthetic BiologyTagsmCherry, GFPExpressionBacterialAvailable SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
pHL 1405
Plasmid#53010PurposepLtetO-1:RhyB, pCon:DsrA, pLtetO-1:hfq, pLlacO-1:sodB::gfpDepositorInsertsRhyB sRNA
DsrA sRNA
hfq
sodB
UseSynthetic BiologyTagsgfpExpressionBacterialMutationcontains -56 to +141 relative to start codonPromoterpCon, pLlacO-1, and pLtetO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1075
Plasmid#53004PurposepCon:fhlA::mCherry, pLlacO-1:sodB::gfpDepositorInsertsfhlA
sodB
UseSynthetic BiologyTagsGFP and mCherryExpressionBacterialMutationcontains −309 to +96 bp relative to start codon a…PromoterpCon and pLlacO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1022
Plasmid#52996PurposepLtetO-1:RhyB, pCon:OxyS, pLlacO-1:sodB::gfpDepositorInsertsRhyB sRNA
OxyS sRNA
sodB
UseSynthetic BiologyTagsgfpExpressionBacterialMutationcontains −56 to +141 bp relative to start codonPromoterpCon, pLlacO-1, and pLtetO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1073
Plasmid#53003PurposepCon:ompC::mCherry, pLlacO-1:sodB::gfpDepositorInsertsmCherry
GFP
UseSynthetic BiologyTagsompC (target mRNA) and sodB (target mRNA)ExpressionBacterialMutationcontains −56 to +141 bp relative to start codon a…PromoterpCon and pLlacO-1Available SinceMay 16, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1394
Plasmid#53009PurposepLtetO-1:RhyB, pCon:T7(st3) (133bps)::mCherry, pLlacO-1:sodB::gfpDepositorInsertsRhyB sRNA
T7(st3) (133bps)
sodB
UseSynthetic BiologyTagsgfp and mCherryExpressionBacterialMutationcontains −56 to +141 bp relative to start codonPromoterpCon, pLlacO-1, and pLtetO-1Available SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
pHL 1303
Plasmid#53008PurposepLtetO-1:sodB::mCherry, pLlacO-1:ompC::gfpDepositorInsertssodB
ompC
UseSynthetic BiologyTagsGFP and mCherryExpressionBacterialMutationcontains -56 to +141 relative to start codon and …PromoterpLlacO-1 and pLtetO-1Available SinceMay 14, 2014AvailabilityAcademic Institutions and Nonprofits only -
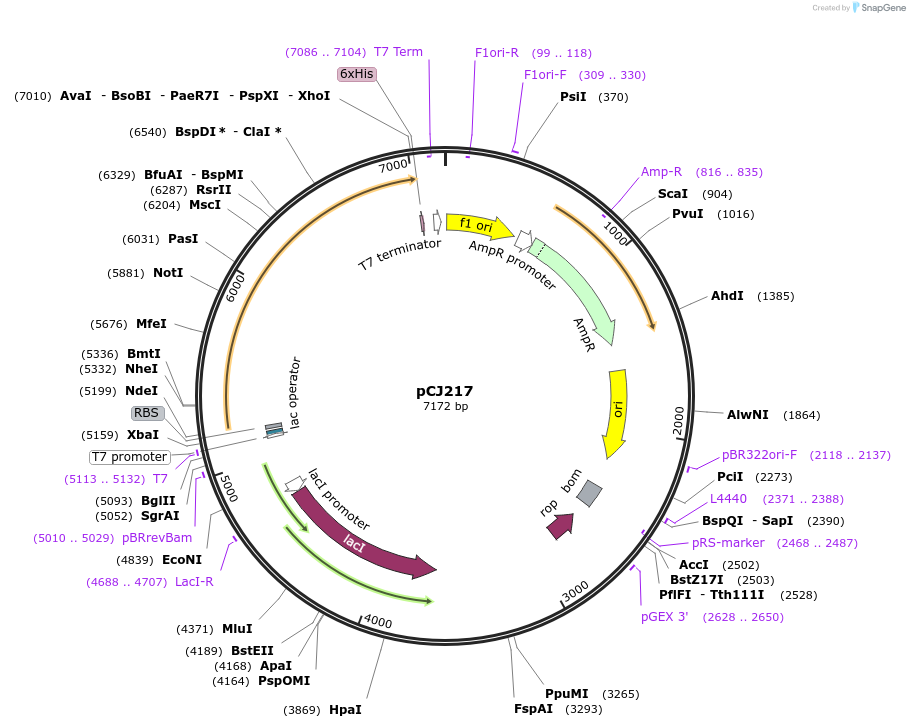
pCJ217
Plasmid#162685PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating two point mutations, G489C and S530C, to introduce a 6th disulfide bond (from PETase).DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with mutations to introduce a 6th disulfide bond from PETase
TagsHisExpressionBacterialMutationG489C, S530C; codon optimized for E. coli K12PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
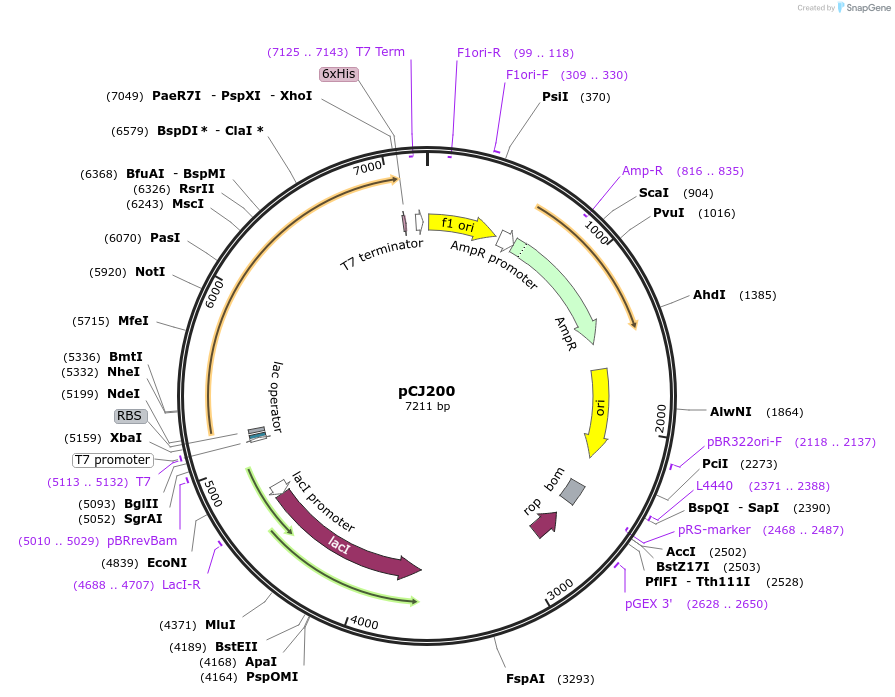
pCJ200
Plasmid#162673PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C and a sequence Tyr75:Phe104 to introduce a 6th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C and a sequence Y75-F104 to introduce a 6th…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
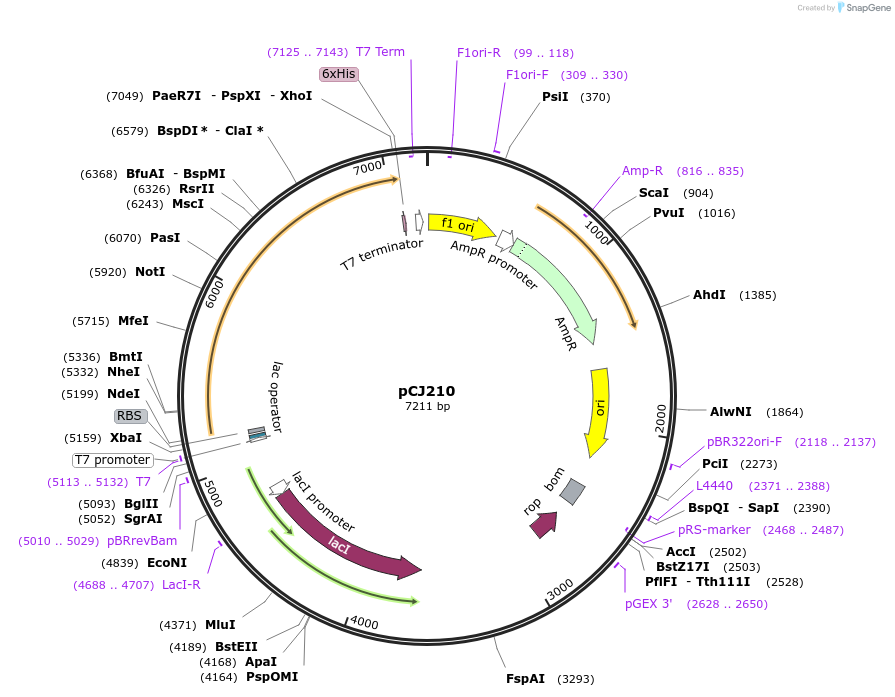
pCJ210
Plasmid#162683PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C, G489C, S530C, and a sequence Tyr75:Phe104 to introduce a 6th and 7th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C, G489C, S530C, and a sequence Y75-F104 to i…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
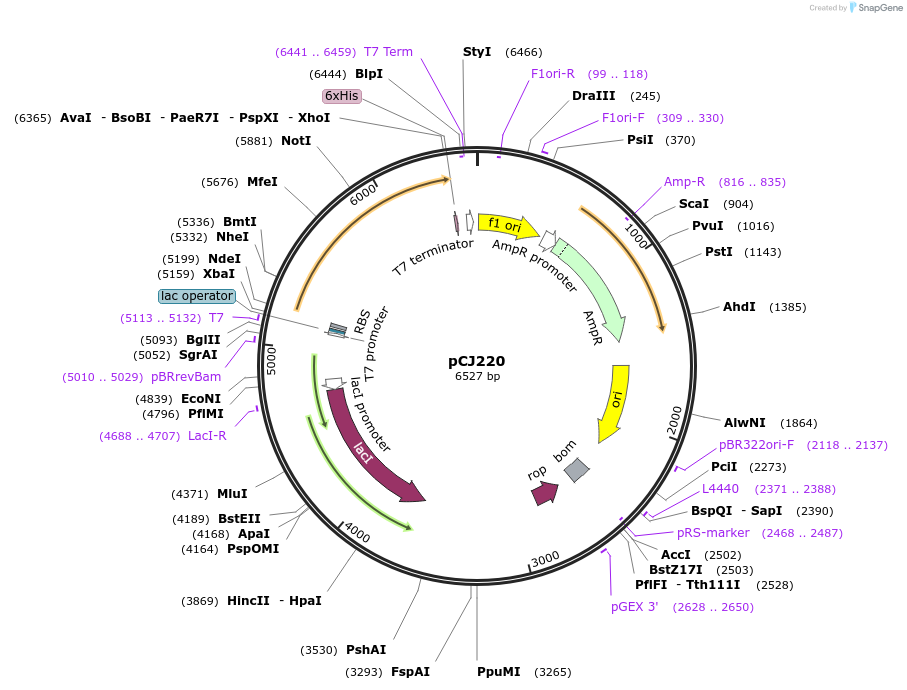
pCJ220
Plasmid#162686PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224W and C529S mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with lid deletion and C224W and C529S mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224W, C529S; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
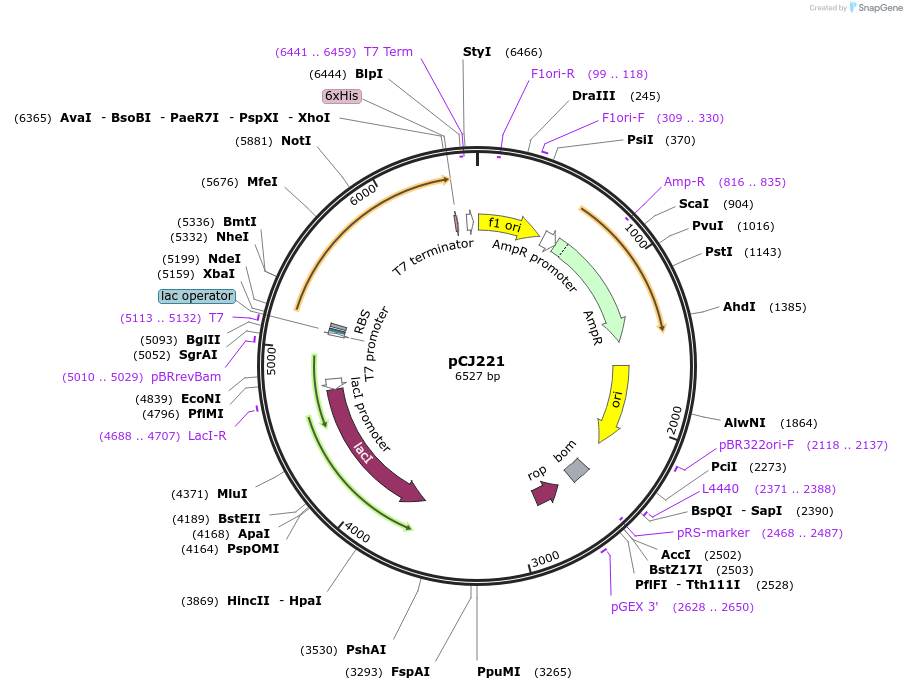
pCJ221
Plasmid#162687PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224H and C529F mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with lid deletion and C224H and C529F mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224H, C529F; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
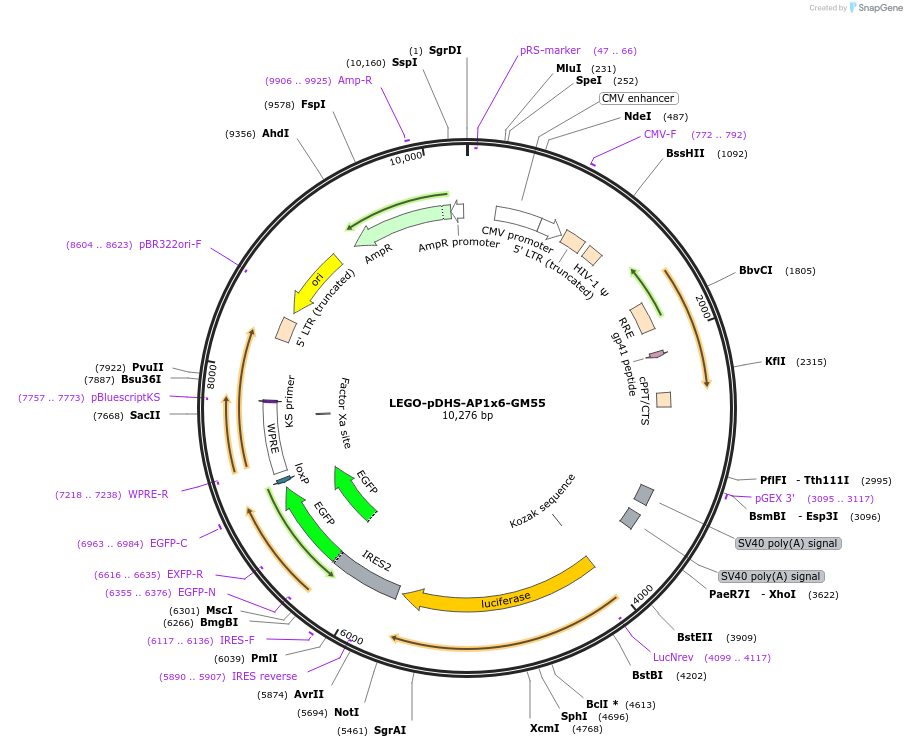
LEGO-pDHS-AP1x6-GM55
Plasmid#217416PurposeLuciferase expression vector with 6 AP-1 sites and the minimal GM-CSF promoter, and a chromatin priming element. Alias: LEGO-GM55-AP-1x6-pDHS(IL-3)DepositorInsertLuciferase, GFP
UseLentiviral and LuciferaseExpressionMammalianPromoter6 AP-1 sites, Human -55 to +28 minimal CSF2 promo…Available SinceMay 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
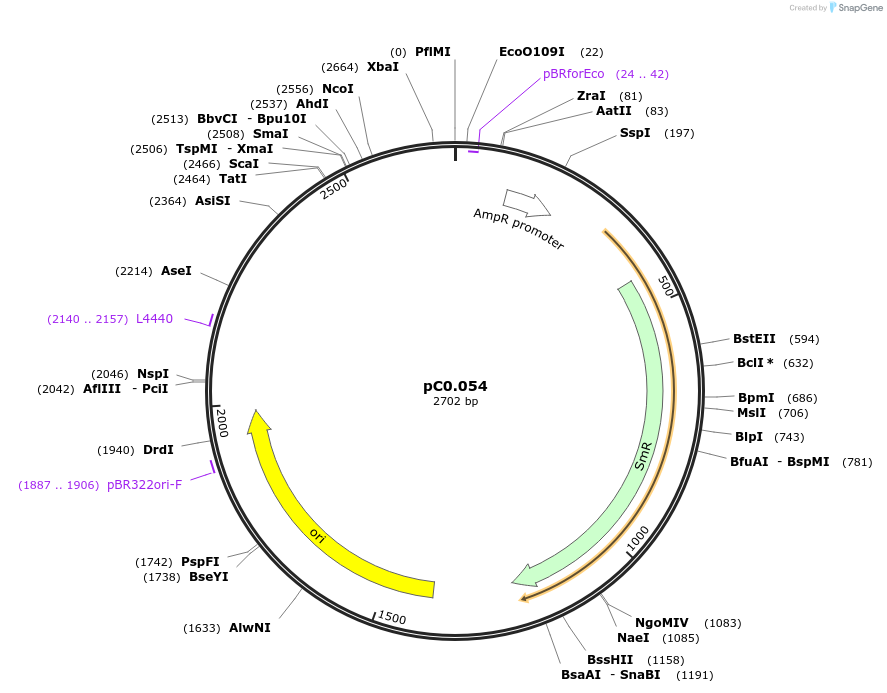
pC0.054
Plasmid#119572PurposeLevel 0 part. PromoterDepositorInsertPpetE
UseSynthetic BiologyMutationRBS* added (22 bp) upstream of ATG. Deletion of n…Available SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
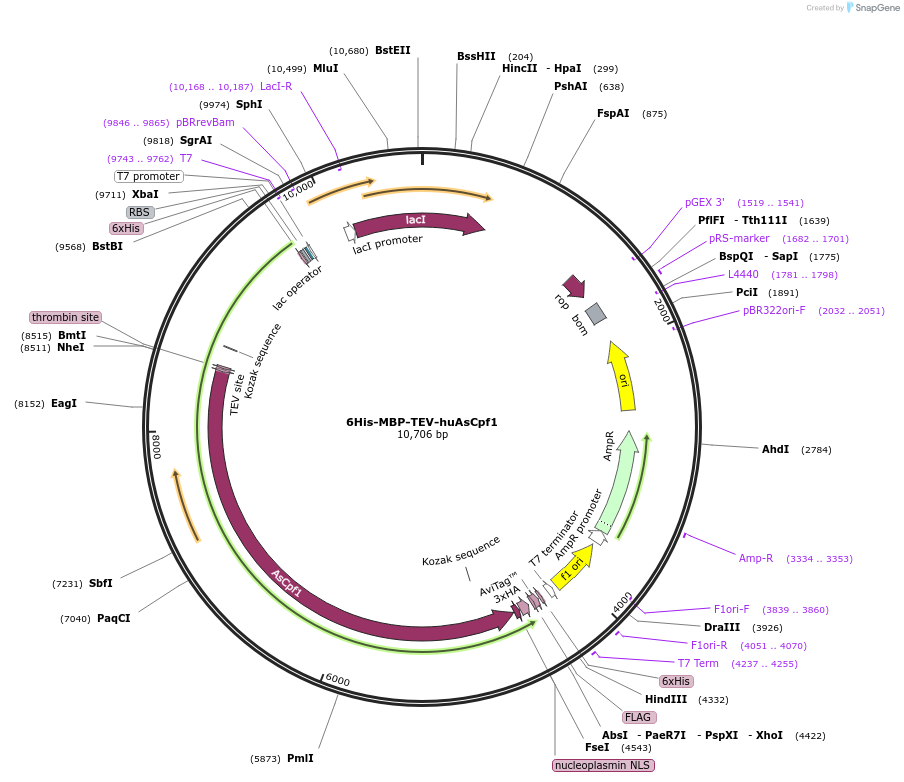
6His-MBP-TEV-huAsCpf1
Plasmid#90095PurposeBacterial expression plasmid for protein purificationDepositorInserthuAsCpf1
Tags3xHA, 6xHis, MBP, Nucleoplasmin NLS, and TEV site…ExpressionBacterialAvailable SinceJune 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
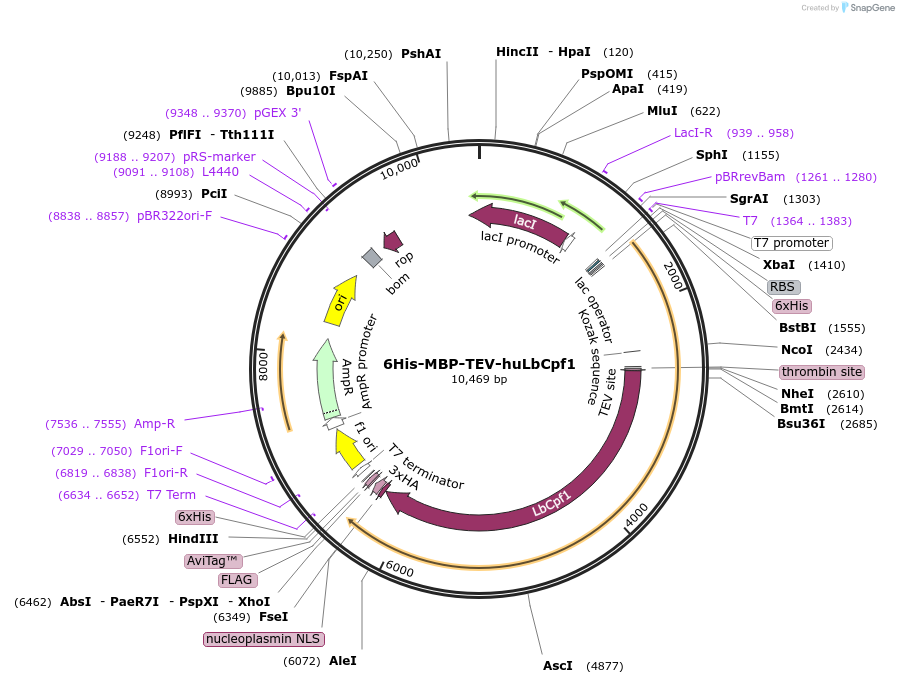
6His-MBP-TEV-huLbCpf1
Plasmid#90096PurposeBacterial expression plasmid for protein purificationDepositorInserthuLbCpf1
Tags3xHA, 6xHis, MBP, Nucleoplasmin NLS, and TEV site…ExpressionBacterialAvailable SinceJune 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
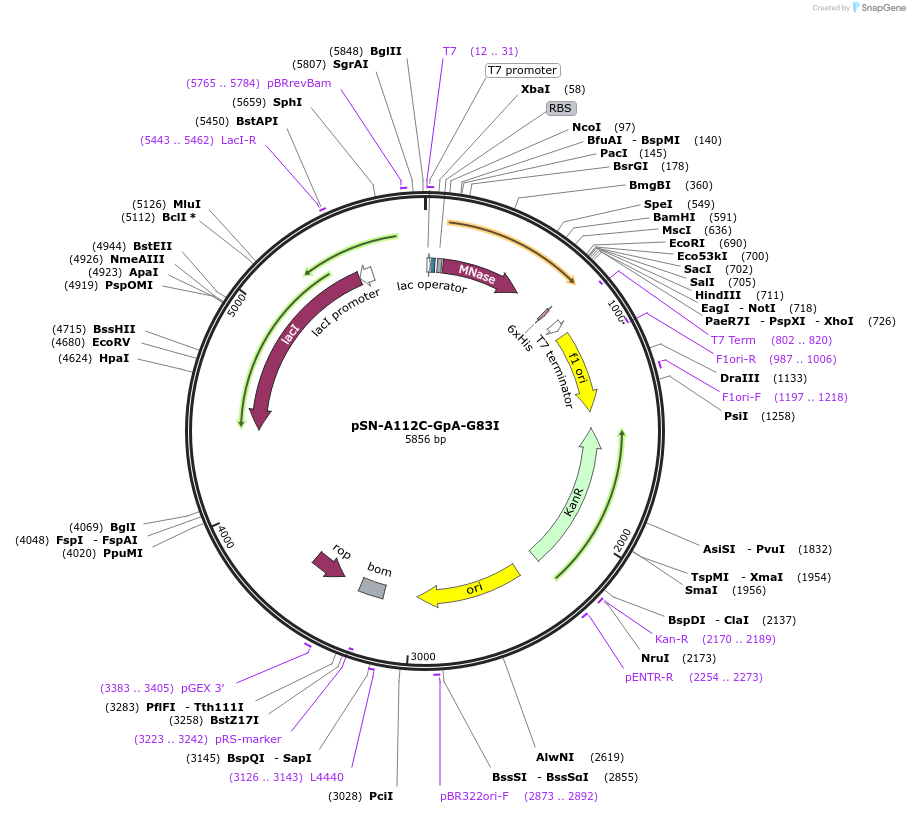
pSN-A112C-GpA-G83I
Plasmid#191239PurposeExpresses nuclease A-H124L from Staphylococcus aureus, with an A112C mutation, fused through a flexible linker to the transmembrane domain of GpA-G83I and a His-tag, for fluorescent studies.DepositorInsertnuc (nuclease A), GYPA (GPA) (E(transmembrane)R)
Tagsnuclease A from S. aureus fused to the TM domain …ExpressionBacterialMutationChanged Alanine 112 to Cysteine in SN, and Glycin…PromoterT7 promoterAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only