We narrowed to 9,847 results for: try
-
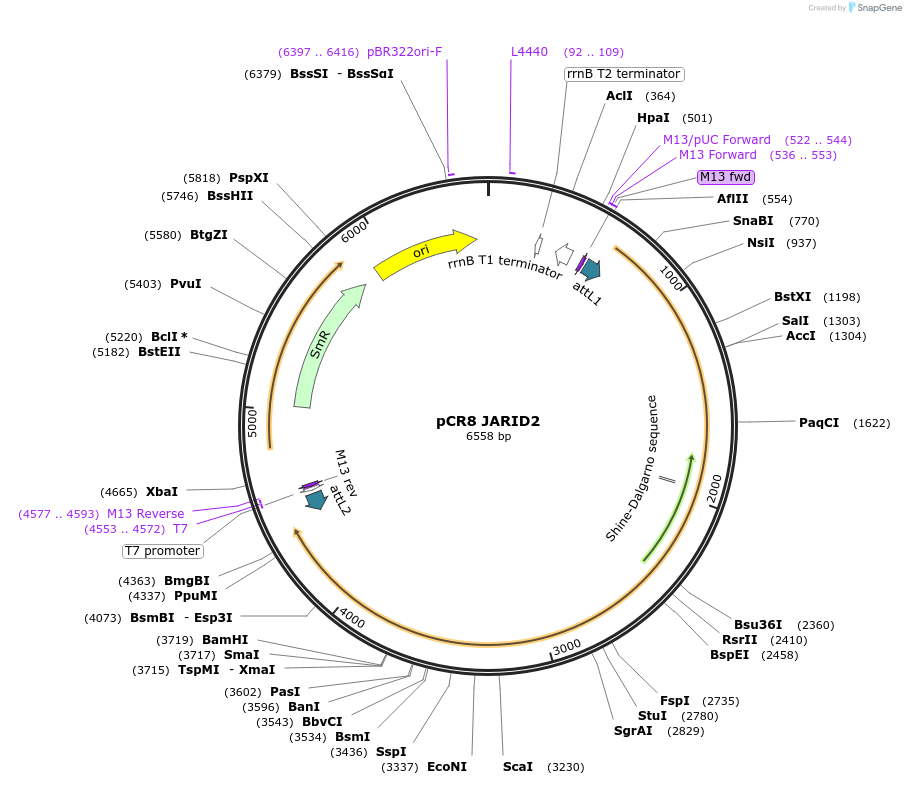
Plasmid#114443PurposeEntry vector for gateway cloning containing human JARID2 coding sequenceDepositorAvailable SinceSept. 6, 2018AvailabilityAcademic Institutions and Nonprofits only
-
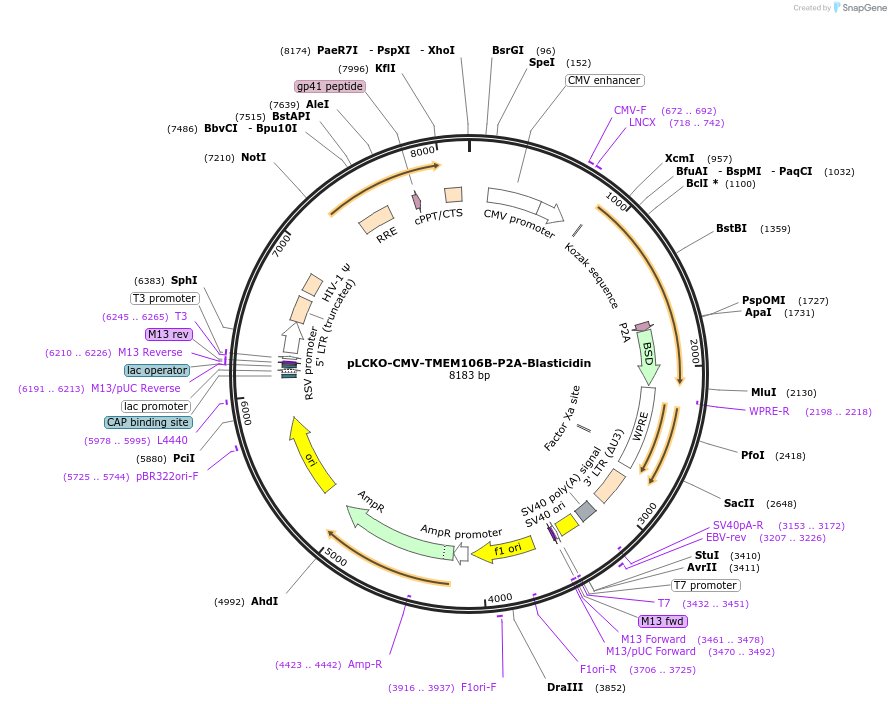
pLCKO-CMV-TMEM106B-P2A-Blasticidin
Plasmid#237478PurposeEncodes full length human TMEM106B protein, along with blasticidin selection marker.DepositorInsertTMEM106B (TMEM106B Human)
UseLentiviralTagsP2A-BlasticidinExpressionMammalianPromoterCMVAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
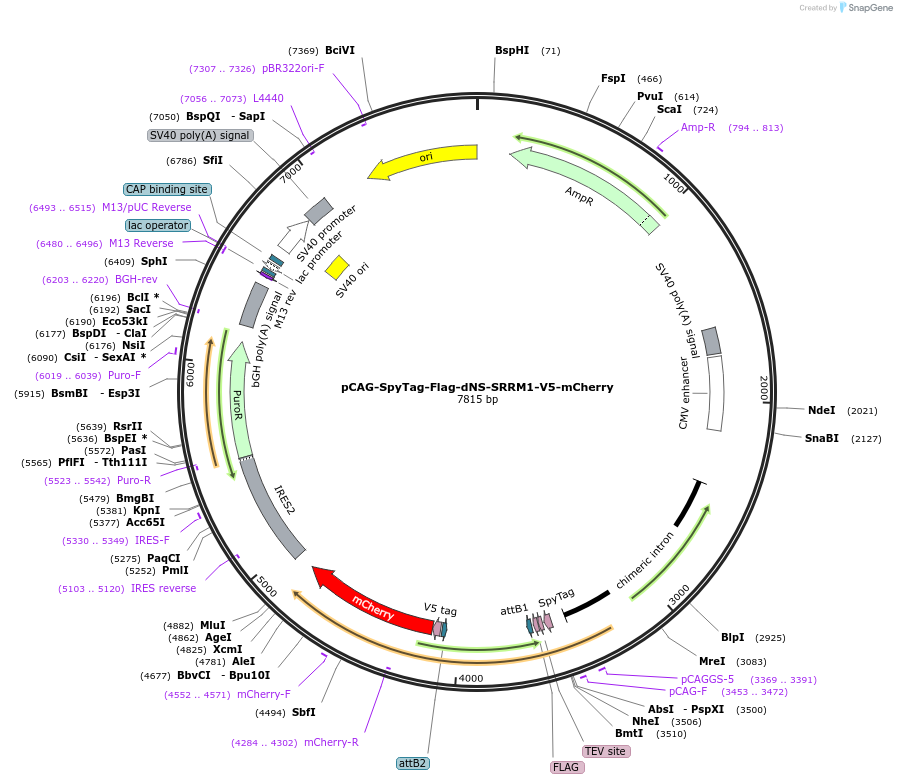
pCAG-SpyTag-Flag-dNS-SRRM1-V5-mCherry
Plasmid#235096PurposeSRRM1 lacking Nuclear Speckle domain (dNS-SRRM1) mCherry fusion protein without MCP domainDepositorInsertpCAG-SpyTag-Flag-dNS-SRRM1-V5-mCherry (SRRM1 Human)
ExpressionMammalianMutationThe SRRM1 entry clone from DNASU was modified usi…PromoterCAGAvailable SinceApril 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
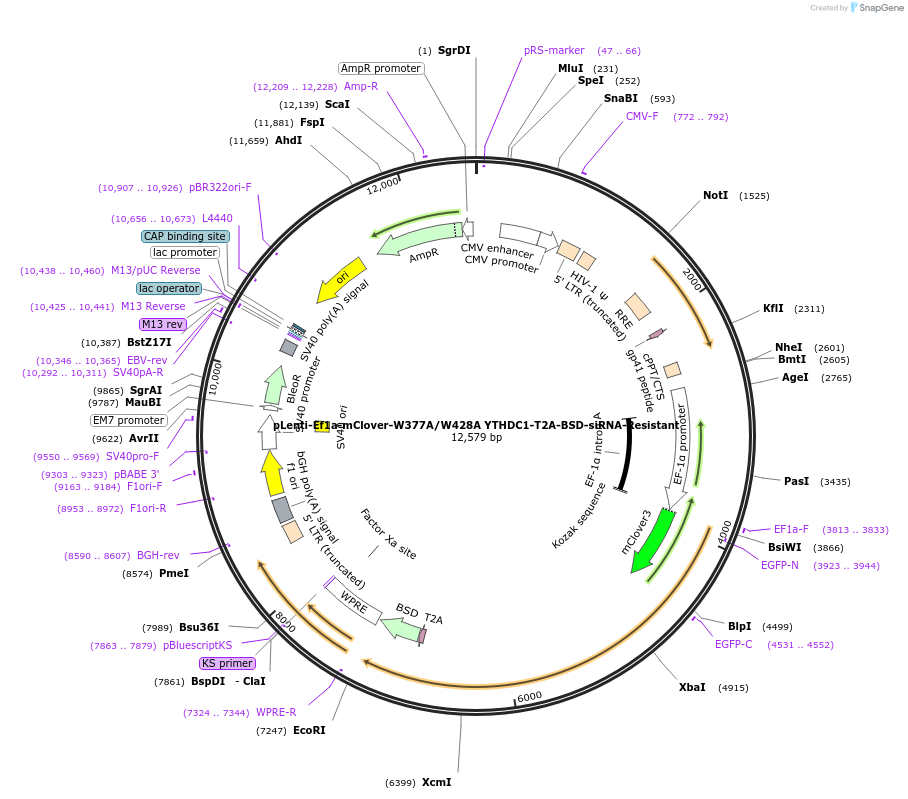
pLenti-Ef1a-mClover-W377A/W428A YTHDC1-T2A-BSD-siRNA-Resistant
Plasmid#177130PurposeGenerate lentivirus to introduce mClover tagged m6A binding null mutant W377A/W428A YTHDC1DepositorInsertYTHDC1 (YTHDC1 Human)
UseLentiviralTagsmClover3MutationW428A/W377A (m6A binding defects), and siRNA resi…PromoterEf1aAvailable SinceJuly 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
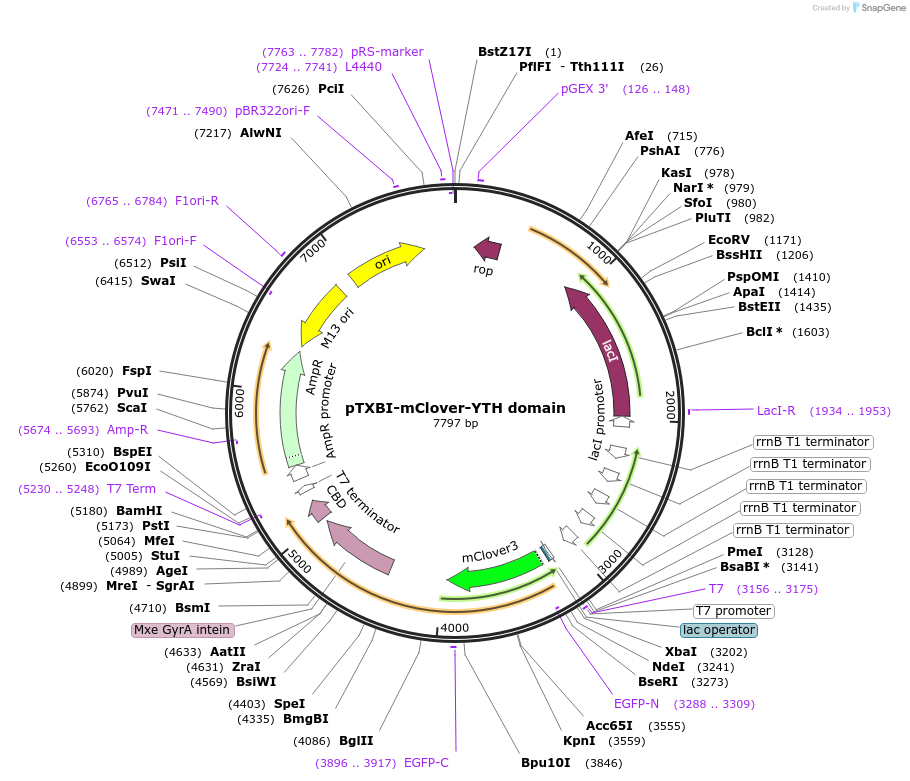
pTXBI-mClover-YTH domain
Plasmid#177123PurposeBacterial expression of YTH domain of YTHDC1 tagged with N-terminal mClover3 and C-terminal Mxe GtrA intein-Chitin binding domain for protein purificationDepositorInsertYTHDC1 (YTH domain) (YTHDC1 Human)
TagsMxe GyrA intein-Chitin binding domain for protein…ExpressionBacterialMutationYTH domain only (IDR1 and IDR2 deletion)PromoterT7Available SinceJan. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
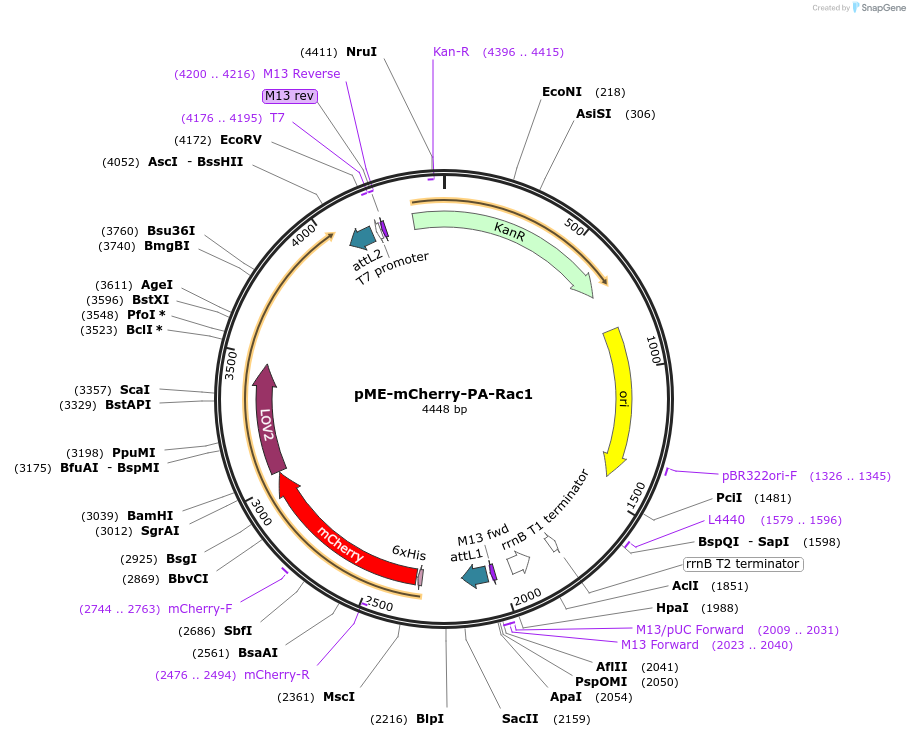
pME-mCherry-PA-Rac1
Plasmid#169042PurposeMiddle entry vector for Gateway cloning that contains an mCherry fused with an optimized photoactivatable Rac1 gene insertDepositorInsertPA-Rac-1 (RAC1 Human, Avena sativa (oat))
UseGateway entry cloning vectorTags6X His and Fused with mCherryMutationRac1 starts at I4 and contains mutations Q61L, E9…Available SinceJan. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
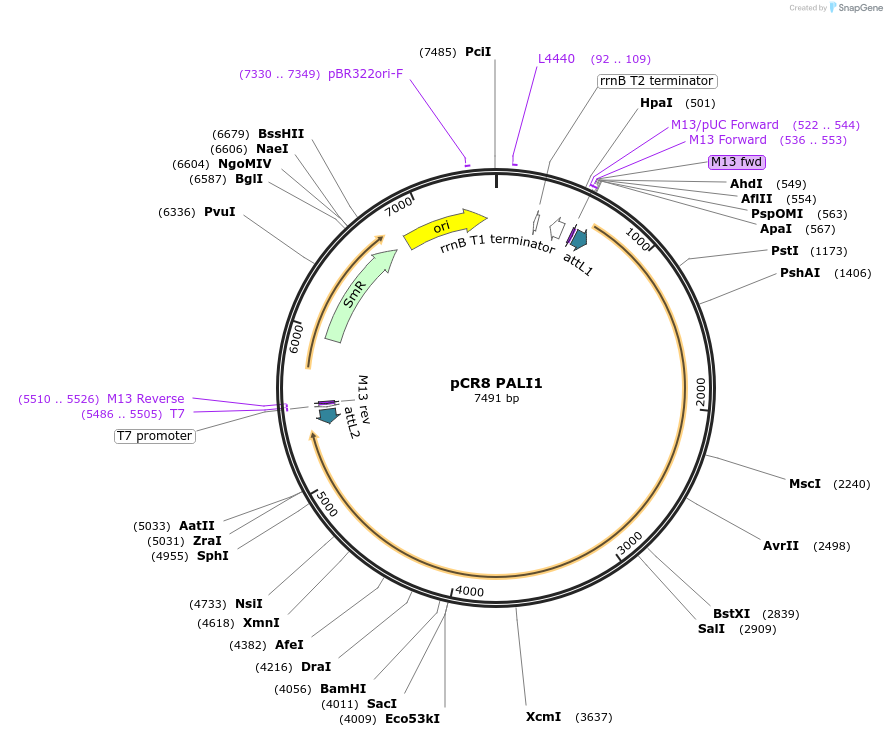
pCR8 PALI1
Plasmid#114442PurposeEntry vector for gateway cloning containing human PALI1 coding sequenceDepositorAvailable SinceSept. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
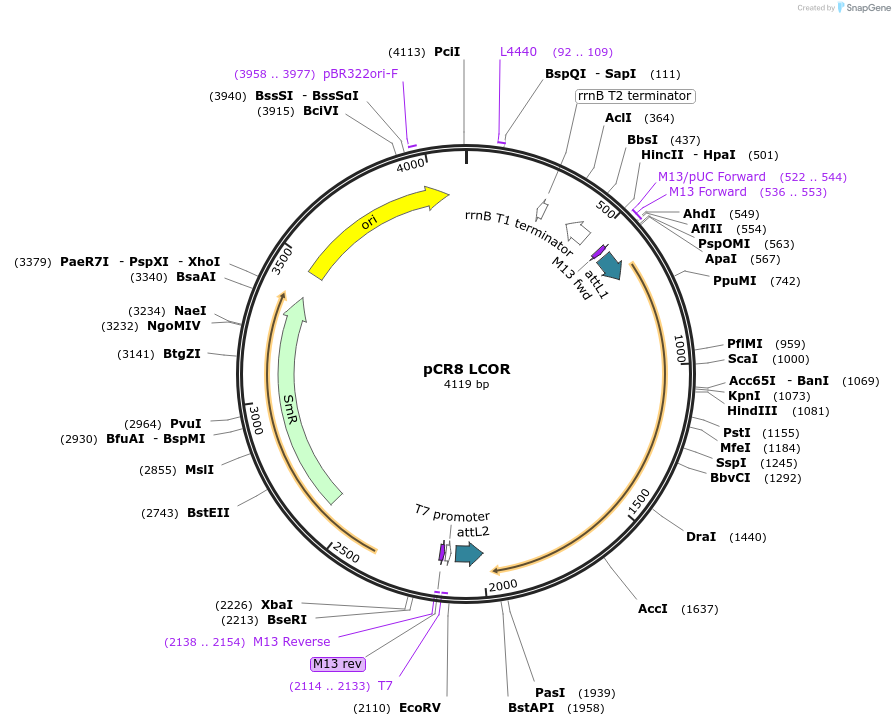
pCR8 LCOR
Plasmid#114441PurposeEntry vector for gateway cloning containing human LCOR coding sequenceDepositorAvailable SinceSept. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
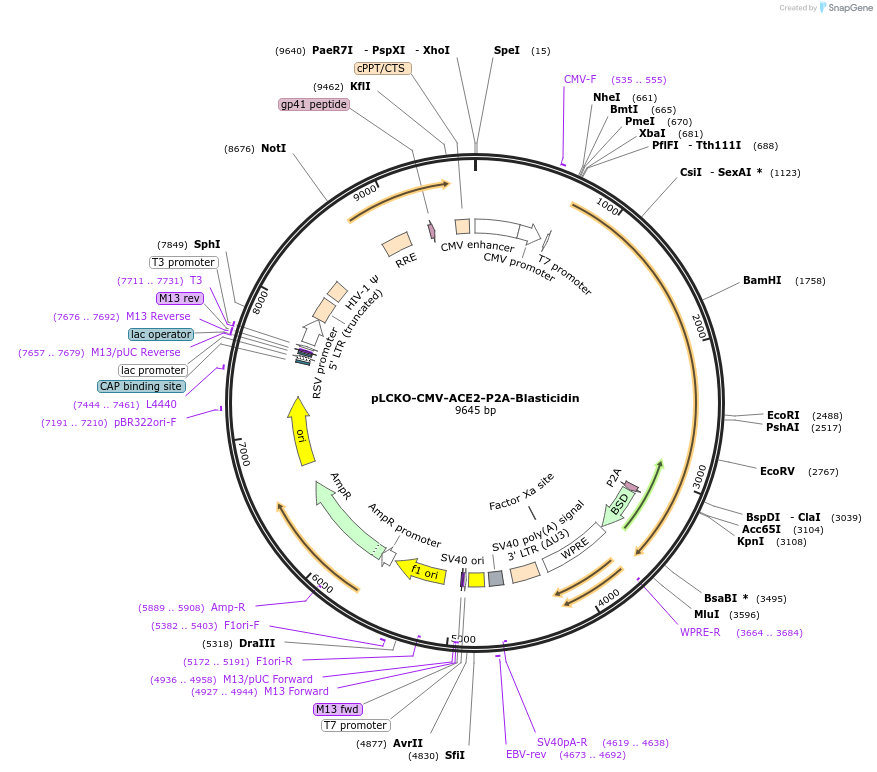
pLCKO-CMV-ACE2-P2A-Blasticidin
Plasmid#237476PurposeEncodes full length human ACE2 protein, along with blasticidin selection marker.DepositorAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
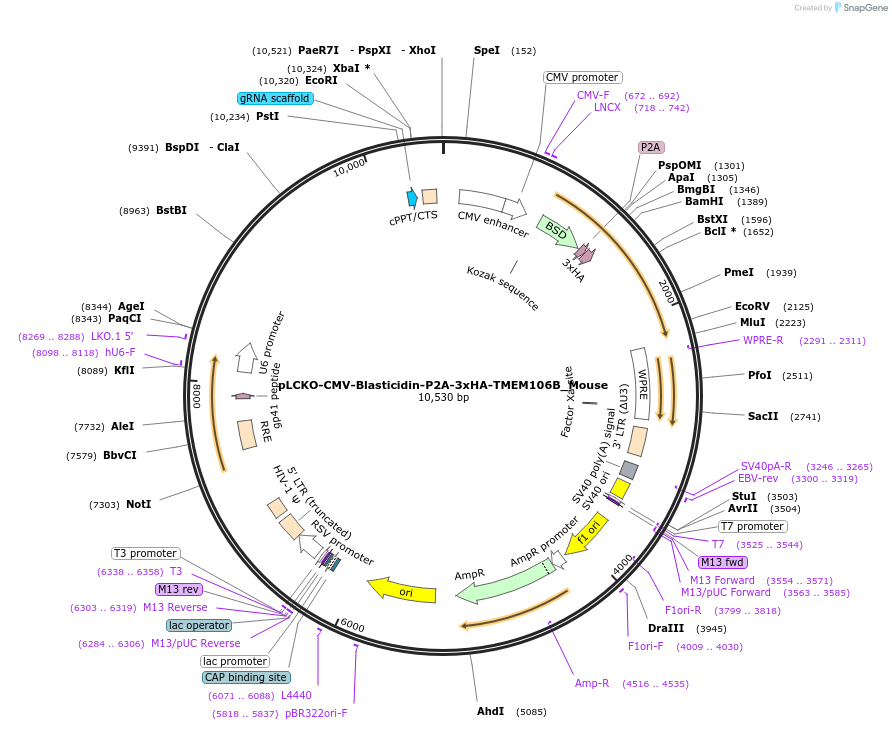
pLCKO-CMV-Blasticidin-P2A-3xHA-TMEM106B_Mouse
Plasmid#238133PurposeThis plasmid encodes the full-length Mouse (Mus musculus) TMEM106B protein fused with a 3xHA tag, along with an N-terminal blasticidin selection marker.DepositorInsertTMEM106B (Tmem106b Mouse)
UseLentiviralTagsBlasticidin-P2A-3xHAExpressionMammalianPromoterCMVAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
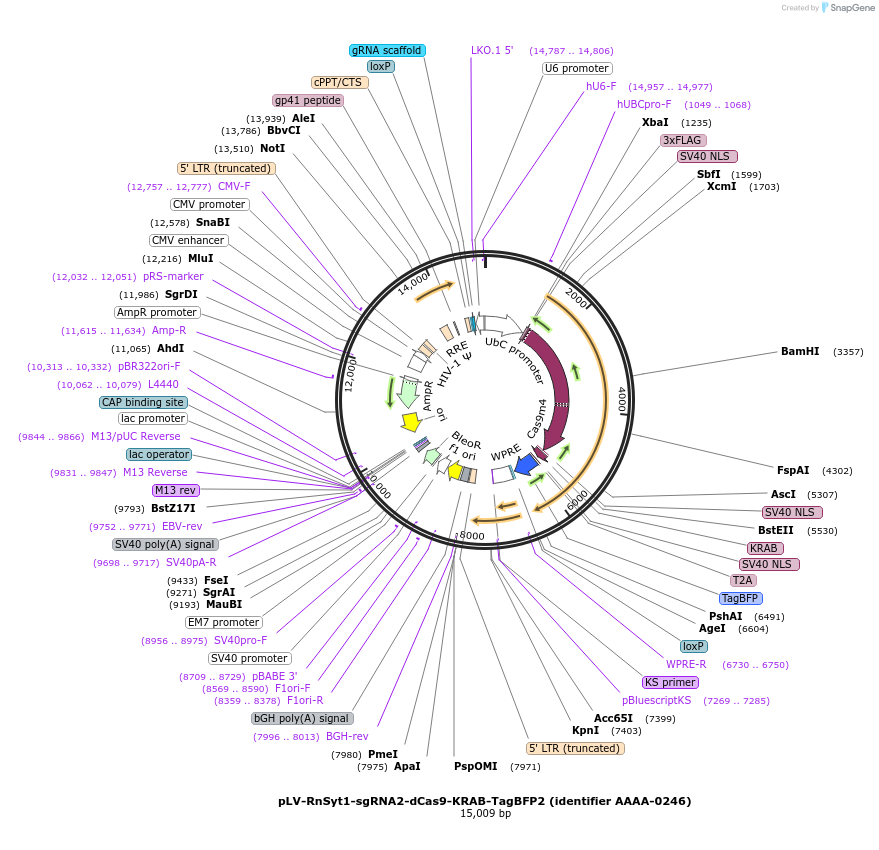
pLV-RnSyt1-sgRNA2-dCas9-KRAB-TagBFP2 (identifier AAAA-0246)
Plasmid#202555PurposeSynaptotagmin-1 sgRNA2 with TagBFP2DepositorInsertCRISPRi single guide RNA sequence against 5’-UTR of rat synaptotagmin-1 (Syt1) gene (insert 5’ - CACCAGTACTCGCGTGCCTCGCACCGG) (Syt1 Rat)
UseCRISPR and LentiviralTags3xFlag (N-terminus of Cas9-KRAB), T2A-TagBFP2Available SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
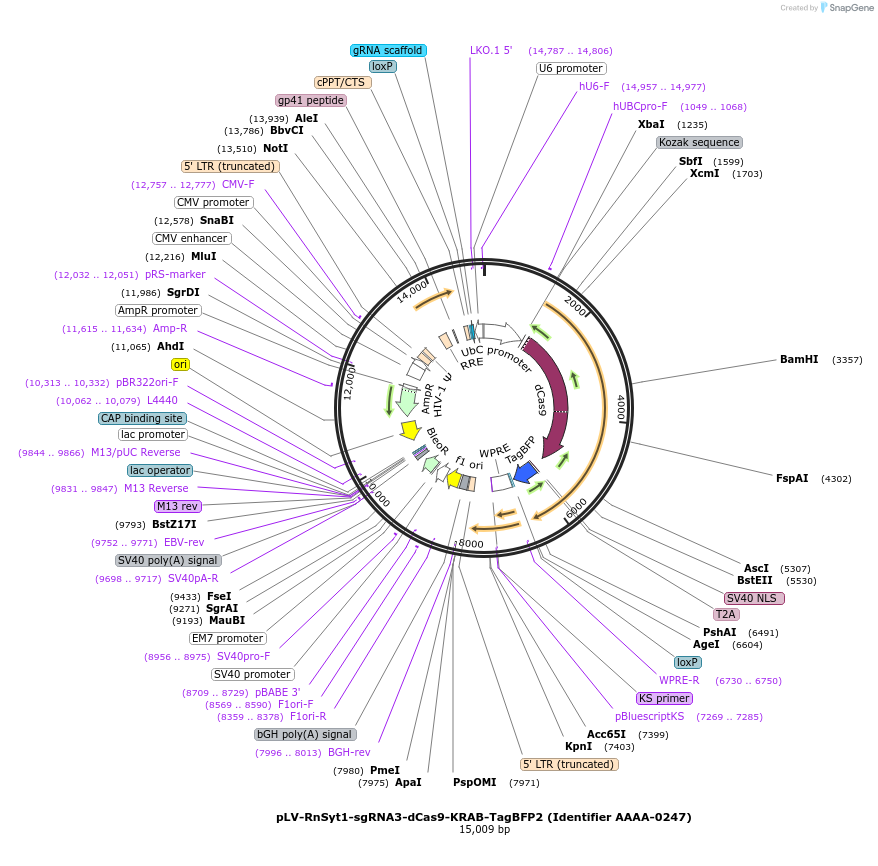
pLV-RnSyt1-sgRNA3-dCas9-KRAB-TagBFP2 (Identifier AAAA-0247)
Plasmid#202556PurposeSynaptotagmin-1 sgRNA3 with TagBFP2DepositorInsertCRISPRi single guide RNA sequence against 5’-UTR of rat synaptotagmin-1 (Syt1) gene (insert 5’ - CACCTCCTCCTGCAGCGGCAGCATCGG) (Syt1 Rat)
UseCRISPR and LentiviralTags3xFlag (N-terminus of Cas9-KRAB), T2A-TagBFP2Available SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
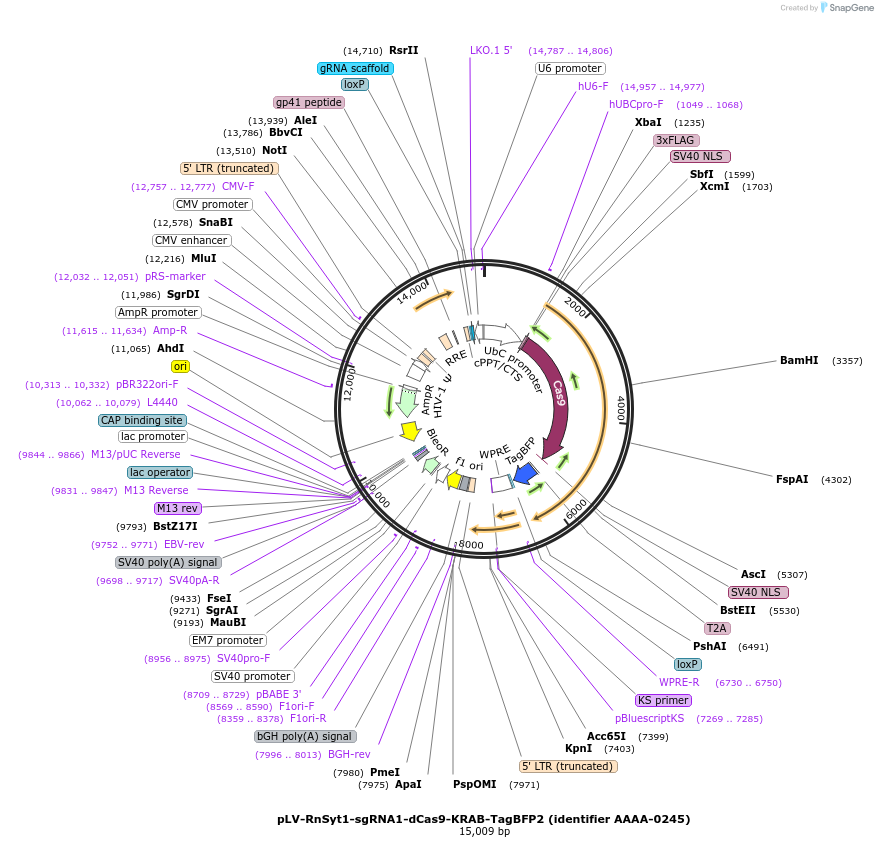
pLV-RnSyt1-sgRNA1-dCas9-KRAB-TagBFP2 (identifier AAAA-0245)
Plasmid#202554PurposeSynaptotagmin-1 sgRNA1 with TagBFP2DepositorInsertCRISPRi single guide RNA sequence against 5’-UTR of rat synaptotagmin-1 (Syt1) gene (insert 5’ -CACCGCGTGCCTCGCACCGGTCCGCGG) (Syt1 R. norvegius (gRNA))
UseCRISPR and LentiviralTags3xFlag (N-terminus of Cas9-KRAB), T2A-TagBFP2Available SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
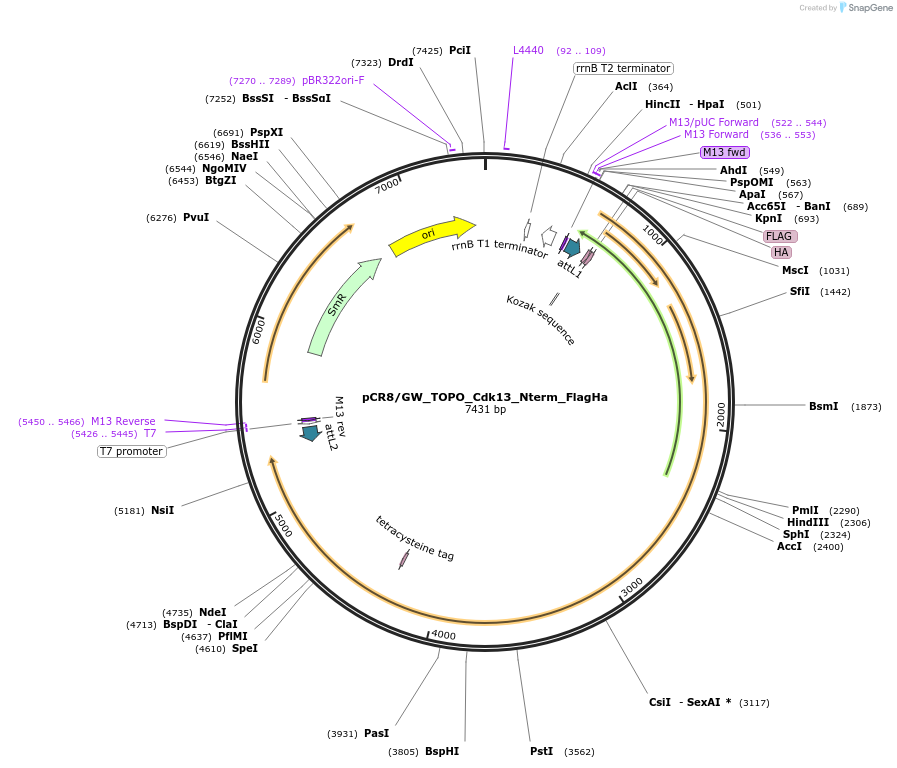
pCR8/GW_TOPO_Cdk13_Nterm_FlagHa
Plasmid#127343PurposeN-terminal Flag- HA- tandem epitope tagged mouse Cdk13 transgene (NP_001074527.1 isoform) cloned into Gateway entry vectorDepositorInsertCyclin Dependent Kinase 13 (Cdk13 Mouse)
UseEntry vector with transgene for gateway cloningTagsFlag- HA- Tandem EpitopesMutationBase pairs 1-1554 of transgene are codon optimize…PromoterNoneAvailable SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
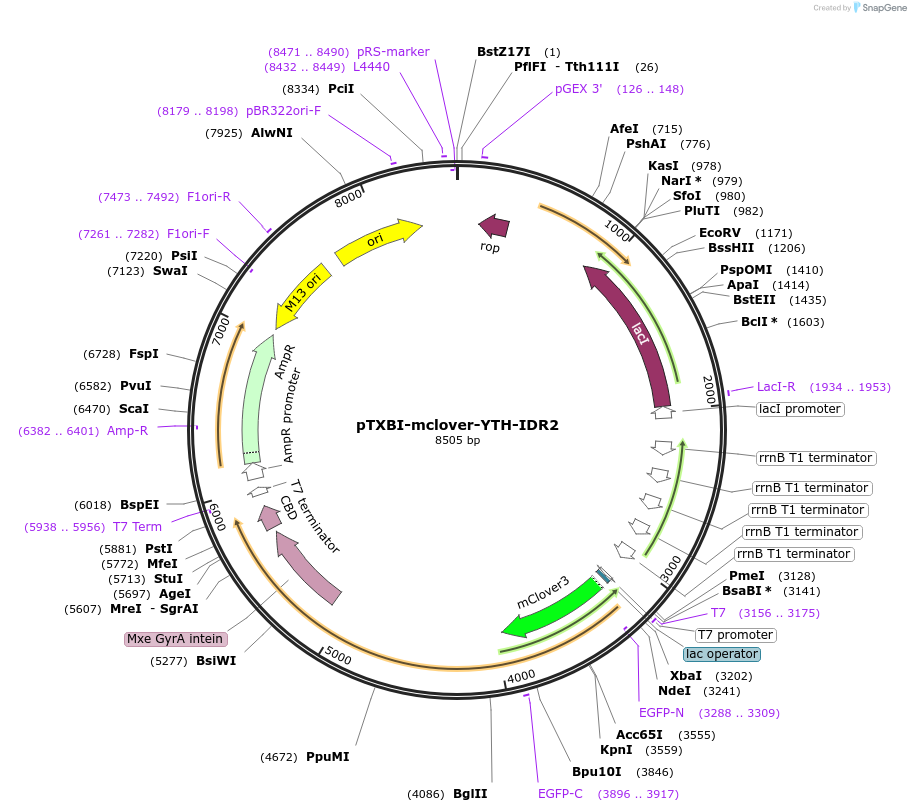
pTXBI-mclover-YTH-IDR2
Plasmid#177122PurposeBacterial expression of YTH-IDR2 fragment of YTHDC1 tagged with N-terminal mClover3 and C-terminal Mxe GtrA intein-Chitin binding domain for protein purificationDepositorInsertYTHDC1 (YTH and IDR2 domains) (YTHDC1 Human)
TagsMxe GyrA intein-Chitin binding domain for protein…ExpressionBacterialMutationYTH and IDR2 domains only (IDR1 deletion)PromoterT7Available SinceNov. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pFOSGE_COF2F04
Plasmid#70901PurposeGateway entry cloneDepositorInsertmod(mdg4) (mod(mdg4) Fly)
UseGateway entry vectorAvailable SinceNov. 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
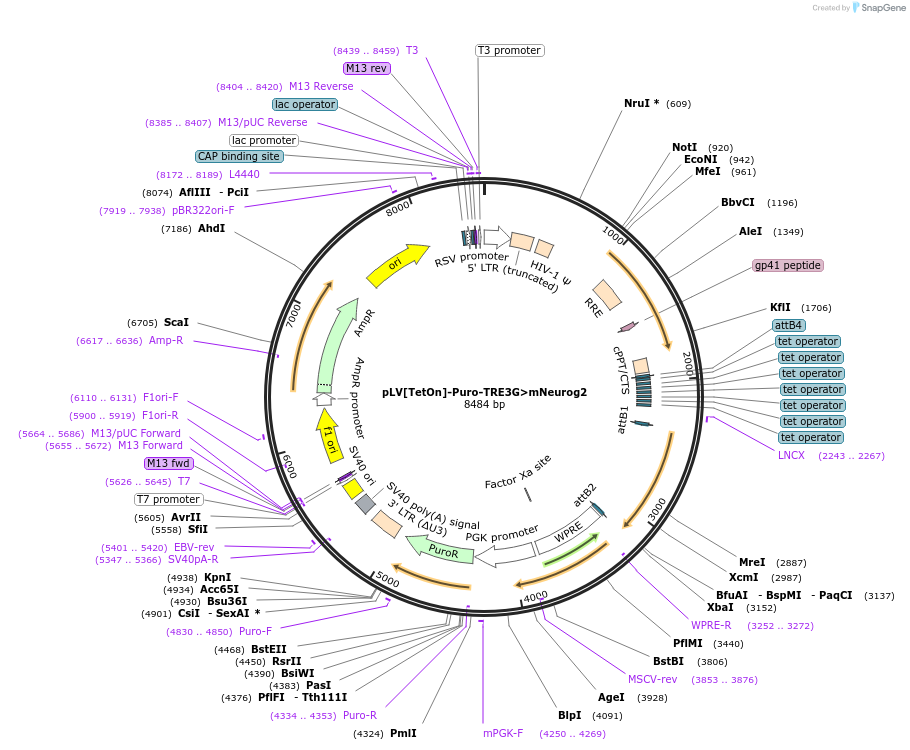
pLV[TetOn]-Puro-TRE3G>mNeurog2
Plasmid#198754PurposeVector encodes the gene for murine neurogenin-2 under control of a Tet-controlled promoter and the puromycin resistance gene under control of a constitutive PGK promoterDepositorAvailable SinceJuly 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
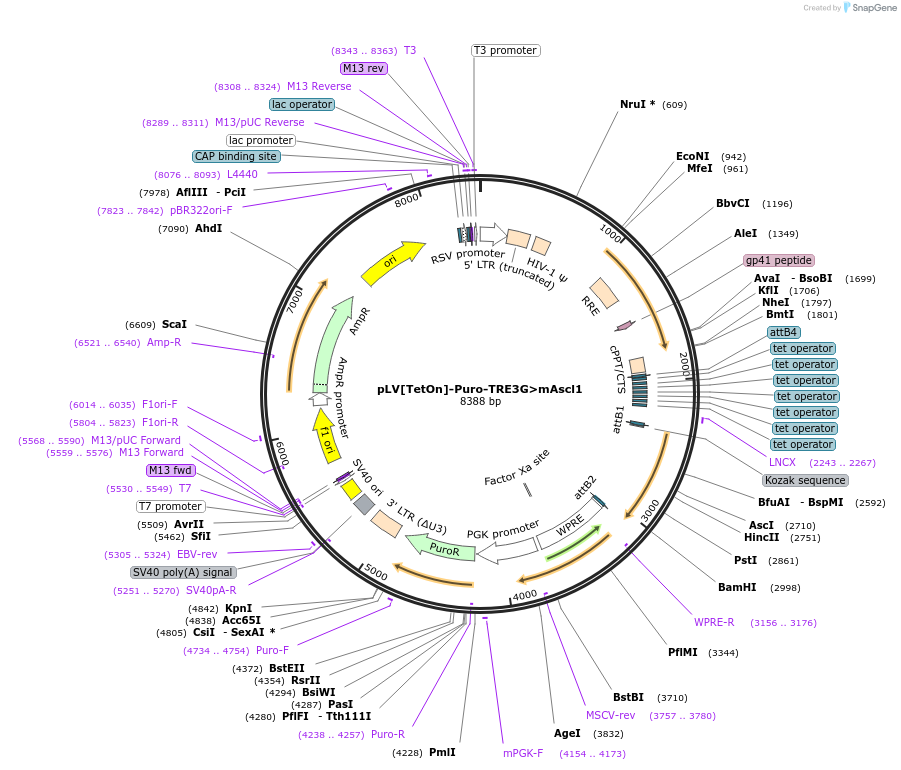
pLV[TetOn]-Puro-TRE3G>mAscl1
Plasmid#198755PurposeVector encodes the gene for murine Ascl1 under control of a Tet-controlled promoter and the puromycin resistance gene under control of a constitutive PGK promoterDepositorAvailable SinceJuly 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
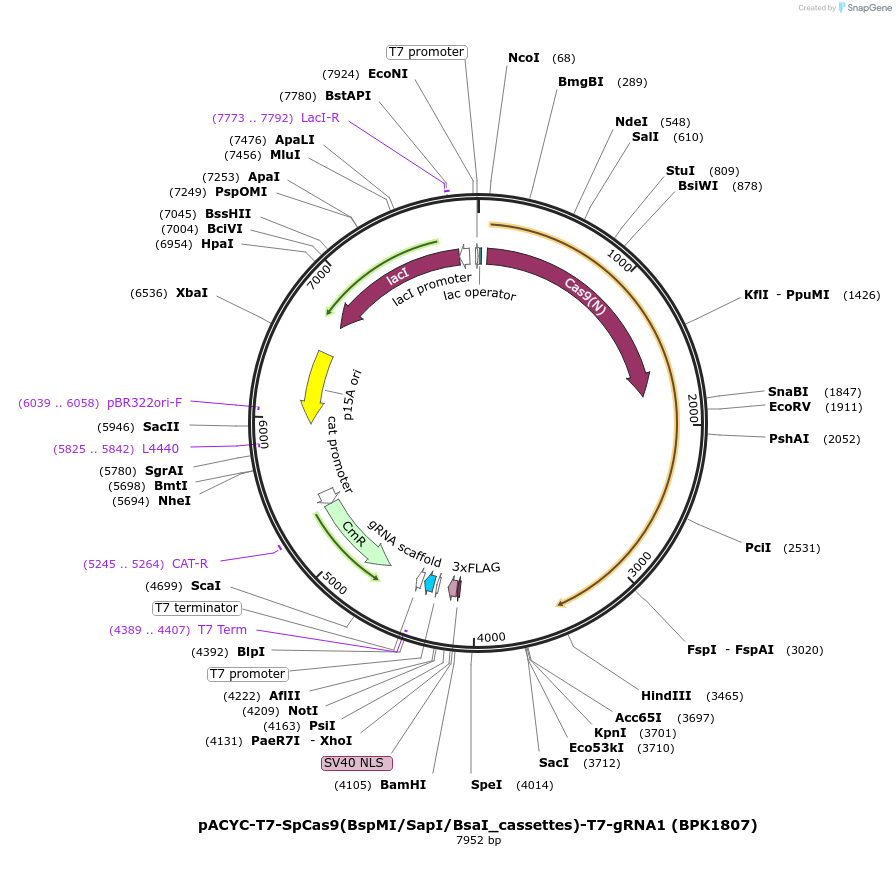
pACYC-T7-SpCas9(BspMI/SapI/BsaI_cassettes)-T7-gRNA1 (BPK1807)
Plasmid#223065PurposeDual pT7 entry plasmid for SpCas9(6AA) library; bacterial expression plasmid with type IIS RE cassettes around D1135/S1136, G1218/E1219, R1335/T1337 (precursor to MMW94). Expresses a gRNA.DepositorInsertentry vector for human codon opt. bacterial expr. plasmid for SpCas9(6AA_NNS) library, with gRNA targeting EGFP site 1
UseCRISPRTagsNLS(SV40)-3xFLAGExpressionBacterialMutationThree regions of SpCas9 encode type IIS restricti…PromoterDual T7Available SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
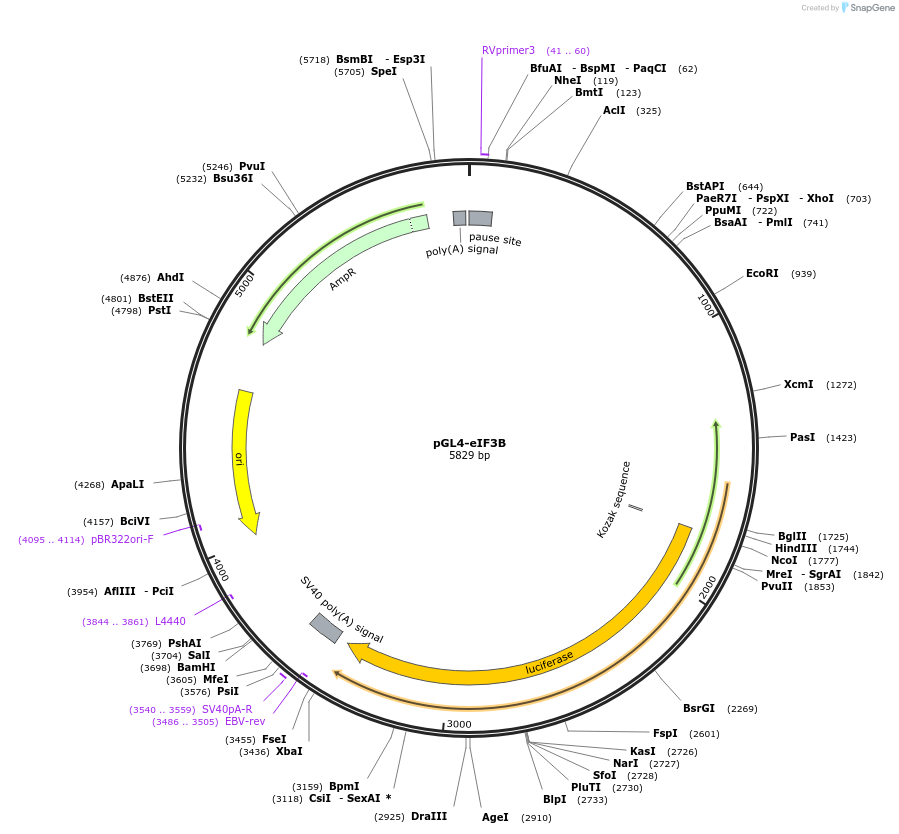
pGL4-eIF3B
Plasmid#133308PurposeExpression of luciferase driven by eIF3B promoter regionDepositorAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only